Abstract
Natural killer (NK) cells mediate innate defense against viral infections, but the mechanisms in place to access their functions as needed during diverse challenges while limiting collateral damage are poorly understood. Recent molecular characterization of effects mediated through infection-induced inhibitory/activating NK receptor-ligand pairs and cytokines are providing new insights into pathways regulating their responses and revealing unexpected consequences for NK cell subset effects, maintenance, proliferation and function through times overlapping with adaptive and long-lived immunity. The observations define flexible pathways for experience-induced “conditioning” and challenge narrowly defined roles for NK cells and innate immunity as first responders with prescribed functions. They suggest that individual experiences as well as genes influence the innate immune resources available to fight off an infection.
Introduction
Natural killer (NK) cells were originally described as lymphocytes of the innate immune system capable of eliminating tumor and infected cells without prior antigen exposure [1]. This was quickly followed by the demonstration of the first NK cell activating receptor, the receptor for Fc portions of immunoglobulin molecules, CD16, and the ability of NK cells to interface with these components of adaptive immunity to mediate antibody-dependent cellular cytotoxicity (ADCC) [1]. NK cells are now known to be proficient at many important tasks [2] but are still best appreciated for their non-redundant roles in defense against some viruses. Plentiful studies in the mouse and human demonstrate NK cell-dependent protective effects during infections with coxsackievirus, human immunodeficiency virus (HIV), hepatitis C virus (HCV), influenza virus, and poxvirus, but most importantly herpesviruses [3-5]. The mechanisms for delivery of their direct antiviral effects include killing of infected target cells and production of interferon γ (IFNγ), and absence of the cells prior to infection increases early replication of viruses sensitive to NK cell-mediated defense [5,6]. There is compelling evidence that their antiviral effects are regulated by a repertoire of germ-line encoded NK cell receptors (NKRs) recognizing ligands on virus-infected cells and by innate cytokine responses induced during infections. Despite extensive work in many laboratories, the molecular details of the pathways controlling NK cell responses have been elusive because of the many closely related inhibitory and activating receptors as well as the pleotropic and sometimes paradoxical effects of cytokines.
Exciting new molecular characterization of the NK cell receptor-ligand pairs and cytokine effects modified in response to viral infections are filling in significant gaps in knowledge. In addition to providing details on the pathways controlling NK cell contributions to defense and those protecting from potential immune-mediated pathology, the new information is revealing unexpected flexibility in NK cell responses based on experience-induced “conditioning”. In particular, much progress has been made in identifying infection-induced changes in the expression of ligands for NK cell activating and inhibitory receptors, in the role for activating receptors in expanding and sustaining NK cell subsets for extended periods, and in their responses to innate cytokines. The observations have implications for understanding the regulation of NK cell functions not only during early innate responses to viruses but also during periods overlapping with adaptive and long-term immunity. Moreover, they suggest that individual variations in resistance to infection are based on experiences as well as genes.
NK Cell Receptors
The NK cell receptors (NKRs) delivering inhibitory signals were first appreciated because of their ability to receive negative “self” signals from class 1 major histocompatibility (MHC1) molecules expressed on normal tissues [7,8]. It is now clear that there are complex groups of NKRs with different genetic and structural characteristics, representatives of both inhibitory and activating receptor in each group, and individual NK cells expressing combinations of both inhibitory and activating receptors (Fig. 1). In total, the NKRs are thought to provide NK cells with a system for surveying the extracellular environment such that the integration of resulting positive and negative signals determines the overall nature of their responses with a balance towards inhibition under normal conditions but a shift to activation under conditions of infection or stress [8,9].
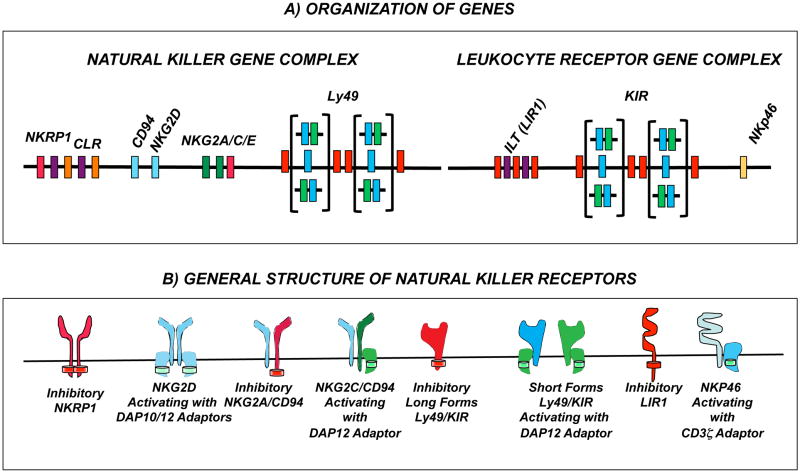
Figure 1. NK cell receptor genes and their products.
Only receptors mentioned in the text are represented. (A) NK cell receptor genes reside principally in two genomic regions named the natural killer gene complex or the lymphoctye receptor complex. The natural killer gene complex (NKC) on human chromosome 12p12-p13 and mouse chromosome 6 encodes receptor molecules of the the C-type lectin-like family. Within the Ly49 gene family presents the most extensive inter-specific and inter-individual variation in terms of gene content. Whereas in rodents Ly49 define haplotypes with different gene number and extensive allelic polymorphism, in humans, there is a single Ly49 gene which corresponds to the pseudogene, LY49L. Other C-type receptor genes (including the single CD94 gene as well as the NKG2, NKRP1 and CLR gene families are more conserved. The leukocyte receptor complex, on human chromosome 19q13.4 and mouse chromosome 7, encodes immunoglobin-like receptors. Within this complex, the KIR gene family is highly variable between species and individuals. Humans carry polymorphic KIR haplotypes that also vary in gene number. Rodents lack KIR genes but carry ILT (known as paired-iimunoglobin-receptor (Pir) genes in mice) and Nkp46 gene homologues. (B) Natural killer receptor products of the genes mediate inhibitory or activating signals, the balance of which determines NK cell activity. Irrespective of their family, inhibitory NK cell receptors (in red) bear an immunotyrosin inhibitory motif (ITIM, red disk) in the cytoplasmic domain. Activating receptors (other colors) associate with adaptor proteins that carry immunotyrosine activating motifs (ITAM, green disks). NKG2D forms homodimers expressed in virtually all NK cells. The preferred adaptor protein is DAP10 but in mice NKG2D can also associate with DAP12. Activating NKG2C/CD94, Ly49 and KIR receptors associate to the signaling adaptor DAP12 whereas NKP46 uses CD3ζ.
