The pheromone response in Candida albicans is mediated by the Ste2 receptor. Intracellular (IC) loop 3 and C-terminal tail regions of Ste2 are required for signaling, whereas the large IC1 region is dispensable. Heterologous expression of receptors from asexual species can also drive signaling in C. albicans, allowing functional pheromone-receptor couples to be analyzed.
Abstract
Candida albicans is an important human fungal pathogen in which sexual reproduction is under the control of the novel white–opaque switch. Opaque cells are the mating-competent form, whereas white cells do not mate but can still respond to pheromones, resulting in biofilm formation. In this study, we first define the domains of the α-pheromone receptor Ste2 that are necessary for signaling in both white and opaque forms. Both cell states require the IC loop 3 (IC3) and the C-terminal tail of Ste2 for the cellular response, whereas the first IC loop (IC1) of Ste2 is dispensable for signaling. To also address pheromone-receptor interactions in related species, including apparently asexual Candida species, Ste2 orthologues were heterologously expressed in Candida albicans. Ste2 receptors from multiple Candida clade species were functional when expressed in C. albicans, whereas the Ste2 receptor of Candida lusitaniae was nonfunctional. Significantly, however, expression of a chimeric C. lusitaniae Ste2 receptor containing the C-terminal tail of Ste2 from C. albicans generated a productive response to C. lusitaniae pheromone. This system has allowed us to characterize pheromones from multiple Candida species and indicates that functional pheromone-receptor couples exist in fungal species that have yet to be shown to undergo sexual mating.
INTRODUCTION
Candida species are the most prevalent human fungal pathogens and the fourth most common cause of microbial bloodstream infections in the United States (Wisplinghoff et al., 2004). The primary culprit is Candida albicans, a hemiascomycete yeast related to Saccharomyces cerevisiae, although these species diverged more than 100 million years ago (Gargas and Taylor, 1995; Hedges, 2002). C. albicans is a natural component of human microbiota, but under certain circumstances it can become a life-threatening opportunistic pathogen, particularly in immunocompromised individuals (Edmond et al., 1999; Ruhnke and Maschmeyer, 2002). A number of species related to C. albicans have now been grouped together in a so-called Candida clade (see Supplemental Figure S1A), including the important human pathogens Candida tropicalis and Candida parapsilosis (Butler et al., 2009). In addition, the clade includes species such as Lodderomyces elongisporus and Candida lusitaniae, which are emerging human pathogens.
Like all Candida species, C. albicans was originally designated an asexual species, but studies over the past decade have established a highly elaborate mating cycle that shows fundamental differences compared with that of S. cerevisiae (Soll, 2009; Alby and Bennett, 2010; Butler, 2010). In particular, C. albicans a and α cells must undergo a heritable change in their state to become mating competent. C. albicans cells typically exist in the white state, where they are round and form dome-shaped colonies, but they can stochastically switch to the opaque state, where cells are more elongated and form darker, flatter colonies (Slutsky et al., 1987). This phenotypic switch regulates the sexual program, as only opaque cells are competent for mating (Miller and Johnson, 2002). White and opaque cells differ in their expression of ∼1300 genes, including several opaque-specific genes directly implicated in mating signaling (Lan et al., 2002; Tsong et al., 2003; Tuch et al., 2010).
Once cells have switched to the opaque state, the regulation of mating between C. albicans a and α cells occurs via pheromone signaling between the two mating types. MTLa cells secrete a pheromone and recognize α pheromone via the Ste2 receptor, while MTLα cells secrete α pheromone and recognize a pheromone via the Ste3 receptor (Magee et al., 2002; Bennett et al., 2003). Pheromone-receptor signaling induces a mitogen-activated protein kinase (MAPK) cascade, leading to the formation of mating projections (shmoos) and, ultimately, cell–cell fusion (Magee et al., 2002; Bennett et al., 2003, 2005; Yi et al., 2008). However, instead of a complete sexual cycle involving meiosis, the tetraploid mating products of C. albicans undergo concerted chromosome loss as part of a parasexual mechanism to return to the diploid state (Bennett and Johnson, 2003; Alby and Bennett, 2010).
Notably, pheromone treatment of C. albicans white a or α cells does not activate a mating response, but instead increases gene expression of factors involved in cell adhesion and biofilm development (Daniels et al., 2006; Yi et al., 2008; Sahni et al., 2009). It is hypothesized that biofilm formation by white cells provides an optimal environment for pheromone signaling between opaque cells, thereby increasing the efficiency of mating events in the host (Daniels et al., 2006). It was also revealed that pheromone signaling in white and opaque cells occurs via the same receptor-mediated MAPK pathway. However, whereas the Cph1 (Ste12) transcription factor was activated in opaque cells, the Tec1 transcription factor mediated the pheromone response in white cells (Yi et al., 2008; Sahni et al., 2010).
In comparison with C. albicans, much less is known about mating in other Candida species. For example, mating has never been observed in C. tropicalis or C. parapsilosis (Butler et al., 2009; Bennett, 2010; Butler, 2010). Surprisingly, L. elongisporus has no MTL locus but has been reported to undergo self-mating (homothallism) owing to observations of asci formation (Recca and Mrak, 1952; van der Walt, 1966; Butler et al., 2009). C. lusitaniae was recently shown to be fully sexual, and undergoes meiosis, despite lacking several highly conserved meiosis genes (Reedy et al., 2009; Sherwood and Bennett, 2009). With the singular exception of C. albicans, pheromone-induced mating responses have not been described in Candida clade organisms, and little is therefore known about pheromone-receptor signaling in these species.
Pheromone receptors belong to the largest family of G protein–coupled receptors (Naider and Becker, 2004). These receptors consist of seven transmembrane (TM) domains and contain distinct regions important for pheromone recognition and interaction with a heterotrimeric G protein (Dosil et al., 2000; Palczewski et al., 2000; Naider and Becker, 2004; Yi et al., 2009; Jones and Bennett, 2011). In S. cerevisiae Ste2, the extracellular (EC) loops 2 and 3, as well as TM domains 1, 5, and 6, have all been implicated in recognition of α pheromone (Lee et al., 2001; Lin et al., 2003; Naider and Becker, 2004). In addition, the IC C-terminus of S. cerevisiae Ste2 is necessary for pheromone induction of shmoos, but not for mating or cell cycle arrest (Konopka et al., 1988). Additional studies show that the C-terminal tail plays an important role in promoting the formation of receptor/G-protein preactivation complexes, and such complexes might enhance the rate of G-protein signaling (Dosil et al., 2000).
Within the Candida clade, an alignment of Ste2 sequences reveals that they share a conserved IC3 region, but are dissimilar in other potential signaling regions, including IC1 and the C-terminal tail (Figure S1B). Significantly, recent studies indicated that C. albicans Ste2 contained an unusually large IC1 region, and that this region was critical for the pheromone response in white cells, but was dispensable for pheromone signaling in opaque cells (Yi et al., 2009). Thus, signaling through the Ste2 receptor appeared to be distinct in the two cell types, suggesting that this was key to defining the distinct cellular responses by white and opaque cells. Paradoxically, the large IC1 region in C. albicans Ste2 was not present in the Ste3 receptor, and yet white α cells also formed biofilms in response to pheromone (Daniels et al., 2006). In addition, in contrast to pheromone signaling in S. cerevisiae, partial truncation of the C. albicans Ste2 C-terminus was reported to completely block pheromone-induced signaling in both white and opaque phenotypes (Konopka et al., 1988; Yi et al., 2009).
In this work, we first address the mechanism of pheromone signaling through the C. albicans Ste2 receptor. We show that loss of IC3, or removal of the entire cytoplasmic tail of Ste2, prevents pheromone signaling in both white and opaque cells. However, in contrast to published reports, the IC1 region of C. albicans Ste2 is shown to be dispensable for the pheromone response in white and opaque cells, and we therefore propose that pheromone-receptor signaling is similar in the two cell states. In addition, we heterologously expressed Ste2 receptors from C. tropicalis, C. parapsilosis, C. lusitaniae, and L. elongisporus in C. albicans. Most of these receptors result in productive signaling in response to their native pheromone in both white and opaque cells. Chimeric Ste2 receptors were also constructed that consist of Candida species Ste2 fused to the C-terminal tail of C. albicans Ste2. Signaling via the Ste2 chimeras was significantly enhanced in comparison with the native Ste2 genes. Furthermore, we found that the C. lusitaniae Ste2 receptor was nonfunctional when expressed in C. albicans, whereas the chimeric Ste2 receptor could respond to the native pheromone from this species. These results demonstrate that engineered strains expressing chimeric pheromone receptors represent powerful tools for the identification of canonical fungal pheromones. They also establish that “asexual” Candida species, such as C. tropicalis and C. parapsilosis, express functional pheromone-receptor pairs.
RESULTS
The IC3 domain and C-terminal tail of Ste2 are critical for the pheromone response in C. albicans opaque cells
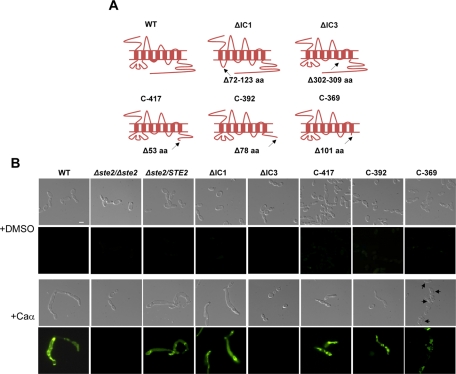
To determine the regions of C. albicans Ste2 required for the response to α pheromone, we constructed mutant Ste2 alleles lacking different IC domains. In particular, IC1 and IC3 deletions lacking either the first or third IC loop, respectively, were tested. These deletions removed amino acid residues 72–123 (ΔIC1) or 302–309 (ΔIC3), and are identical to those tested previously (Figure 1; Yi et al., 2009). The IC1 region of C. albicans Ste2 is of particular interest, since it is considerably larger than that of Ste2 from S. cerevisiae or other Candida clade species, and is rich in glutamine and asparagine amino acids, suggestive of protein–protein interactions (Yi et al., 2009; Figure S1B). Three truncations of the C-terminal cytoplasmic tail were also constructed, in which the last 53, 78, or 101 amino acids were removed to generate C-417, C-392, and C-369 mutants, respectively (full-length Ste2 is 470 amino acids [aa]). These three cutoff points were chosen because 1) C-417 is similar to a truncation mutant previously characterized in C. albicans (Ste2p-CTerΔ1; Yi et al., 2009), 2) C-392 is analogous to a truncated version of S. cerevisiae Ste2 (1–326 aa) that has been studied for its pheromone response (Konopka et al., 1988), and 3) C-369 has had the entire cytoplasmic tail following the final TM domain removed. All constructs were transformed into the native STE2 locus in a Δste2/Δste2 MTLa strain derived from C. albicans strain SC5314 (see Materials and Methods). The parental Δste2/Δste2 strain also contained a green fluorescent protein (GFP) reporter, FUS1-GFP, to monitor activation of these genes in response to pheromone (Alby et al., 2009).
FIGURE 1:
Schematic of the C. albicans Ste2 receptor, including TM domains, IC and EC loops, and mutant receptors lacking these regions. Analysis of pheromone responses in C. albicans strains expressing mutant Ste2 receptors. (A) The Ste2 receptor has seven TM domains that result in three IC and three EC loops and an IC C-terminal cytoplasmic tail. The position and number of disrupted amino acids are indicated by arrows in the Ste2 receptors. (B) Response of wild-type and Ste2 mutant strains to pheromone. Images of mating projections (shmoos) in C. albicans opaque strains were taken after 24 h of α-pheromone treatment. Isolates are expressing a FUS1-GFP reporter construct that is expressed in opaque cells responding to pheromone. The ΔIC1, C-417, and C-392 derivatives of Ste2 form shmoos and activate FUS1 expression when challenged with α pheromone, while receptors lacking the IC3 region or the entire C-terminal tail (C-369) are defective in shmooing and FUS1 expression. Arrows in C-369 indicate very short mating projections formed in response to pheromone. Scale bar: 5 μm.
Consistent with previous studies (Yi et al., 2009), we found that the third cytoplasmic loop (IC3) of C. albicans Ste2 is critical for the response to α pheromone by opaque a cells. Thus, ΔIC3 deletion mutants do not form polarized mating projections (shmoos) or express mating genes in response to synthetic α pheromone, and conjugation with opaque α cells is completely abolished (Figure 1B and Table 1). In contrast to loss of IC3, deletion of the IC1 region of Ste2 did not significantly affect the response to pheromone. Thus, the percentage of cells forming polarized mating projections (96%) was similar to that of wild-type cells (97%), as was mating-projection size (30 μm vs. 31 μm in ΔIC1 and wild-type, respectively; Figure 1B and Table 1). Mating efficiency was partially impacted by loss of IC1, as mating was reduced in ΔIC1 mutants (22.7%) compared with the wild-type and STE2-complemented strains (57.2% and 50.1%, respectively).
TABLE 1:
Shmoo formation and mating in C. albicans strains expressing mutant Ste2 receptors.
| Strain | Pheromone | Shmoo percentage | Projection size (μm) | Mating (%) |
|---|---|---|---|---|
| WT | − | 0 | 0 | 0 |
| WT | + | 97.6 ± 1.5 | 31.1± 6.7 | 57.2 ± 12.5 |
| Δste2/Δste2 | − | 0 | 0 | 0 |
| Δste2/Δste2 | + | 0 | 0 | 0 |
| Δste2/STE2 | − | 0 | 0 | 0 |
| Δste2/STE2 | + | 96.5 ± 2.2 | 27.8 ± 9.1 | 50.1 ± 22.1 |
| ΔIC1 | − | 0 | 0 | 0 |
| ΔIC1 | + | 96.4 ± 1.5 | 29.9 ± 5.6 | 22.7 ± 10.1 |
| ΔIC3 | − | 0 | 0 | 0 |
| ΔIC3 | + | 0 | 0 | 0 |
| C-417 | − | 0 | 0 | 0 |
| C-417 | + | 43.2 ± 4.8 | 8.9 ± 3.9 | 1.5 ± 0.6 |
| C-392 | − | 0 | 0 | 0 |
| C-392 | + | 61.4 ± 6.1 | 7.7± 2.4 | 3.6 ± 2.3 |
| C-369 | − | 0 | 0 | 0 |
| C-369 | + | 5.1 ± 1.7 | 2.4 ± 1.0 | 0.019 ± 0.005 |
Quantification of the pheromone response (shmooing) and mating frequency of opaque cells from C. albicans MTLa strains expressing different Ste2 receptors. Shmoo percentage was determined by the addition of α pheromone for 24 h in Spider medium and analysis of polarized mating projections by microscopy. One thousand cells were analyzed to determine the average size of the mating projections. Mating frequency was determined by coincubation of the Ste2-expressing opaque a cells with wild-type opaque α cells for 48 h and plating onto selective media to calculate a/α formation (see Materials and Methods). Each data point is the mean ± the SD from two independent experiments with at least three replicates.
In comparing C-terminal truncated Ste2 alleles, it was noted that opaque a cells expressing C-417 and C-392 (53 and 78 aa deleted, respectively) exhibited a robust mating response to α pheromone, albeit with a lower percentage of shmoos (43.2% and 61.4%, respectively) and shorter conjugation tubes (8.9 and 7.7 μm) than wild-type (97% shmooing, 31-μm projections). C-417 and C-392 exhibited a significantly reduced mating frequency, however, with only 1.5% and 3.6% of a cells, respectively, undergoing mating with α cells (Figure 1B and Table 1). The C-369 mutant, which lacks the entire C-terminal tail, showed much greater defects in projection formation (5.1%) and shmoo size (2.4 μm), and exhibited an extremely low mating frequency (0.019%; Figures 1B and S2 and Table 1). In addition, halo assays (measuring cell cycle arrest in response to pheromone) showed that ΔIC3 and C-369 mutants lost the ability to undergo cell cycle arrest, whereas C417 and C-392 mutants showed diminished arrest (Figure S3).
These results reveal that the C-terminal cytoplasmic tail and the conserved IC3 region of Ste2 play important roles in mediating the response to α pheromone by opaque a cells. However, in contrast to previous studies (Yi et al., 2009), we show that deletion of the entire cytoplasmic tail (101 aa) is necessary to prevent efficient mating-projection formation, as partial deletion of the C-terminal tail still permits active pheromone signaling.