The NKRs can be segregated into two main classes based on their distinct extracellular structure: the Ig-like and the C-type lectin-like receptors [10]. The majority of Ig-like receptors, including the killer Ig-like receptors (KIRs) in humans, map to the leukocyte receptor cluster (LRC) present on human chromosome 19p and mouse chromosome 7. Families of C-type lectin-like receptor genes, including Ly49 in mouse and CD94, NKG2A/C/E, NKRP1 and NKG2D receptors in human and mouse, reside in a region known as the natural killer gene complex (NKC) on human chromosome 12p or mouse chromosome 6 [11]. The NK cell repertoire, and to a certain extent the ability of NK cells to recognize specific target cells, stems from genetic polymorphism as well as different combinations and expression levels of NKRs. The KIR receptors in humans subserve the function of the murine Ly49 receptors. Both sets of receptors belong to highly polygenic and polymorphic families with different individuals or strains inheriting genetic variation in the individual genes as well as different numbers of genes. These receptors are expressed on overlapping subsets of NK cells in a stochastic fashion [12,13]. By contrast, CD94 and NKG2 genes have limited polymorphism and are orthologous genes in mouse and human [14,15]. Essentially all NK cells express NKG2D whereas CD94/NKG2A and CD94/NKG2C receptors are expressed on overlapping subsets of NK cells. Unlike the KIR and Ly49 receptors that are stably maintained once expressed, CD94/NKG2 receptors can be modulated by cytokines in the environment [16]. With few exceptions, NKRP1 genes are conserved and expressed in the majority of NK cells [13].
Inhibitory KIR and Ly49 NKRs have been more extensively characterized than their activating counterparts, especially with regard to ligand-binding specificity and function. These receptors individually possess a high binding affinity for particular polymorphic MHC1 molecules (HLA-A/B/C in humans; H2-D/K/L in mice). The CD94/NKG2A heterodimers are also inhibitory and bind to non-classical MHC1 molecules (human HLA-E and mouse Qa1) [16]. Inhibitory receptors signal via immunoreceptor tyrosine-based inhibition motifs (ITIMs) located in their cytoplasmic tails. Inhibitory signals are predominant under normal conditions, when the majority of target cells express MHC1 molecules. When target cells are transformed or infected, surface expression of MHCI molecules can be abrogated; the absence of inhibitory NKR ligation allows the NK cell to kill its target (“missing-self” recognition).
In addition to a requirement for release from dominant inhibitory signaling, delivery of effective NK cell killing requires the ligation of activating NKRs. Activating NKR lack ITIMs, but they associate with membrane-bound adaptor molecules, such as DAP12 (Ly49, KIR, mouse NKG2D, NKG2C/E), FcεRγ, and CD3ζ (CD16, NKp46), that bear immunoreceptor tyrosine-based activation motifs (ITAMs) [8,13,17]. The phosphorylation of ITAM leads to the recruitment of the tyrosine kinases, Zap70 and Syk. In addition, human NKG2D, and a mouse NKG2D length variant, associate with DAP10. The signaling motif in DAP10 is a YINM sequence, with phosphorylation leading to the binding of PI3K and Grb2 for signaling [18]. In the human, a KIR2DL4 receptor can have both inhibitory and stimulating activity. KIR2DL4 has a cytoplasmic tail containing a single ITIM, and a charged amino acid in the transmembrane domain that interacts with FcεRγ [19]. Some activating NKRs have no or attenuated affinity for MHC1 molecules. Others bind stress-induced molecules or non-self peptides thought to be associated with MHC1 molecules. Remarkably, the activating NKG2D receptors are evolutionary conserved and inherited by all individuals in a species. Likewise, activating CD94/NKG2C and NKp46 receptors are conserved and broadly inherited. Thus, in contrast to the diversity of receptors inherited for the Ly49 and KIR genes, the NKG2D, CD94/NKG2C, and NKp46 receptors provide opportunities for accessing responses in different individuals under different conditions of infections.
Viral infection-induced receptor-ligand pairs in the mouse
Some activating NKRs are able to directly recognize virus-infected cells. For example, Ly49H from C57BL/6 resistant mice was the first NKR demonstrated to be a viral resistance gene (cmv1) and mediates its effects by recognizing the mouse cytomegalovirus (MCMV) m157 viral protein, which is highly homologous to MHC1 and expressed only on MCMV-infected cells [20-23] (Fig. 2; Table 1). Ligation of Ly49H by m157 results in the release of cytolytic granules, cytokines, and chemokines, as well as a robust NK cell proliferation [24], and although only short of half of the NK cells in strains of mice having the Ly49h gene basally express the molecule, the proportions are dramatically increased during the infection [25]. Investigation of this activating receptor-ligand pair has made a number of key discoveries concerning the role for activating receptors in accessing NK cell responses to viral infection possible, but combinations of activating receptors stimulated directly by viral products may be rare because of their negative effects on viral replication and the ability of viruses to be selected with modified genetic material under immune pressure. Another example might be the influenza hemagglutinin (HA) glycoprotein binding to NKp46 [26].
Figure 2. Receptor regulation of NK cells during viral infections.
The inhibitory receptors are mostly appreciated for their ability to recognize classical and non-classical major histocompatibility class 1 (MHC1) molecules in balancing towards dampened NK cell stimulation under normal conditions, but there are a few examples of viral products accessing these receptors to avoid NK cell-mediated defense. There is a growing list of examples of activating receptors, from both highly diverse and highly conserved gene famlies in human and mouse, recognizing ligands expressed on virus-infected cells. A few of these directly recognize viral products. Most indirectly recognize either MHC1 or non-classical MHC1 along with virus-induced changes on infected cells or stress molecules induced in virus-infected cells. (SeeTable 1 for detailed examples.)