Transcriptional response to pheromone in ΔIC1, ΔIC3, and C-terminal truncation mutants of Ste2
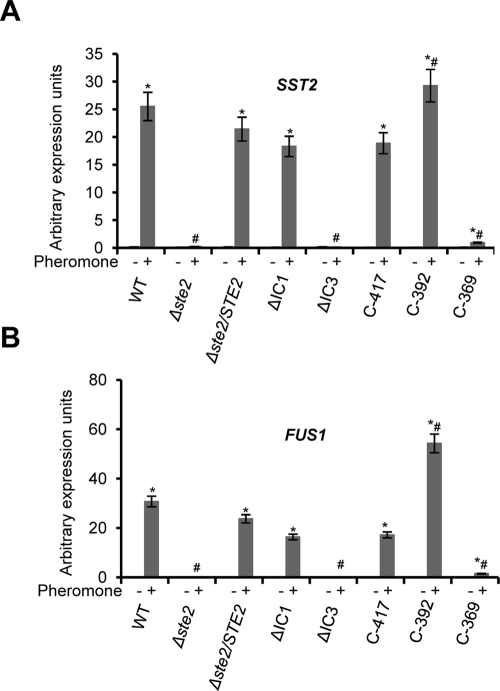
To determine the transcriptional response of Ste2 alleles to α pheromone, quantitative PCR (qPCR) of the mating-specific genes SST2 and FUS1 was performed in the presence or absence of pheromone. As shown in Figure 2, pheromone-induced gene expression was observed in the wild-type strain but not in Δste2/Δste2 or ΔIC3 mutants. Deletion of the IC1 region had only a minor effect on signaling through Ste2, as induction of FUS1/SST2 expression was almost as high as that in the STE2-complemented strain. In addition, C-417 and C-392 mutants that showed only modest defects in shmoo formation also showed either slightly reduced gene induction (C-417) or twofold increased gene induction (C-392) relative to the STE2-complemented strain (Figure 2). Interestingly, a similar phenotype was previously reported for an S. cerevisiae C-terminal truncation mutant, ste2-T326, which displayed < 2% of normal mating projections, but was 10-fold more sensitive to α pheromone in agglutination and cell division arrest assays (Konopka et al., 1988). It is possible that increased receptor numbers are partially responsible for the increased transcriptional response observed in C. albicans C-392 strains, as analysis of Ste2-GFP fusion proteins revealed that GFP levels in both C-417-GFP– and C392-GFP–expressing strains were approximately twofold higher than the wild-type strain when responding to α pheromone (Figure 3).
FIGURE 2:
Quantitative RT-PCR indicating expression of the mating genes SST2 and FUS1 in response to pheromone. Deletion of the IC3 region of Ste2 or the entire C-terminal tail blocked pheromone induction of mating genes in opaque cells. Quantitative gene expression of SST2 (A) and FUS1 (B) was compared when each strain was treated with C. albicans α pheromone (+) or DMSO (−). *, p < 0.05 vs. DMSO. Expression in response to α pheromone was also compared between each mutant allele and wild-type STE2 (#p < 0.05 vs. wild-type). Each value is presented as the mean ± SD.
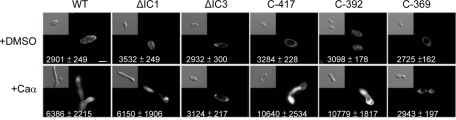
FIGURE 3:
Analysis of Ste2-GFP fusion protein expression during the response to pheromone in strains expressing mutant Ste2 receptors. Microscopy was used to analyze Ste2-GFP expression in the presence or absence of C. albicans α pheromone. Ste2 localized to the cell surface without pheromone induction, while pheromone treatment stimulated both increased expression and internalization of wild-type, ΔIC1, C-417, and C-392 receptors, but not ΔIC3 or C-369. The GFP intensity is indicated at the bottom of each image. Values are given as the mean ± SD. Numbers are an average of five replicates. Scale bar: 5 μm.
In contrast to the relatively modest phenotypes seen with C-417 and C-392, C-369 mutants showed almost a complete loss in pheromone-induced gene expression (Figure 2). C-369 mutants displayed low receptor expression and, unlike wild-type Ste2, did not show significantly increased receptor levels when cells were challenged with α pheromone (Figure 3). These results suggest that pheromone signaling in opaque cells is highly impaired in the C-369 mutant, in part due to compromised expression of this receptor (Figure 3).
The role of IC1, IC3, and the C-terminus of Ste2 in pheromone signaling in white cells
Unlike opaque cells, white cells responding to pheromone do not form shmoos or undergo mating, but up-regulate genes that cause enhanced biofilm formation (Yi et al., 2008). It has been reported that different domains of Ste2 are involved in pheromone signaling in white and opaque cells, with the IC1 region specifically required for the pheromone response in white cells (Yi et al., 2009). We therefore tested each of our mutant Ste2 receptors for their ability to mount a response to pheromone in white cells.
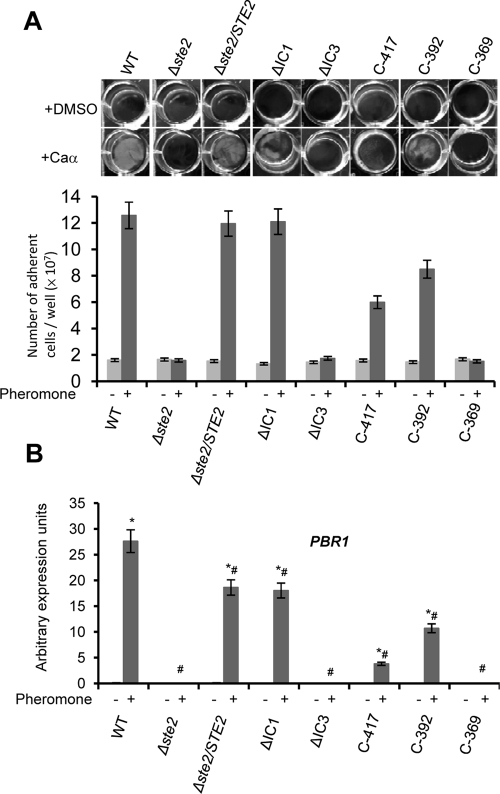
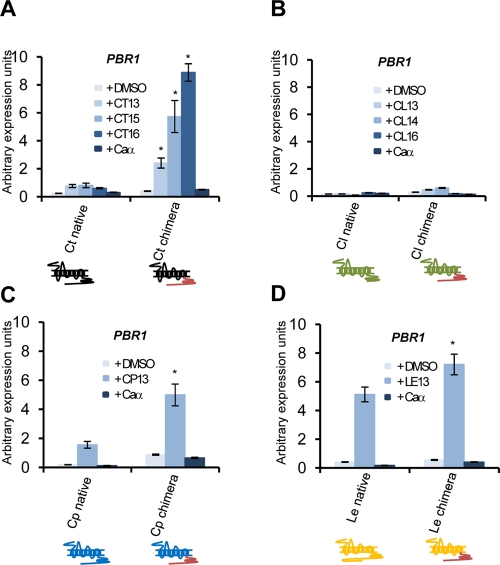
Figure 4 shows biofilm formation in white cells responding to pheromone and quantitative expression of PBR1, a gene up-regulated by pheromone signaling in white cells (Sahni et al., 2009). First, as seen in opaque cells, deletion of the IC3 region of Ste2 abolished all white-cell responses to pheromone, including both biofilm formation and PBR1 expression. Similarly, complete deletion of the C-terminal tail in the C-369 mutant strain prevented any detectable response by white cells. Partial deletion of the C-terminal tail in strains expressing C-392 and C-417 resulted in an intermediate response by white cells, as both biofilm development and PBR1 expression were reduced (Figure 4).
FIGURE 4:
Analysis of the pheromone response in C. albicans white cells expressing different STE2 alleles. We show that removal of the IC3 region or the entire cytoplasmic tail of Ste2 leads to a complete loss of pheromone-induced gene expression and biofilm development in white cells. (A) Images of white cells adhering to plastic when treated with pheromone. White MTLa cells were incubated in Lee's medium and treated with α pheromone in 12-well culture dishes for 24 h. Nonadherent cells were removed by washing, and the remaining adherent cells were photographed and quantitated. (B) Quantitative gene expression of PBR1, a gene induced in white cells in response to pheromone. Expression was analyzed and compared between α pheromone and the DMSO control (*, p < 0.05 vs. DMSO) or compared with the response in wild-type cells upon addition of pheromone (#, p < 0.05 vs. wild-type). Expression levels were normalized to the PAT1 gene. Values are the mean ± SD from two independent experiments with at least three replicates.
Previous analysis of an IC1 deletion mutant in C. albicans Ste2 indicated the white-cell response to pheromone was abolished, although the response by opaque cells was unaffected (Yi et al., 2009). In contrast to the former study, we found that white cells expressing ΔIC1 produced a robust biofilm in the presence of pheromone, similar to that formed by the wild-type control (Figure 4A). Quantitative reverse transcriptase PCR (RT-PCR) confirmed that deleting the IC1 region did not affect PBR1 gene induction in response to α pheromone (Figure 4B). Pheromone-induced biofilm formation of ΔIC1 mutants also showed no difference compared with the control strain, even when responding to lower concentrations of pheromone (unpublished data). These data indicate that the IC1 region of C. albicans Ste2 does not play a selective role in the white-cell pheromone response.
Curiously, in S. cerevisiae, the IC1 region of Ste2 only affected signaling when the C-terminal tail was also truncated (Chinault et al., 2004). We therefore tested a C. albicans Ste2 allele in which the IC1 region was deleted in the C-392 construct. This receptor behaved similarly to the C-392 allele when expressed in both white and opaque cells (unpublished data). Thus, even when tested in combination with a partial loss of the C-terminal tail, there is no significant role for the IC1 region of C. albicans Ste2 in signaling.