Table 1. NK Cells in Viral Infection: Evidence for Activating Receptors and Ligands.
| Receptor Class | Species | Virus Family | Virus | Receptor Involvement | Ligand | References |
|---|---|---|---|---|---|---|
Single Molecule Associated with Activating Adaptor Highly Polygeneic Recognition Molecule |
Mouse | Herpesviridae | MCMV | Ly49H | m157 | [21-24] |
| Ly49P | H-2Dk (+ m04) | [29] | ||||
| Ly49L | H-2k (+ m04) | [32] | ||||
| Ly49P1 | [32] | |||||
| Human | Retroviridae | HIV | KIR3DS1 | HLA-B (Bw4-80I) | [47-50] | |
| Herpesviridae | EBV | KIR2DS1 | HLA-C | [55] | ||
|
| ||||||
 Genetically-Conserved Recognition Molecule |
Mouse | Orthomyxoviridae | Influenza | NKp46 | Hemagglutinin (HA) | [26] |
| Human | Orthomyxoviridae | Influenza | NKp46 | HA | [52] | |
| NKp30 | HA | [51] | ||||
| Flaviviridae | HCV | NKp30, NKp46 | ? | [46,92] | ||
| Herpesviridae | HSV | NKp30, NKp44, NKp46 | ICP10? | [54] | ||
| Herpesviridae | KSV | NKp80 | activation-induced C-type lectin | [57] | ||
| Poxviridae | vaccinia | NKp30, NKp44, NKp46 | ? | [56] | ||
| Filovirus/Rhabdoviridae | Ebola/Marburg | NKp30 | ? | [53] | ||
|
| ||||||
Molecule Associated with CD94 and Activating Adaptor Genetically-Conserved Recognition Molecule |
Mouse | Poxviridae | Ectromelia | CD94/NKG2E | Qa-1b (+ ?) | [33] |
| Human | Herpesviridae | HCMV | CD94/NKG2C | HLA-E (+?) | [43] | |
| Bunyavirdea | Puumala hantavirus | CD94/NKG2C | HLA-E (+?) | [42] | ||
| Hepadaviridae | HBV | CD94/NKG2C | ? | [45] | ||
| Flaviviridae | HCV | CD94/NKG2C | ? | [45,46] | ||
|
| ||||||
Homodimer Associated with Activating Adaptor Genetically-Conserved Recognition Molecul |
Mouse | Herpesviridae | MCMV | NKG2D | Stress Molecules (Raeα-ε, H60a-c, MULT1) | [35] |
| Human | Herpesviridae | HCMV | NKG2D | Stress Molecules (MICA-C, ULBP1-6) | [35] | |
| Herpesviridae | KSV | NKG2D | Stress Molecules (MICA-C, ULBP1-6) | [57] | ||
Recent studies have extended the characterization of stimulatory receptor-ligand pairs during MCMV infections to activating NKRs recognizing changes induced by infection and restricted by MHC1 molecules. NK cells have a protective effect in MCMV-infected mice of the MA/My strain, which lacks Ly49H. Under these conditions, resistance is dependent on both the H-2Dk and the NKC loci [27,28], with an activating Ly49P receptor recognizing MCMV-infected cells and in vivo defense depending on the presence of both the viral m04/gp34 protein and H2-Dk [27,29**]. In H2-Dk transgeneic mice, MHC1 controls early viral replication and robust NK cell proliferation; however, the specific activating NKR responsible for these effects has not yet been identified [30*,31*]. Other activating Ly49 receptors recognizing MCMV-infected cells in vitro in an m04-specific and H2-dependent manner include the Ly49P1 receptor from NOD/Ltj mice, Ly49D2 from PWK/Pas mice, and Ly49L from BALB/c mice [32**]. Of these, only Ly49LBALB/c has been validated in vivo with infection of BALB mice having both NK cell and H2k-dependent control of viral spread as compared to H2d or H2b animals. Here, the antiviral effects correlate with expansion of Ly49L+ NK cells and IFNγ secretion, both of which are abrogated during infection with Δm04 MCMV. Thus, there is a developing picture of different combinations of the highly polygeneic and polymorphic Ly49 activating receptors in the mouse with resistance to MCMV depending on the presence of particular polymorphic MHC1 molecules and viral products. The interactions between the viral products and/or their peptides with MHC1 remain to be defined, but the observations fit a model of particular MHC1 molecules presenting peptides to particular activating receptors. They suggest that the interactions may have provided important evolutionary pressure for the diversification of these receptor-ligand combinations.
Another family of receptors, NKG2, has recently been implicated in resistance to ectromelia virus (mouse pox). The CD94/NKG2E heterodimer specifically triggers NK cells for activation via the recognition of the non-classical class 1 molecule, Qa-1b on virus-infected cells, and the combination results in improved survival. Again, whether the virus modifies Qa-1b expression or provides a peptide for Qa-1b presentation remains unknown, but the combination in this case is between a conserved activating receptor and an inherited polymorphic host molecule influenced by the infection [33**]. Another conserved activating receptor, NKG2D, is broadly expressed on all NK cells. It recognizes multiple stress ligands (MICA-C and ULBP1-6 in humans; Raeα-ε, H60a-c, and MULT1 in mice) that are induced during CMV infection [35]. The importance of these conserved receptor-ligand pairs is validated by the demonstration that both mouse and human CMV have mechanisms for blocking the expression of NKG2D ligands on the cell surface to escape NK cell detection [35]. In short, current data indicate that numerous activating receptors allow NK cells to recognize MCMV infection either directly (Ly49H:m157) or indirectly (Ly49P/L:m04:H2-Dk; NKG2D:stress molecules).