Taken together, our results indicate loss of the entire cytoplasmic tail of Ste2 is necessary to abolish the response to α pheromone in opaque and white cells, as partially truncated C-terminal tail mutants are still active at transducing the pheromone signal, albeit at a reduced efficiency. In addition, the IC3 region is critical for the pheromone response, whereas the large IC1 domain of C. albicans Ste2 is dispensable for pheromone signaling in both white and opaque cells.
Comparative analysis of Ste2 alleles when expressed in different strain backgrounds of C. albicans
Previous studies analyzing C. albicans Ste2 function utilized gene fusions between Ste2 and GFP in the P37005 strain background. As we observed contradictory results to these studies using native receptors expressed in SC5314, we investigated whether either the strain background or fusion to GFP could influence Ste2 activity. Again, mutant Ste2 alleles were integrated at the native STE2 locus, and constructs were compared either with or without GFP fused to the C-terminus of Ste2.
In general, we observed similar results when mutant Ste2 receptors were analyzed in SC5314 or P37005 strain backgrounds (compare Table 1 and Figure 4 with Table S1 and Figure S4). In particular, deletion of the IC1 region of Ste2 did not significantly impair pheromone signaling in white or opaque cells in either strain background. We also examined constructs in which GFP was fused to Ste2 from which the IC1, IC3, or the C-terminal tail regions had been deleted. The addition of GFP to these Ste2 alleles had little effect on either pheromone-induced mating in opaque cells or biofilm development in white cells (Table S2 and Figure S5).
These experiments confirm that the IC1 region of C. albicans Ste2 plays no significant role in transduction of the pheromone signal in white or opaque cells. Loss of the IC1 region does not significantly influence pheromone receptor levels and is not necessary for signaling, even in receptors with compromised activity. In addition, regardless of the strain background, partial loss of the Ste2 C-terminal tail reduces, but does not abolish, pheromone signaling, whereas complete deletion of the C-terminal tail blocks signaling in both white and opaque cells.
Heterologous expression of Ste2 receptors from multiple Candida species in C. albicans
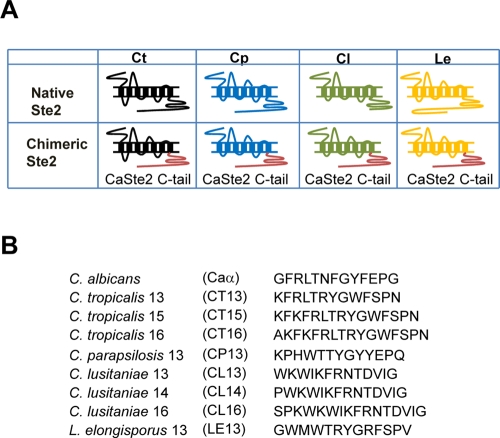
Genome sequencing has identified Ste2 orthologues from multiple Candida clade species, including C. tropicalis, C. parapsilosis, C. lusitaniae, and L. elongisporus (Butler et al., 2009). Of these four species, only C. lusitaniae has been observed to undergo mating, and although spore formation was reported in L. elongisporus, this has yet to be shown to represent a sexual program (Recca and Mrak, 1952; van der Walt, 1966; Butler et al., 2009; Reedy et al., 2009). The Ste2 proteins from these species share only limited homology with C. albicans Ste2; they contain similar IC3 regions, but IC1 domains are small, and the C-terminal tail region is highly variable (Figure S1B). We therefore examined whether Ste2 orthologues from other Candida species can be functionally expressed in C. albicans Δste2/Δste2 strains, and can respond either to their native pheromones or pheromones from other species. Owing to their highly variable C-terminal tail regions and the importance of this domain in signal transduction, we also constructed chimeric Ste2 receptors in which the C-terminal tail region from each species was replaced with the C. albicans Ste2 tail (Figure 5A). The resulting engineered strains allow for the testing of putative α-pheromone peptides from each species (Figure 5B; Alby and Bennett, 2011).
FIGURE 5:
Schematic diagrams of native and chimeric Ste2 receptors from C. tropicalis (Ct), C. parapsilosis (Cp), C. lusitaniae (Cl), and L. elongisporus (Le), and the predicted sequences of α pheromones for these species. (A) Figure showing Ste2 receptors from different Candida clade species. Both native receptors and chimeric receptors were tested by expression in C. albicans, the latter made by replacing the C-terminal tail with that from C. albicans Ste2. (B) Sequences of the predicted α pheromones of related Candida clade species.
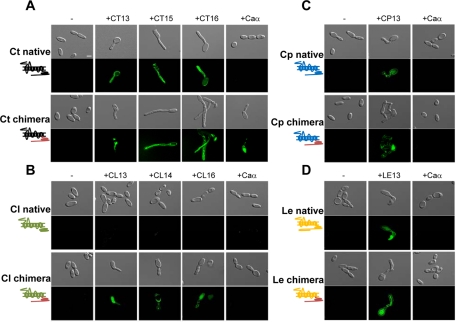
As shown in Figure 6 and Table 2, native Ste2 receptors from C. tropicalis, C. parapsilosis, and L. elongisporus were at least partially active when expressed in C. albicans opaque a cells and challenged with synthetic α pheromones. Significantly, however, chimeric versions of these Ste2 receptors resulted in increased pheromone signaling, as evidenced by more cells undergoing shmooing and longer mating projections (Table 2). For example, expression of the native Ste2 receptor from C. tropicalis resulted in 12–48% of cells forming shmoos, depending on the pheromone tested. In comparison, expression of the chimeric C. tropicalis Ste2 (containing the C-terminal tail of C. albicans Ste2) resulted in 15–73% of cells forming mating projections when challenged with the C. tropicalis pheromones (Figure 6A and Table 2). Similarly, expression of chimeric Ste2 receptors from C. parapsilosis and L. elongisporus resulted in a more efficient response to pheromone than that seen with native Ste2 receptors, as cells formed more shmoos with longer projections (Figure 6, C and D, and Table 2). Interestingly, expression of the C. tropicalis chimeric receptor in C. albicans also enabled a response to C. albicans α pheromone (8% of cells formed shmoos; Figure 6A) and C. lusitaniae α pheromone (Figure S6). This result supports the recent proposal that certain mating pheromones may be able to promote interspecies signaling between different Candida species (Alby and Bennett, 2011).
FIGURE 6:
Images of shmoo formation in C. albicans strains expressing native and chimeric versions of Ste2 receptors from related Candida clade species. C. albicans Δste2 strains were engineered to express native or chimeric Ste2 receptors from C. tropicalis (A), C. lusitaniae (B), C. parapsilosis (C), or L. elongisporus (D). Each strain containing a FUS1-GFP reporter gene was challenged with either α pheromone for that species (see Figure 5B) or with C. albicans α pheromone (Caα) for 24 h. Top, native Ste2; bottom, chimeric Ste2. Scale bar: 5 μm.
TABLE 2:
Analysis of shmoo formation in strains engineered to express Ste2 receptors from multiple Candida clade species.
| Straina | Pheromone | Shmoo percentage | Projection size (μm) |
|---|---|---|---|
| Ct native | − | 0 | 0 |
| Ct native | +CT13 | 12.8 ± 2.9 | 6.9 ± 2.8 |
| Ct native | +CT15 | 37.1 ± 5.6 | 14.1 ± 6.1 |
| Ct native | +CT16 | 48.1 ± 6.1 | 12.6 ± 4.7 |
| Ct native | +Caα | 0 | 0 |
| Ct chimera | − | 0 | 0 |
| Ct chimera | +CT13 | 15.3 ± 5.3 | 15.8 ± 5.8 |
| Ct chimera | +CT15 | 54.2 ± 6.4 | 23.1 ± 10.1 |
| Ct chimera | +CT16 | 73.1 ± 8.5 | 23.8 ± 11.1 |
| Ct chimera | +Caα | 8.0 ± 2.5 | 8.0 ± 2.9 |
| Cp native | − | 0 | 0 |
| Cp native | +CP13 | 34.0 ± 13.3 | 8.7 ± 3.5 |
| Cp native | +Caα | 0 | 0 |
| Cp chimera | − | 0 | 0 |
| Cp chimera | +CP13 | 74.0 ± 9.0 | 14.9 ± 7.7 |
| Cp chimera | +Caα | 0 | 0 |
| Cl native | − | 0 | 0 |
| Cl native | +CL13 | 0 | 0 |
| Cl native | +CL14 | 0 | 0 |
| Cl native | +CL16 | 0 | 0 |
| Cl native | +Caα | 0 | 0 |
| Cl chimera | − | 0 | 0 |
| Cl chimera | +CL13 | 9.0 ± 3.3 | 7.3 ± 3.7 |
| Cl chimera | +CL14 | 14.6 ± 3.2 | 8.5 ± 3.2 |
| Cl chimera | +CL16 | 10.2 ± 2.1 | 8.0 ± 2.4 |
| Cl chimera | +Caα | 0 | 0 |
| Le native | − | 0 | 0 |
| Le native | +LE13 | 36.8 ± 4.3 | 9.5 ± 5.1 |
| Le native | +Caα | 0 | 0 |
| Le chimera | − | 0 | 0 |
| Le chimera | +LE13 | 65.0 ± 8.4 | 13.6 ± 3.9 |
| Le chimera | +Caα | 0 | 0 |
Percentage of mating projections and projection size was measured in strains incubated in Spider medium and treated with pheromones for 24 h. At least 1000 cells were measured for each condition, using a Zeiss Observer Z1 microscope. Each data point is the mean ± the SD from two independent experiments with at least three replicates.
aCt, C. tropicalis; Cp, C. parapsilosis; Cl, C. lusitaniae; Le, L. elongisporus.