Like their activating counterparts, inhibitory NKRs can recognize infected cells in numerous ways. The viral product m157 is recognized by an inhibitory Ly49 receptor [23]. Several others are triggered (directly or indirectly) in the presence of m04/gp34. Most susceptible strains, including BALB, show improved viral control when infected with Δm04 MCMV [36**]. One reason for this could be the role of m04/gp34 in preventing “missing-self” recognition by NK cells because the protein is known to escort MHC1 molecules to the surface of infected cells to maintain a basal level of expression throughout the infection. This basal level of MHC1 expression is sufficient to trigger several inhibitory receptors (e.g., Ly49A). During infections with Δm04 MCMV, MHC1 ligands are depleted and viral titers significantly decrease. Conversely, the presence of m04/gp34 during MCMV infection impairs NK cell in vivo expansion and ex vivo killing of cognate target cells as compared to Δm04 MCMV infection. Effects of the inhibitory Ly49C receptor can also be observed during MCMV infections. Ly49H+ NK cells bearing Ly49C have decreased IFNγ secretion and proliferation compared to Ly49H+Ly49C- NK cells [37]. In studies of the H2q allele impact on MCMV resistance in H-2Dk transgenic mice, H-2Dk animals are only marginally more resistant than their non-transgenic counterparts in the presence of H2q. Indeed, a gene dosage effect is observed on the inhibitory action of H2q [31]. This suggests that inhibitory NKR triggering by H2q dominates over activating NKR triggering by H2-Dk on MCMV-infected cells. However, it is also possible that inhibitory receptors benefit proper NK cell responses. For example, in the context of MA/My resistance Ly49G+ NK cells are found to preferentially expand and secrete IFNγ upon MCMV infection, while their depletion increases viral titers. It remains to be clarified if the improved NK cell antiviral response of Ly49G+ NK cells results from MHC1-dependent education [30,38] or from MHC1-independent mechanisms at play during infection [39].
Viral infection-induced receptor-ligand pairs in the human
Human viral infections are infrequently studied in the acute phase due to logistical difficulties in obtaining samples. However, there are some informative studies. Peripheral blood NK cell numbers are reduced in the acute phase of influenza H1N1 infection and in acute hepatitis C virus (HCV) infection the numbers of cytotoxic CD56dim NK cells are also reduced [40,41*]. Conversely, in acute Puumala hantavirus infection, NK cell numbers are elevated up to two-fold [42**]. The expansion is associated with increased proliferation, increased plasma levels of IL-15, and increased expression of NKG2C. Hantavirus infected endothelial cells upregulate HLA-E, and both IL-15 and HLA-E are required to drive NK cell proliferation. The increases in NK cells are long-lived and more prominent in CMV-seropositive individuals. Because CMV seropositivity is associated with increased levels of NKG2C-positive NK cells [43], CMV may “prime” NKG2C NK cells to respond to hantavirus. The expansion may occur by upregulating HLA-E through expression of an HLA-E binding peptide [44], and/or an as of yet unidentified virus-induced peptide [42**]. Upregulation of NKG2C has also been observed in chronic hepatitis B virus (HBV) and HCV infections [45,46*]. This may therefore be a common theme of viral imprinting on the NK cell repertoire through NKG2C activation.
Human NK responses can be driven by either conserved activating receptors such as NKG2C or by the polymorphic KIR system. The activating KIR3DS1 alone or in combination with its HLA-Bw480I ligands is associated with a beneficial outcome of HIV infection [47-49]. This immunogenetic observation is supported by in vitro assays of KIR3DS1-positive NK cell effects for limiting viral replication in HIV-infected HLA-Bw480I-positive targets [50]. Taken together, the studies in the human indicate that activating receptor-ligand pairs provide protection against acute infections, stimulate increases in NK cell numbers or particular subsets, and result in long-term benefit to the host. The extended periods of increases in NK cells expressing the conserved NKG2C receptor following different viral infections suggest that this receptor might provide protection to most members of a species if its ligands are induced. Conversely, because the KIR activating receptors are polymorphic and polygenic and the MHC1 molecules are polygenic, any requirement for these receptor-ligand pairs would allow opportunities for viruses to pass through “holes” in the genetic repertoire of subgroups in a diverse population while others mount effective defense.
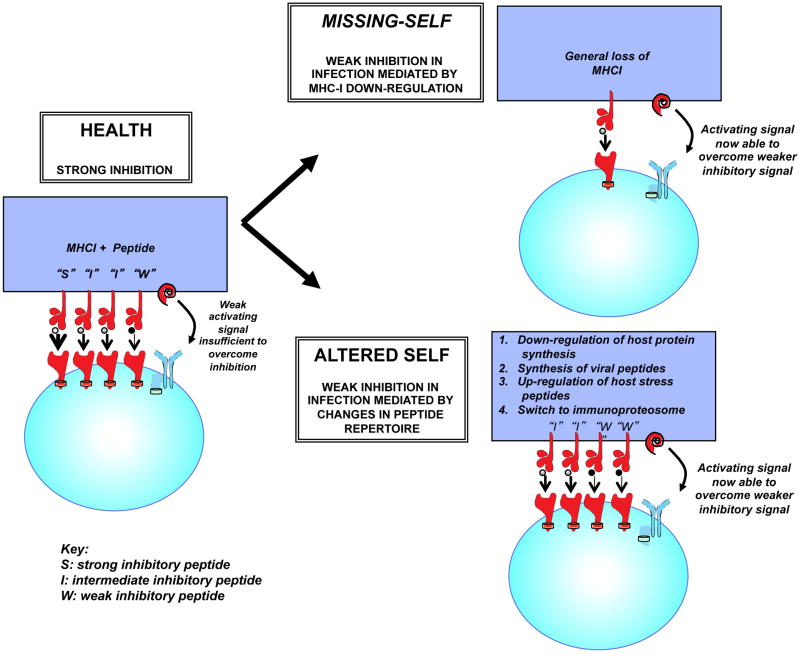
NK cell activation occurs when target cells upregulate ligands for activating receptors that are present. In addition to those discussed above, the list of human NK activating receptors associated with recognition of viruses is growing, and candidates are in place for influenza virus [51,52], Ebola [53], herpes simplex [54], Epstein Barr [55], vaccina virus [56], and Kaposi's sarcoma-associated viruses (KSV) [57]. NK cell activation, however, also may occur when constitutive inhibitory signals are lost, such as by MHC1 down-regulation on infected cells [58,59]. An important role for inhibitory receptors has been shown in HIV infection in which an allelic hierarchy of KIR3DL1:HLA-Bw4 interactions can determine the rate of HIV disease progression [60,61]. Similarly in HCV infection, KIR2DL3 and its group 1 HLA-C (HLA-C1) ligands are protective against chronic infection and are associated with a beneficial response to IFNα based therapies [62-65]. The KIR2DL3:HLA-C1 combination is a relatively weak inhibitory interaction when compared to other inhibitory KIR:HLA assortments. Critically, it has been shown that HLA class 1 down-regulation is not necessary to modulate this inhibitory interaction, and that changing the peptide content of the HLA molecule is a more efficient mechanism of removing an inhibitory signal on an NK cell [66**] (Fig. 3). This may be important for viral infections without specific mechanisms to downregulate MHC1, i.e. “missing self”, and likened to sensing “altered-self”. The DRiP hypothesis suggests that peptides loaded onto MHC1 are in a large part derived from unfolded recently synthesized polypeptides or defective ribosomal products “DRiPs” [67]. Thus, viral infection may change the MHC1 peptide repertoire more rapidly than would be anticipated from degradation of mature viral proteins, thereby permitting a rapid NK cell response. This may be important for HIV infection, as a peptide variant that escapes T cell recognition also abrogates KIR3DL1 binding, and could thus lead to NK cell activation [68]. Furthermore HIV can adapt to the NK cell response by mutating to increase the inhibition of KIR2DL2+ NK cells [69**]. Hence, resistance might be linked to an inhibitory receptor, and this would require particular combinations of inhibitory receptors and MHC1 molecules.