In contrast to the other Ste2 receptors, expression of C. lusitaniae Ste2 in C. albicans produced a strain that was nonresponsive to its native α pheromone. We therefore tested whether cells expressing the chimeric C. lusitaniae Ste2 receptor could now respond to pheromone. Strikingly, we observed significant shmooing (9–12%) in the chimeric strain when it was challenged with C. lusitaniae 13-mer, 14-mer, or 16-mer pheromones (Figure 6B and Table 2). These results provide further evidence for the critical role of the C-terminal tail of Ste2 for transduction of the pheromone signal and initiation of polarized growth.
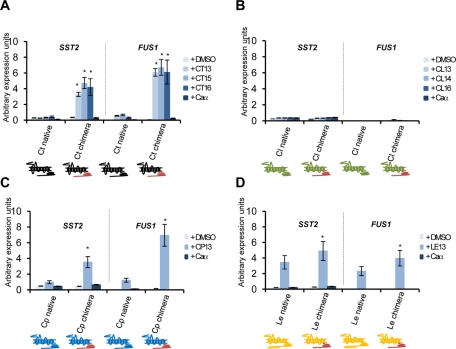
To investigate downstream signaling events in these strains, we analyzed expression of the mating-specific genes SST2 and FUS1 by using RT-PCR. As expected based on their increased morphological response, the chimeric Ste2 receptors from C. tropicalis, C. parapsilosis, and L. elongisporus resulted in higher pheromone-induced gene expression than the corresponding native Ste2 receptors (Figure 7). For example, expression of the chimeric receptor of C. tropicalis induced SST2 and FUS1 ∼10-fold more than the native C. tropicalis receptor in response to C. tropicalis pheromones (Figure 7A). In contrast, no mating-gene expression was observed at 4 h in the strain expressing the C. lusitaniae chimeric Ste2 receptor, despite evidence of shmooing in this strain (Figure 7B). We therefore performed the study over a longer time course and analyzed gene expression 24 h after C. lusitaniae pheromone treatment. In these prolonged experiments, C. lusitaniae pheromones were successful in inducing significant SST2/FUS1 gene expression in the engineered strain (Figure S7).
FIGURE 7:
Quantitative RT-PCR expression of mating genes in C. albicans strains expressing Ste2 receptors from alternative Candida clade species. Gene expression of SST2 and FUS1, two genes induced in mating opaque cells, was analyzed. Strains were treated with 10 μg/ml pheromone for 4 h in Spider medium. Strains expressed native or chimeric Ste2 from (A) C. tropicalis, (B) C. lusitaniae, (C) C. parapsilosis, or (D) L. elongisporus, in C. albicans. *, p < 0.05 native vs. chimeric Ste2. Expression levels were normalized to the PAT1 gene. Values are presented as the mean ± SD from two independent experiments with at least three replicates.
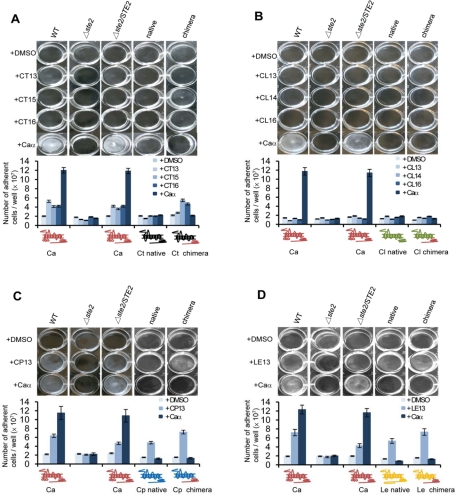
Heterologous expression of Candida Ste2 receptors in C. albicans white cells promotes cell adhesion and biofilm formation
Ste2 receptors from most Candida species do not contain the large IC1 domain found in C. albicans Ste2 (and its sister species Candida dubliniensis; Figure S1B). We therefore tested whether these Ste2 receptors from alternative species are able to induce cell adhesion and biofilm formation when expressed in C. albicans white cells. Ste2 receptors from C. tropicalis, C. parapsilosis, and L. elongisporus, but not C. lusitaniae, were able to induce biofilm formation in C. albicans white cells when challenged with the corresponding pheromone. Both native and chimeric receptors promoted pheromone-induced biofilm formation, although biofilm mass was greater in strains expressing the chimeric receptor, indicating the C-terminal tail from C. albicans Ste2 further enhances pheromone signaling in white cells in a manner similar to that observed in opaque cells (Figure 8).
FIGURE 8:
Pheromone induction of biofilm formation in C. albicans white cells expressing Ste2 receptors from other Candida clade species. Cell adhesion and biofilm development were analyzed in C. albicans white cells expressing Ste2 receptors from (A) C. tropicalis, (B) C. lusitaniae, (C) C. parapsilosis, or (D) L. elongisporus. Assays were performed in Lee's medium in 12-well plastic plates in the presence or absence of C. albicans α pheromone, or α pheromones from the respective species (see Figure 5B). Quantification of the number of adherent white cells is shown in the images. Each experiment was performed using at least two independent isolates and three experimental replicates. Values are represented as the mean ± SD.
Gene expression of the PBR1 gene in white cells was also monitored, and again was highest in strains expressing the chimeric form of C. tropicalis, C. parapsilosis, and L. elongisporus Ste2 (Figure 9). Although biofilms were not observed in strains expressing the C. lusitaniae Ste2 receptor, low-level expression of the PBR1 gene was induced after pheromone treatment for 24 h (Figure S8). It is therefore apparent that the weak induction of pheromone signaling in this strain is not sufficient for significant biofilm development (Figure 8B).
FIGURE 9:
Pheromone induction of PBR1 in white cells expressing Ste2 receptors from different Candida species. Expression of the PBR1 gene was analyzed in C. albicans white cells expressing native or chimeric Ste2 receptors from (A) C. tropicalis, (B) C. lusitaniae, (C) C. parapsilosis, or (D) L. elongisporus. PBR1 expression was quantitated after treatment of strains with 10 μg/ml pheromone for 4 h. The chimeric receptors significantly induced PBR1 gene expression over the native Ste2 receptors in three of the four species tested. *, p < 0.05 native vs. chimeric response. Expression levels were normalized to the PAT1 gene. Values are presented as the mean ± SD from two independent isolates with at least three experimental replicates.
These results provide independent confirmation that the large IC1 region of C. albicans Ste2 is not required for either the white or opaque pheromone response, as Ste2 receptors from other Candida species lack this unique region but are still productive in pheromone signaling in both cell types. In contrast, the C-terminal cytoplasmic tail of C. albicans Ste2 is important for efficient transduction of the pheromone signal in white and opaque cells.
Ste2 chimeras as powerful tools to investigate pheromone-receptor interactions in Candida species
The apparent lack of sexual reproduction in several Candida species is a significant hurdle to the use of classical genetics in these organisms. A common first step in addressing whether a species has a sexual cycle is to challenge the organism with synthetic pheromone and to monitor gene expression changes or to look for a morphological response. For example, in the case of C. parapsilosis, this species was recently shown to be nonresponsive when exposed to synthetic pheromone, and it is not clear at what step the pheromone signaling pathway is compromised (Sai et al., 2011). The ability to heterologously express Ste2 receptors in C. albicans now allows us to validate the activity of synthetic pheromones from multiple Candida species and to demonstrate productive coupling between pheromone and receptor for the first time.
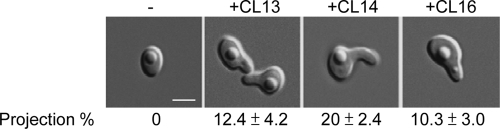
To confirm that the specificity of pheromone-receptor interactions is not affected by heterologous expression in C. albicans, we further examined pheromone signaling in C. lusitaniae. This species has a complete sexual cycle, including cell–cell conjugation, meiosis, and sporulation (Reedy et al., 2009), but shmoo formation in response to pheromone has not been investigated. We therefore addressed whether the synthetic pheromones used in this study are also able to activate shmoo formation in C. lusitaniae strains. As shown in Figure 10, synthetic pheromones corresponding to CL13, CL14, and CL16 were able to induce C. lusitaniae a strains to form mating projections with 12%, 20%, and 10% efficiency, respectively. The ability of synthetic peptides to induce mating projections in C. albicans strains expressing the C. lusitaniae chimeric receptor was very similar, with CL14 again proving to be the most active (Table 2). Furthermore, neither the chimeric C. lusitaniae Ste2 expressed in C. albicans, nor C. lusitaniae itself, was able to respond to synthetic pheromones from other Candida species (Figure S6; unpublished data).
FIGURE 10:
Synthetic C. lusitaniae pheromones are able to induce a mating response in C. lusitaniae strains. The pheromone response of C. lusitaniae MTLa cells was tested by incubating cells on PDA together with the addition of synthetic pheromone. 13-mer, 14-mer, and 16-mer pheromones were tested, and each was found to induce shmoo formation. Images were taken after 24 h treatment. Scale bar: 2 μm.
These results demonstrate that pheromone-receptor interactions defined by heterologous expression studies in C. albicans can be used to identify bona fide pheromones from other Candida species. We therefore propose that this system provides an invaluable tool for the identification of active fungal pheromones and productive pheromone-receptor couples. These validated pheromones can subsequently be used with confidence to investigate the existence of sexual programs in related species, such as C. tropicalis, C. parapsilosis, and L. elongisporus.
DISCUSSION
In this work, we first identify the IC domains in the C. albicans Ste2 pheromone receptor that are important for downstream signaling events. C. albicans is unique in that it can exist in two phenotypic states; the opaque state is the mating-competent form and responds to pheromone by up-regulation of mating genes and formation of polarized mating projections (Miller and Johnson, 2002). In contrast, the white state does not undergo mating but can still respond to pheromone by increased expression of cell adhesins that promote biofilm formation (Yi et al., 2008). We demonstrate that IC loop 3 (IC3) and the C-terminal tail of the Ste2 receptor are critical for the pheromone response, as the loss of either of these regions abolished the response in both white and opaque cells. Thus, opaque cells no longer underwent the morphological response (shmooing), the transcriptional response (expression of SST2/FUS1), or cell–cell conjugation, while white cells no longer formed biofilms or showed expression of PBR1. In S. cerevisiae, the conserved IC3 region is a multifunctional domain that plays a similarly pleiotropic role, controlling receptor activation, ligand discrimination, and receptor internalization (Stefan and Blumer, 1994).
In contrast to an earlier report, we show that removal of the first IC region (IC1) of C. albicans Ste2 does not influence pheromone signaling in either the white or opaque state. Previously it was suggested that the IC1 region of Ste2 was essential for the pheromone response in white a cells but dispensable for signaling in opaque cells (Yi et al., 2009). The authors concluded that the pheromone signal transduction machinery must be distinct in white and opaque cells of C. albicans. This result was surprising, in part because white α cells of C. albicans also form biofilms when challenged with pheromone, yet the corresponding pheromone receptor, Ste3, does not contain a large IC1 region (Yi et al., 2009). We now show that the IC1 region does not play a decisive role in pheromone signaling, and that white cells expressing Ste2 lacking this domain still efficiently form biofilms in the presence of pheromone (Figure 4).
An important difference between our initial experiments and those previously described was that the earlier study utilized Ste2-GFP fusion receptors expressed in the P37005 strain background (Yi et al., 2009), while we tested native Ste2 receptors in the SC5314 strain background. We therefore addressed whether fusion with GFP or the C. albicans strain background could account for the observed differences in pheromone signaling. We found the addition of the GFP protein had little effect on any of the phenotypes analyzed here, and while P37005 forms pheromone-induced biofilms more efficiently than SC5314, results were qualitatively similar in both strain backgrounds (compare Figure 4 with Figures S4 and S5). Furthermore, independent support that IC1 is not required for pheromone signaling in white cells has come from analysis of Ste2 receptors from other Candida species heterologously expressed in C. albicans. For example, expression of C. tropicalis or C. parapsilosis Ste2 could still promote pheromone-induced biofilm formation in C. albicans white cells, despite the fact that these receptors do not contain the large IC1 region characteristic of C. albicans Ste2. Taken together, these experiments establish that IC1 is not an essential part of the C. albicans pheromone-signaling pathway, either in white or opaque cells.
In S. cerevisiae, the function of IC1 in Ste2 receptor signaling is unclear. Substitution of the IC1 domain does not have a significant effect on signaling unless part of the C-terminal tail of the pheromone receptor is also removed (Chinault et al., 2004). To test whether the IC1 region of C. albicans Ste2 similarly plays a redundant role in pheromone signaling, we constructed a mutant Ste2 allele lacking both IC1 and the C-terminal 78 amino acids (C-392). However, loss of IC1 did not further compromise the activity of the C. albicans C-392 receptor. Thus, while it is intriguing that the IC1 region of C. albicans Ste2 is rich in glutamines and asparagines, given that such regions can promote protein–protein interactions (Perutz et al., 1994; Michelitsch and Weissman, 2000; Yi et al., 2009), our studies have not detected any significant role for IC1 in C. albicans pheromone signaling.
Our studies also demonstrate that complete removal of the C-terminal tail of C. albicans Ste2 (101 aa in C-369) essentially abolishes pheromone signaling, while partial deletion of the tail (removal of 53 or 78 aa in C-417 and C-392, respectively) does not substantially inhibit signaling (either in white or opaque cells). However, despite having only a moderate effect on signaling, mating with α cells was reduced by more than an order of magnitude in C-417– and C-392–expressing strains. These results differ from those seen in S. cerevisiae, where a partial truncation of the C-terminal tail, ste2-T-326, caused cells to be highly defective in formation of mating projections but did not adversely affect mating or cell cycle arrest (Konopka et al., 1988). Partial C-terminal tail truncations therefore have contrasting phenotypes in S. cerevisiae and C. albicans; in S. cerevisiae, these mutations have a negative effect on shmooing but little effect on mating, whereas in C. albicans they cause a significant decrease in mating but relatively little effect on the formation of mating projections. These results provide additional support for the model that polarized growth (shmooing) and mating (cell conjugation) are separable processes in yeast (Strickfaden and Pryciak, 2008). Interestingly, studies on the S. cerevisiae ste2-T-326 mutant have also revealed it lacks a motif for receptor internalization and is defective in orientating mating projections toward a pheromone source (Vallier et al., 2002). The supersensitivity of ste2-T326 mutants to pheromone is therefore thought to be due to a defect in recovery from pheromone treatment, rather than the observed increase in receptor numbers (Konopka et al., 1988). In comparison, loss of the entire cytoplasmic tail of C. albicans Ste2 appeared to compromise both receptor expression and downstream receptor signaling.
We further investigated pheromone-receptor signaling by heterologous expression of Ste2 receptors from four other Candida clade species in C. albicans, in large part because pheromone-receptor interactions have yet to be investigated for any of these species. C. lusitaniae (Cl) is a fully sexual species that undergoes both mating and meiosis (Reedy et al., 2009), L. elongisporus (Le) is capable of forming spores, yet mating in this species has not been observed (Recca and Mrak, 1952; van der Walt, 1966; Butler et al., 2009), while C. parapsilosis (Cp), and C. tropicalis (Ct) are both reported to be obligate asexual species (Figure S1A; Butler et al., 2009). The Ste2 receptors from these four alternative species have highly conserved IC3 domains but do not contain the large IC1 region specific to C. albicans Ste2; they also show C-terminal tail sequences that vary significantly in both their length and sequence (Figure S1B). Expression of the native Ste2 receptors from three of the Candida clade species (Ct, Cp, and Le) in C. albicans resulted in engineered strains that could respond to synthetic pheromones from each species. In several cases, alternative forms of the pheromone were tested, as it was not clear how proteolytic processing of the prepropheromone would generate the mature pheromone peptide. For example, in C. tropicalis, peptides of 13, 15, and 16 amino acids were analyzed, and all were found to be active, although the 15-mer and 16-mer generated the strongest response. In this manner, we show that it is possible to test and validate putative pheromones from each species by using the engineered C. albicans strains.
Due to the highly variable C-terminal domain between different Ste2 receptors, we also examined the consequences of replacing the native C-terminal tail of Ste2 with that from C. albicans. These chimeric Ste2 receptors were consistently more active than native receptors, again indicating that the C-terminal tail is important for transducing the pheromone signal. This fact was most striking with C. lusitaniae Ste2, as this receptor was completely nonresponsive to pheromone when expressed in C. albicans, whereas expression of the chimeric version of the receptor resulted in cells forming mating projections and expressing mating-specific genes when challenged with C. lusitaniae pheromone(s). We confirmed that the specificity of pheromone-receptor signaling was maintained in these engineered strains by validating the synthetic C. lusitaniae pheromones on the native species. These results establish that manipulation of Ste2 receptor domains is an effective means for analysis of pheromone-receptor interactions and their downstream signaling pathways.