Figure 3. Two potential mechanisms for losing inhibitory signals from KIR receptors.
NK cells are regulated by a balance between activating and inhibitory receptors. In health this balance favors inhibition of NK cells, because the inhibitory signal dominates the activating signal. During viral infections the inhibitory signal derived from MHC class I may be lost by down-regulation of cell surface MHC class I, thus favoring NK cell activation: “Missing-self model”. However multiple mechanisms exist to change the peptide repertoire of cell surface MHC class I. If this change favors presentation of weak inhibitory “antagonist” peptides then these peptides can efficiently disrupt the inhibitory signal due to strong and intermediate inhibitory peptides, and thus also favor NK cell activation: “Altered-self model”.
Synergy between innate receptor mediated and cytokine responses may be an important factor in the ability of the host to eradicate viral infections. With the exception of CD16, activating NK cell receptors function as co-receptors and require stimulation of at least two receptors or one receptor plus cytokine stimulation [70*,71]. Moreover, cytokines can influence expression of target cell ligands to regulate NK cell delivery of antiviral effects with interferons (IFNs) having profound effects on protein/peptide synthesis and MHC1 expression [72]. In HCV infection, a SNP upstream of the IFNλ3 gene (IL-28B), a type III IFN, is strongly associated with clearance of infection [73**,74*,75*,76*,77*]. KIR2DS3 and IFNλ3 synergize on a genetic level to render individuals particularly susceptible to chronic HCV infection [78*]. However, KIR2DL3:HLA-C1 and this polymorphism do not provide synergistic protective effects [79]. The paradox of HCV infection is that the protective IFNλ3 SNP and clearance of HCV infection are both associated with lower levels of interferon stimulated genes [80]. This, combined with the observation that successful treatment of HCV infection in liver transplants assorts with the donor liver IFNλ3 genotype [81], implies that instead of synergizing at the level of the NK cell, these two genetic mechanisms act at discrete places during the innate immune responses; IFNλ3 acting at a direct intracellular level to inhibit virus replication within infected cells and KIR acting at the level of NK cell activation to enhance this arm of immune defense.
NK receptors for proliferation, maintenance and function following infection
Early interest in NK receptors was driven by efforts to understand the regulation of innate NK cell-mediated killing of virus-infected target cells, but delivery of activating signals can also lead to proliferation. Because NK cells are relatively abundant in many tissues, and high proportions basally express or are induced by cytokines to express different activating receptors, the advantages of NK cell proliferative responses have remained obscure. Interestingly, however, NK cells numbers or subset frequencies can be decreased during particular conditions of viral infections. As noted above, reductions are observed during acute infections with influenza or HCV [40,41], but they have also been reported during symptomatic primary infections with Epstein Barr virus (EBV) [82] and during chronic infections with HCV and HIV [83-87]. Thus, NK cell loss can accompany viral infection. Given that NK cells have the potential to mediate a wide range of important biological effects [1,6], such loss can significantly alter the resources available for fighting off viruses. Interestingly, activating receptor-ligand pairs, depending on the repertoire of highly polygeneic and polymorphic KIR receptors and a particular MHC1 molecule inherited by an individual, are associated with recognition of EBV and HIV [50,55]. In the case of HIV, the combination is coupled with NK cell proliferation and increased cell yields [88] as well as with beneficial long-term outcomes, including protection against opportunistic infections [47-49]. The ability of activating receptors to stimulate proliferation suggests that they might have important biological consequences for maintaining NK cells during an infection as well as delivering direct antiviral effects.
Mouse studies examining the consequences of Ly49H deficiency as compared to the absence of cytotoxicity due to a deficiency in perforin 1 have separated the biological importance of an activating receptor's role in proliferation from any function it has in killing [89**]. The ligand for Ly49H, m157, is induced, and resistance to infection is elevated with NK cell proliferation stimulated during MCMV infections of mice expressing Ly49H. The absence of either Ly49H or perforin results in elevated levels of early MCMV replication. There are, however, profound differences in NK cell proliferation and yields, with the populations disappearing under Ly49H-deficient, but Ly49H+ NK cells dramatically expanding under perforin-deficient, conditions. The critical role for Ly49H in supporting proliferation in the absence of cytotoxicity is shown under double deficient conditions. These observations indicate that if an activating receptor is missing and/or if the virus does not induce a ligand for an activating receptor, NK cells would fail to limit early infection such that under the resulting circumstances of sustained viral replication, maintenance of and access to NK cells would not be sustained for extended periods of time (Fig. 4). By protecting from events paralleling those of “exhaustion” in T cell responses to viral infections [90], inhibitory receptors might help balance the magnitude of the proliferative response to obtain optimal numbers and frequencies of cells, but this putative role in maintaining NK cell subsets has yet to be examined. Thus, although there is much to be learned about controlling the magnitude of NK cell proliferation, the results to date demonstrate that signaling through activating receptors is important for sustaining the populations into later periods of infection.