We recently reported that the Ste2 receptor of C. albicans is activated not only by its cognate pheromone but by pheromones from C. tropicalis, C. parapsilosis, and L. elongisporus and these events could activate self-mating of C. albicans opaque cells or biofilm formation by white cells (Alby and Bennett, 2011). This is despite the fact that some of these pheromones share little homology with C. albicans pheromone (e.g., C. parapsilosis pheromone shares only five out of 13 residues with that of C. albicans). Such interspecies signaling is again noted in this study, as pheromones from one species were occasionally active on receptors from another species. For example, the pheromone receptor of C. tropicalis was shown to respond to pheromones from both C. albicans and C. lusitaniae (Table 2 and Figures 6 and S6). These observations suggest that interspecies signaling between two different Candida species might take place if conditions compatible to mating in both species can be found.
Taken together, our results reveal the regions of the Ste2 receptor that are critical for pheromone signaling in Candida and that heterologous expression of native or chimeric receptors in C. albicans can be used to analyze pheromone-receptor interactions from multiple Candida clade species. We are therefore able to establish that C. tropicalis and C. parapsilosis, two apparently asexual species, have functional pheromone-receptor interactions, and these results increase the likelihood that cryptic mating cycles have yet to be discovered for these species. Our results set the stage for future studies that will further explore the mechanism of pheromone signaling and the potential identification of sexual programs in other Candida species.
MATERIALS AND METHODS
Media and reagents
Media used in the laboratory were prepared as described previously (Bedell and Soll, 1979; Liu et al., 1994). Pheromone peptides were resuspended at a concentration of 10 mg/ml in 10% dimethyl sulfoxide (DMSO).
Plasmids and strains
STE2 deletion strains were made as described previously (Alby et al., 2009). Strains and oligos used in this study are listed in the Tables S3 and Table S4. An addback of the wild-type STE2 gene was made by amplification of the promoter and open reading frame (ORF) using oligos 443/504 and cloning the gene into pSFS2A (Reuss et al., 2004), using ApaI/XhoI to generate pRB19. The construct was linearized with BsaBI, which cuts in the STE2 promoter, and transformed in C. albicans. To construct different mutant alleles of Ste2, we performed fusion PCR reactions to combine different regions of the STE2 gene as follows.
For Ste2(ΔIC1), PCRs were performed with oligo pairs to amplify the promoter of STE2 together with the first part of the STE2 gene up to the IC1 region (oligos 443/620) and also the remainder of the STE2 gene downstream of the IC1 region (oligos 618/504). The two PCR fragments were combined by a fusion PCR reaction with oligos 443 and 504, and the resulting product was cut with ApaI/XhoI and cloned into pSFS2A (Reuss et al., 2004). This construct was linearized with BsaBI and transformed into Δste2/Δste2 strains CAY996, CAY997, or CAY1234.
A similar construct was made for Ste2(ΔIC3) by combining PCR products amplified with oligos 443/619 and 621/504 by subsequent fusion PCR with oligos 443/504. The IC3 product was cloned into pSFS2A with ApaI/XhoI and linearized for transformation with BsaBI. The C-terminal truncation allele Ste2(369) was made by PCR amplification, using oligos 443/622 and 639/504, and combining the products by fusion PCR with oligos 443/504. Similarly, the C-terminal truncation allele Ste2(392) was made by PCR, using oligos 443/623 and 640/504 and combining the products by PCR with oligos 443/504. For both C-terminal alleles, products were digested with ApaI/XhoI, cloned into pSFS2A, and subsequently linearized for transformation with BsaBI. The C-terminal truncation allele Ste2(417) was made by digestion of the STE2 addback plasmid with EcoNI, filling in of the overhang with Klenow (New England Biolabs, Ipswich, MA), and religation to insert an extra base into the ORF. This resulted in a stop codon being in-frame three codons downstream from the original EcoNI site. To construct IC1 and C-terminal double-deletion regions of Ste2 receptor, plasmids pSFS-ΔIC1 and pSFS-C392 were digested with BsaBI/SmaI. The 1.5-kb fragment encompassing part of Ste2 and its promoter from pSFS-ΔIC1 was then cloned into BsaBI/SmaI sites of pSFS-C392 to create pSFS-ΔIC1-ΔC392.
Heterologous STE2 alleles from alternative Candida clade species were expressed in C. albicans under the control of the native STE2 promoter as follows. The C. albicans STE2 promoter was first PCR-amplified, using oligos 443/444 or 458/444, which introduced an ApaI or KpnI site, respectively. The C. lusitaniae STE2 gene was PCR-amplified with oligos 446/447 and fused with the 458/444 product by PCR, using oligos 458/447. This was cloned into pSFS2A, using KpnI and XhoI to generate pRB1. The C. parapsilosis STE2 gene was PCR-amplified with oligos 449/460 and fused with the 458/444 product by PCR, using oligos 458/460. The product was cloned into pSFS2A, using KpnI and SalI to generate pRB27. The C. tropicalis STE2 gene was PCR-amplified with oligos 452/453 and fused with the 443/444 product by PCR, using oligos 443/453. The L. elongisporus STE2 gene was PCR-amplified with oligos 455/456 and fused with the 443/444 product by PCR with oligos 443/456. Both C. tropicalis and L. elongisporus STE2 constructs were cloned into pSFS2A, using ApaI and XhoI to generate pRB18 and pRB17, respectively. All STE2 constructs were linearized by BsaBI prior to transformation into Δste2/Δste2 strains RBY1107 or CAY1478.
Chimeric STE2 alleles were also constructed by fusion PCR. C. lusitaniae chimeric STE2 was generated by PCR of pRB1, using oligos 458/505, and PCR of pRB19, using oligos 502/504. These two products were fused by PCR, using oligos 458/504, and cloned into pSFS2A, using KpnI/XhoI. Chimeric C. parapsilosis STE2 was constructed by PCR of pRB27 with oligos 458/506 and PCR of pRB19, using oligos 502/503. These products were fused by PCR, using oligos 458/503, and cloned into pSFS2A, using KpnI/SalI. Chimeric C. tropicalis STE2 was constructed by PCR of pRB18, using oligos 443/507, and PCR of pRB19, using oligos 502/504. Products were fused by PCR, using oligos 443/504, and cloned into pSFS2A, using ApaI/XhoI. Chimeric L. elongisporus STE2 was generated by PCR of pRB17, using oligos 443/508, and PCR of pRB19, using oligos 502/504. Products were fused by PCR, using oligos 443/504, and cloned into pSFS2A, using ApaI/XhoI.
GFP tagging of several STE2 alleles was performed by addition of GFP to the 3′ end of STE2. All constructed plasmids were transformed into P37005 or SC5314 (Lockhart et al., 2002; Bennett et al., 2003) after the DNA was linearized with BsaBI. To construct pSFS-STE2-GFP plasmid, oligos (443/883) were used to amplify STE2 and its endogenous promoter by PCR. The GFP gene was amplified from pNIM1 (Park and Morschhauser, 2005), using oligos (874/822). In a second round of PCR, a fusion STE2-GFP product was amplified with primers 443/822, the product was digested with ApaI/XhoI and ligated into pSFS2A (Reuss et al., 2004) to create pSFS-STE2-GFP. The linearized plasmid was transformed into Δste2/Δste2 mutants to create CAY1677 (SC5314) and CAY1701 (P37005).
Similarly, for plasmids pSFS-ΔIC1-GFP, pSFS-ΔIC3-GFP, pSFS-C417-GFP, pSFS-C392-GFP, and pSFS-C369-GFP, oligos (443/883, 443/883, 443,884, 443/885, 443/886, respectively) were used to PCR amplify STE2 from plasmids containing pSFS-ΔIC1, pSFS- ΔIC3, pSFS-C417, pSFS-C392, and pSFS-C369, respectively. These PCR products were fused with GFP, using PCR with primers 443/822. The fusion PCR product was digested with ApaI/XhoI and ligated into pSFS2A. The plasmid was linearized with BsaBI and transformed into ste2 mutants RBY1107 or RBY1108. White cells were also switched to the opaque stage, as listed in Table S3. To ascertain these Ste2 domain mutant derivatives were correct, plasmid constructs were sequenced before the transformations.
Pheromone response assays
Overnight cultures of C. albicans opaque cells in Spider medium (Liu et al., 1994) at room temperature were grown to an OD600 of 2.0. Culture (0.5 ml) was added into fresh Spider medium (2.5 ml), and treated with or without pheromones at a final concentration of 10 μg/ml, and incubated at room temperature for another 24 h. The cells were imaged with a Zeiss Observer Z1 microscope (Carl Zeiss Microscopy, LLC). To test the pheromone response in C. lusitaniae, cells were grown as patched colonies on potato dextrose agar (PDA) at 30ºC overnight. Cells were then treated with 3 μl of 10 mg/ml C. lusitaniae pheromones and incubated at room temperature for 24 h before being collected for imaging.
Halo assays
Halo assays were performed as previously described, with slight modifications (Schaefer et al., 2007). Briefly, overnight cultures of opaque cells were grown in synthetic complete dextrose (SCD) medium at room temperature. The cells were washed with sterile water twice. Approximately 4 × 105 cells were plated onto solid Spider medium. Two microliters of α pheromone (10 or 1 mg/ml) or 10% DMSO was spotted onto the cells. Images were taken after 2 d incubation at room temperature.
Quantitative mating assays
Quantitative mating assays were performed as previously described (Bennett and Johnson, 2006). Briefly, overnight cultures of opaque cells with different auxotrophic markers were grown in SCD medium at room temperature. Approximately 2 × 107 cells of each strain were mixed together and placed on a nitrocellulose filter on Spider medium and incubated at 25ºC for 48 h. Cells were removed from the filter, resuspended in water, sonicated to disperse clumps, and plated onto selective media to determine the mating frequency, as previously described (Booth et al., 2010; Alby and Bennett, 2011). Mating products were also confirmed by using primers directed against OBPa and OBPα genes that identify MTLa and MTLα loci, respectively.
Biofilm assays
Biofilm assays were performed to quantitate cells adhering to plastic plates, similar to previous studies (Yi et al., 2008, 2009). Overnight cultures of white cells were grown in Spider medium at room temperature and 5 × 107 cells were resuspended in Lee's medium and added to 12-well culture dishes (Costar; Corning Life Sciences, Lowell, MA). Peptide pheromones were added at a final concentration of 10 μg/ml, and cultures were incubated at room temperature for another 24 h. The plastic wells were gently washed with phosphate-buffered saline (PBS) and imaged. Adherent cells were also removed from the well and quantified at OD600. Each experiment was performed using two independent isolates with at least three replicates.
Quantitative PCR
Cell cultures of opaque and white cells were grown as described in Pheromone Response Assays and Biofilm Assays. Cells were induced with pheromones at a final concentration of 10 μg/ml and incubated at room temperature for 4 h or 24 h. Total RNA was isolated following the RiboPure-Yeast Kit protocol (Applied Biosystems, Bedford, MA). RNA was treated with DNaseI (Applied Biosystems) to eliminate DNA contamination and reextracted with phenol/chloroform. cDNA was synthesized using GoScript Reverse Transcriptase (Promega, Madison, WI). Quantitative PCR was performed in a 7300 Real Time PCR System (Applied Biosystems). The signal from each experimental sample was normalized to expression of the PAT1 gene, as previously described (Bennett and Johnson, 2006). Expression values were averaged from at least three independent experiments.
Supplementary Material
Acknowledgments
We thank Shail Kabrawala for assistance with preparing media, Kevin Alby for helpful discussions, and members of the Bennett lab for comments on the manuscript.
Abbreviations used:
- DMSO
dimethyl sulfoxide
- EC
extracellular
- GFP
green fluorescent protein
- IC
intracellular
- MAPK
mitogen-activated protein kinase
- ORF
open reading frame
- PBS
phosphate-buffered saline
- PDA
potato dextrose agar
- qPCR
quantitative PCR
- RT-PCR
reverse transcriptase PCR
- SCD
synthetic complete dextrose
- TM
transmembrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0749) on October 12, 2011.
REFERENCES
- Alby K, Bennett RJ. Sexual reproduction in the Candida clade: cryptic cycles, diverse mechanisms, and alternative functions. Cell Mol Life Sci. 2010;67:3275–3285. doi: 10.1007/s00018-010-0421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K, Bennett RJ. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci USA. 2011;108:2510–2515. doi: 10.1073/pnas.1017234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell GW, Soll DR. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun. 1979;26:348–354. doi: 10.1128/iai.26.1.348-354.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ. Coming of age–sexual reproduction in Candida species. PLoS Pathog. 2010;6:e1001155. doi: 10.1371/journal.ppat.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol Microbiol. 2006;62:100–119. doi: 10.1111/j.1365-2958.2006.05367.x. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Miller MG, Chua PR, Maxon ME, Johnson AD. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol Microbiol. 2005;55:1046–1059. doi: 10.1111/j.1365-2958.2005.04466.x. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth LN, Tuch BB, Johnson AD. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature. 2010;468:959–963. doi: 10.1038/nature09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G. Fungal sex and pathogenesis. Clin Microbiol Rev. 2010;23:140–159. doi: 10.1128/CMR.00053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinault SL, Overton MC, Blumer KJ. Subunits of a yeast oligomeric G protein-coupled receptor are activated independently by agonist but function in concert to activate G protein heterotrimers. J Biol Chem. 2004;279:16091–16100. doi: 10.1074/jbc.M311099200. [DOI] [PubMed] [Google Scholar]
- Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 2006;25:2240–2252. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosil M, Schandel KA, Gupta E, Jenness DD, Konopka JB. The C terminus of the Saccharomyces cerevisiae α-factor receptor contributes to the formation of preactivation complexes with its cognate G protein. Mol Cell Biol. 2000;20:5321–5329. doi: 10.1128/mcb.20.14.5321-5329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Gargas A, Taylor JW. Phylogeny of Discomycetes and early radiations of the apothecial Ascomycotina inferred from SSU rDNA sequence data. Exp Mycol. 1995;19:7–15. doi: 10.1006/emyc.1995.1002. [DOI] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Jones SK Jr, Bennett RJ. Fungal mating pheromones: choreographing the dating game. Fungal Genet Biol. 2011;48:668–676. doi: 10.1016/j.fgb.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka JB, Jenness DD, Hartwell LH. The C-terminus of the S. cerevisiae α-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci USA. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BK, Khare S, Naider F, Becker JM. Identification of residues of the Saccharomyces cerevisiae G protein-coupled receptor contributing to α-factor pheromone binding. J Biol Chem. 2001;276:37950–37961. doi: 10.1074/jbc.M103579200. [DOI] [PubMed] [Google Scholar]
- Lin JC, Parrish W, Eilers M, Smith SO, Konopka JB. Aromatic residues at the extracellular ends of transmembrane domains 5 and 6 promote ligand activation of the G protein-coupled α-factor receptor. Biochemistry. 2003;42:293–301. doi: 10.1021/bi026766o. [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol Microbiol. 2002;46:1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x. [DOI] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Naider F, Becker JM. The α-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides. 2004;25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Palczewski K, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers—their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recca J, Mrak EM. Yeasts occurring in citrus products. Food Technol. 1952;6:450–454. [Google Scholar]
- Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol. 2009;19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Ruhnke M, Maschmeyer G. Management of mycoses in patients with hematologic disease and cancer—review of the literature. Eur J Med Res. 2002;7:227–235. [PubMed] [Google Scholar]
- Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol. 2010;8:e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni N, Yi S, Daniels KJ, Srikantha T, Pujol C, Soll DR. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 2009;5:e1000601. doi: 10.1371/journal.ppat.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai S, Holland LM, McGee CF, Lynch DB, Butler G. Evolution of mating within the Candida parapsilosis species group. Eukaryot Cell. 2011;10:578–587. doi: 10.1128/EC.00276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D, Cote P, Whiteway M, Bennett RJ. Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryot Cell. 2007;6:907–918. doi: 10.1128/EC.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RK, Bennett RJ. Fungal meiosis and parasexual reproduction—lessons from pathogenic yeast. Curr Opin Microbiol. 2009;12:599–607. doi: 10.1016/j.mib.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Blumer KJ. The third cytoplasmic loop of a yeast G-protein-coupled receptor controls pathway activation, ligand discrimination, and receptor internalization. Mol Cell Biol. 1994;14:3339–3349. doi: 10.1128/mcb.14.5.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Pryciak PM. Distinct roles for two Gα-Gβ interfaces in cell polarity control by a yeast heterotrimeric G protein. Mol Biol Cell. 2008;19:181–197. doi: 10.1091/mbc.E07-04-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier LG, Segall JE, Snyder M. The alpha-factor receptor C-terminus is important for mating projection formation and orientation in Saccharomyces cerevisiae. Cell Motil Cytoskeleton. 2002;53:251–266. doi: 10.1002/cm.10073. [DOI] [PubMed] [Google Scholar]
- van der Walt JP. Lodderomyces, a new genus of the Saccharomycetaceae. Antonie Van Leeuwenhoek. 1966;32:1–5. doi: 10.1007/BF02097439. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell. 2008;19:957–970. doi: 10.1091/mbc.E07-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Pujol C, Daniels KJ, Srikantha T, Ma N, Soll DR. A Candida albicans-specific region of the α-pheromone receptor plays a selective role in the white cell pheromone response. Mol Microbiol. 2009;71:925–947. doi: 10.1111/j.1365-2958.2008.06575.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.