Figure 4. Receptor-mediated regulation of NK cell subset proportions and numbers during viral infection.
The importance of stimulating NK cells through activating receptors to modify the proportions of subsets expressing particular activating receptors and/or sustain their numbers under conditions of extended viral infection is becoming clear. It requires the induction of a ligand by the viral infection and the presence of an activating receptor for the ligand on NK cells. Because the absence of stimulation through activating receptors can result in profound decreases in total NK cell numbers or subsets (red triangles, pointing down) (A) whereas the stimulation through activating receptors can sustain the cells but change the proportions expression particular activating receptors (green triangles, pointing up) (B), the responses have consequences for availability of innate resources in fighting off infection. The example depicted is based on expression of a highly polymorphic and polygenic activating receptor in mouse and human, i.e. Ly49 or KIR. Any requirement for these receptors would allow opportunities for viruses to pass through “holes” in the genetic repertoire of subgroups in a diverse population while others mount effective defense. Use of conserved activating receptors, such as CD94/NKG2C, might act to prime for responses to several viruses and enhance defense against subsequent primary viral infections. Thus, the mechanism of changing the proportions or numbers of NK cells during conditions of extended viral infections is an example of how NK cells are conditioned by experience.
What do NK cells do if they are maintained? In addition to mediating defense that can be strictly assigned to periods of innate immunity, NK cells help bridge innate and adaptive immunity in part by producing cytokines, particularly IFNγ, or mediating cytotoxicity to shape downstream adaptive responses. In the case of acute HCV infection of humans, there is NK cell activation of cytotoxic and IFNγ production that does not correlate well with control of viremia [41], but a detected degranulation does correlate with the magnitude of HCV-specific T cells [91]. Surprisingly, the sustained Ly49H+ NK cells that proliferate into periods overlapping with adaptive immunity during MCMV infections of perforin-deficient mice make IL-10 to regulate the magnitude of adaptive immune responses and protect from CD8 T cell-mediated pathology [89]. Increased proportions of NK cells with particular activating receptors producing IL-10 are also observed during chronic HCV infections in human [92]. The regulatory effects of the NK cell IL-10 production can protect from T cell-mediated pathology but have also been suggested to interfere with protective T cell responses against the virus. Thus, a consequence of NK cell expansion and maintenance is providing access to the populations for mediating immunoregulatory as well as direct antiviral effects.
Finally, there are indications that NK cells activated in responses to viral infections and/or vaccination with viral ligands can become long-lived cells with the memory characteristics of specificity and heightened recall during secondary challenges [93**,94**,95]. The first and best studied of these long-lived NK cells are the Ly49H+ cells elicited in response to MCMV infection [93**]. Their importance as memory populations under immunocompetent conditions remains to be determined, as most of the work has relied heavily on immunodeficient conditions to demonstrate the trait, and as the fitness of the populations in the context of memory T cells has not been tested [96]. Nevertheless, the demonstration of the induction of such long-lived populations through activating receptor-ligand pairs, may provide an explanation for the HCMV conditioning, with expansion of NKG2C NK cells, for enhanced resistance to hantavirus infections in humans [42**,43]. It also provides evidence for the influence of experience on an individual's innate immune resources.
Induction of innate cytokines and regulation of NK cell responses
The importance of innate cytokines in regulating NK cell functions was first realized with the demonstration that the antiviral type 1 IFNs, IFNαβ, were potent inducers of elevated NK cell-mediated lysis [1]. Since that time, it has becomes clear that many viral infections induce complex innate pro-inflammatory cytokine cascades, including in addition to type 1 IFNs, biologically active IL-12p70, IL-18 and IL-15 [97]. In terms of overall kinetics of production, cellular sources, and receptor-sensing for production, these responses have been best characterized following MCMV challenges of mice or mouse cells [98-104]. Not all viruses are the same, however, and infections of mice with lymphocytic choriomeningitis virus (LCMV), an agent inducing high systemic levels of type 1 IFN production for expended periods of time, elicit little to no detectable IL-12 [98,105-107]. In general innate cytokine responses are conserved in the human, but they differ substantially amongst extant human viral infections [97]. In acute HIV infection, there is a rapid increase in IFNα and IL-15, with a more delayed increase in serum IL-18 and IL-12p70 [108*]. The cytokine response to HCV infection is delayed following infection and is attenuated further in response to HBV; as compared to hepatitis A virus (HAV), HBV stimulates very weak IFNα and IFNλ responses [109**]. Experimental infections in chimpanzees, with comparisons of HBV and HCV [110,111] or HAV and HCV [112], suggest that there may be a hierarchy of viruses in terms of their ability to result in “stealth” infections below detection for induction of type 1 IFN responses, with HBV and HAV being “stealthier” than HCV.
Interestingly, three of the innate cytokines commonly induced by viruses, type 1 IFNs, IL-12 and IL-15, bind to different receptors using subsets of Janus kinases (Jaks) and signal transducers and activators of transcription (STATs) to stimulate biological responses in cells [113,114]. There are seven total STAT molecules, 1 through 6, with two 5s. Once activated by phosphorylation, STAT complexes translocate to a cell's nucleus, bind to DNA regulatory sequences in promoters of genes, and modify transcriptional control of the target genes. Much of the early work defining cytokine-signaling pathways focused on identifying individual STATs preferentially used by the different cytokines to assign their roles in eliciting specific responses. The type 1 IFNs, IFNαβ, use STAT1 and 2 as a major pathway for induction of antiviral effects, but they have a wide range of other functions. Some of these are immunoregulatory and some are paradoxical [113]. Their activation of STAT1 also stimulates elevated STAT1 expression [115,116] and promotes anti-proliferative effects [117,118] as well as elevated NK cell cytotoxicity and IL-15 expression [99]. The type 1 IFNs appear to be unusually promiscuous in their choice of STATs and have been shown to conditionally active all STATs including STAT4 [113]. STAT4 is well characterized as an important intermediary in IL-12 induction of IFNγ.
Mouse studies dissecting the roles of individual cytokines for eliciting NK cell responses to viral infections have shown that although the type 1 IFNs are key for inducing elevated NK cell cytotoxicity, and the cytokines induce IL-15 to support modest levels of NK cell proliferation at very early times after infection [99], biologically active IL-12 is required for a strong systemic NK cell IFNγ response [99,119] (Fig. 5A). The ability to assign individual cytokines to the early NK cell responses observed during viral infections of mice, i.e., type 1 IFN for cytotoxic function, IL-15 for proliferation, and IL-12 for IFNγ, is surprising given the overlapping effects that can be induced by each of these factors. In addition, it can be contrasted to the linked stimulation of all three responses through activating receptors [24], the role of activating receptor in supporting extended proliferation [25] and maintenance [89**], as well as the observed synergism between activating receptor stimulation for IL-15-dependent [120] and under immunodeficient conditions, IL-15-independent NK cell proliferation [121]. The observations suggest that the NK cells themselves and/or the conditions of infections are limiting the responses that each of the cytokines can induce.
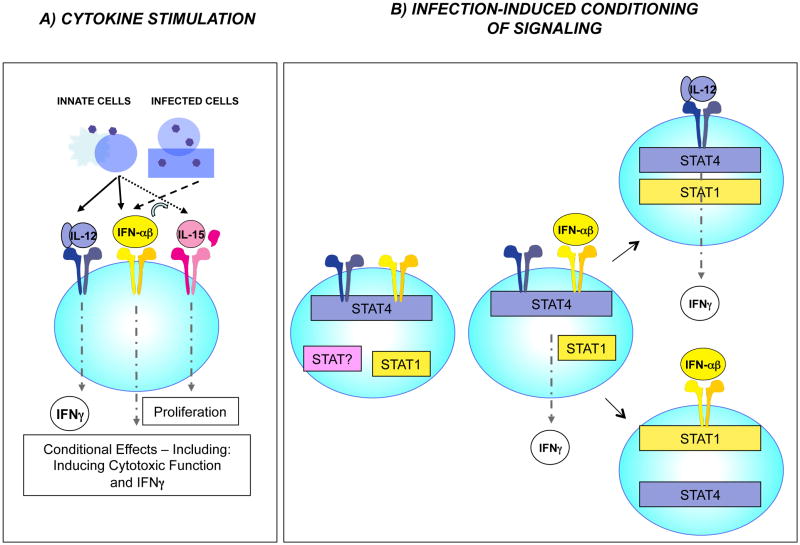
Figure 5. Cytokine regulation of NK cell responses to viral infection.
Many studies of NK cells in isolation and under immunodeficient conditions have demonstrated the roles for different cytokines, including the type 1 IFNs (IFNαβ), IL-12, and IL-15, as well as activating receptors in stimulating NK cell IFNγ production, elevated cytotoxic function, and proliferation. (A) At times of peak innate cytokine responses to infections in immunocompetent mice, however, the factors have key non-overlapping effects. Under these conditions, type 1 IFN is required for induction of systemic elevated cytotoxicity, IL-12 for IFNγ, and IL-15 for proliferation. At the very earliest times after infection, local type 1 IFN induction of NK cell IFNγ can be observed, but it is tightly regulated. The observations suggest that the conditions of infections are limiting NK cell responses to individual cytokines. (B) Studies characterizing NK cell expression of the signal transducers of transcription, STAT1 and STAT4, show that the cytokine effects are regulated in part by high-level basal expression of STAT4 and dynamic regulation of STAT1 levels, with type 1 IFN induction of STAT1 acting to limit type 1 IFN access to STAT4. The balance is important in allowing early type 1 IFN, while protecting from the detrimental consequences of unregulated, induction of IFNγ. Availability of IL-12 provides an alternative pathway to IFNγ. The mechanism shaping the effects of type 1 IFNs is an example of how NK cells are rapidly conditioned by experience during viral infection.
Studies evaluating the role of type 1 IFN signaling pathways in regulating NK cell responses have shown that in comparison to other immune cells from uninfected mice, NK cells basally express high levels of STAT4 and respond to type 1 IFN exposure with activation of STAT4 [107]. In the absence of STAT1, type 1 IFNs elicit systemic production of IFNγ by NK cells through a STAT4-dependent pathway. The type 1 IFN use of the STAT1-dependent pathway to STAT1 induction, however, is important for protection against deregulated induction of IFNγ and a resulting cytokine-mediated disease. The effect occurs at the level of competition for access to the type 1 IFN receptor with its preference for STAT1 over STAT4. Thus, the basal expression of high STAT4 in NK cells allows access to STAT4 activation by type 1 IFNs immediately after exposure whereas STAT1 induction by type 1 IFN provides a built-in regulatory pathway to limit the availability of STAT4 and protect from immune-mediated pathology. Interestingly, type 1 IFN access to STAT4 is inversely correlated with total STAT1 protein levels in healthy donors as compared to those chronically infected with HCV [122**]. Thus, the conditioning of NK cell cytokine responses through regulation of STAT levels is likely to be common to both species. The results suggest that the strict requirement for IL-12 in NK cell IFN-γ responses is a consequence of needing a different cytokine to access STAT4 once STAT1 levels are increased.
Although systemic NK cell IFNγ responses require IL-12, type 1 IFNs can access STAT4 and induce NK cell IFNγ production at the first site of infection, i.e., the peritoneal cavity, and this contributes to promoting an antiviral state [123**]. The pathway appears to be available at this site because of tightly regulated type 1 IFN production over a narrow period of time and delayed induction of STAT1 in NK as compared to other cells in the compartment. The regulation of type 1 IFN access to STAT4 (Fig. 5B) is an example of how fine-tuning of NK cell responses is important for shaping their effects as needed during the changing conditions of infection and of how the immune system is equipped to do this. It is another example of how “experience” modifies innate immunity. In this case, the changes are happening under acute conditions during periods of innate responses. The longer-term consequences for NK cell “conditioning” have yet to be examined, but may play a role in the polarization of NK cells to cytotoxicity following IFNα-treated of individuals chronically infected with HCV [46] as well as the development of NK cell subsets in the human and mouse preferentially mediating cytotoxicity as compared to cytokine production [124,125].
Conclusions
In summary, although NK cells were first proposed to be key mediators of innate immune responses to viral infections, it is now clear that there are many mechanisms in place to dramatically regulate their functions at the very earliest times after infections as well as to support their continued availability through times overlapping with adaptive immunity. A profound flexibility in responding to virus-induced cytokines and changes in the expression of ligands for activating and inhibitory receptors equips previously unexposed individuals or subsets of individuals in a population with the ability to protect against a wide range of potential infections, but it also results in conditioning of the NK cell populations to reflect experience. In the case of the earliest pathways to activation of NK cell responses elicited by type 1 IFNs, there are predisposed responses in place to allow IFNγ production for early antiviral defense and possibly immuneregulation but rapid induction of molecules limiting the response to protect from continued IFNγ production and resulting cytokine-mediated disease. These changes are important acutely. The long-term consequences of this “conditioning” by experience remain to be determined but may have roles in the development of NK cell subsets preferentially mediating particular functions. Alternative pathways are in place to access certain overlapping responses, including the use of different innate cytokines and/or ligands for activating receptors. In contrast, the pathways to NK cell-mediated effects delivered via the activating receptors not only make killing of virus-infected target cells possible, but they also help maintain NK cells through periods of extended viral infections and change the proportions of NK cells expressing particular activating receptors. This “conditioning” has significant consequences for the resources available to fight off an infection and responses to subsequent primary viral infections. Taken together, the results force the consideration of NK cells in particular and the innate immune system in general as being influenced both stably though genetic variations and dynamically by experience.
Highlights.
Here we review recent data contributing to our understanding of how NK cells mediate innate defense against viral infections. The molecular mechanisms mediated through infection-induced inhibitory/activating NK receptor-ligand pairs and cytokines as well as their effects onto NK cell subset function, maintenance and proliferation through times overlapping with adaptive and long-lived immunity are highlighted.
Acknowledgments
The authors apologize to their colleagues whose work was not cited due to space limitations. Research in the authors' laboratories is funded by the Canadian Institutes of Health Research, Canada, The Wellcome Trust, UK, and the National Institutes of Health, USA. They thank their many current and past laboratory members for their contributions to the work reviewed here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 3.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 4.Orange JS, Ballas ZK. Natural killer cells in human health and disease. Clin Immunol. 2006;118:1–10. doi: 10.1016/j.clim.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Lee SH, Biron CA. Here today--not gone tomorrow: roles for activating receptors in sustaining NK cells during viral infections. Eur J Immunol. 2010;40:923–932. doi: 10.1002/eji.201040304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SH, Miyagi T, Biron CA. Keeping NK cells in highly regulated antiviral warfare. Trends Immunol. 2007;28:252–259. doi: 10.1016/j.it.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Sentman CL, Olsson MY, Karre K. Missing self recognition by natural killer cells in MHC class I transgenic mice. A ‘receptor calibration’ model for how effector cells adapt to self. Semin Immunol. 1995;7:109–119. doi: 10.1006/smim.1995.0015. [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 10.Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 12.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 13.Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, Makrigiannis AP. Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol. 2008;20:321–330. doi: 10.1016/j.smim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Bahram S, Inoko H, Shiina T, Radosavljevic M. MIC and other NKG2D ligands: from none to too many. Curr Opin Immunol. 2005;17:505–509. doi: 10.1016/j.coi.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J Exp Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res. 2006;35:263–278. doi: 10.1385/IR:35:3:263. [DOI] [PubMed] [Google Scholar]
- 17.MacFarlane AWt, Campbell KS. Signal transduction in natural killer cells. Curr Top Microbiol Immunol. 2006;298:23–57. doi: 10.1007/3-540-27743-9_2. [DOI] [PubMed] [Google Scholar]
- 18.Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev. 2009;227:150–160. doi: 10.1111/j.1600-065X.2008.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol. 2005;174:3859–3863. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 20.Scalzo AA, Fitzgerald NA, Simmons A, La Vista AB, Shellam GR. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J Exp Med. 1990;171:1469–1483. doi: 10.1084/jem.171.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 22.Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA, Dubbelde CE, Stone LR, Scalzo AA, Yokoyama WM. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 23.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 24.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 26.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 27.Desrosiers MP, Kielczewska A, Loredo-Osti JC, Adam SG, Makrigiannis AP, Lemieux S, Pham T, Lodoen MB, Morgan K, Lanier LL, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat Genet. 2005;37:593–599. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dighe A, Rodriguez M, Sabastian P, Xie X, McVoy M, Brown MG. Requisite H2k role in NK cell-mediated resistance in acute murine cytomegalovirus-infected MA/My mice. J Immunol. 2005;175:6820–6828. doi: 10.4049/jimmunol.175.10.6820. [DOI] [PubMed] [Google Scholar]
- 29**.Kielczewska A, Pyzik M, Sun T, Krmpotic A, Lodoen MB, Munks MW, Babic M, Hill AB, Koszinowski UH, Jonjic S, et al. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J Exp Med. 2009;206:515–523. doi: 10.1084/jem.20080954. Shows that the activating receptor Ly49P requires both host H2-Dk and viral gp34/m04 to recognize MCMV infected cells. MA/My mice are rendered susceptible by infection with mutant MCMV lacking gp34/m04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Xie X, Stadnisky MD, Coats ER, Ahmed Rahim MM, Lundgren A, Xu W, Makrigiannis AP, Brown MG. MHC class I D(k) expression in hematopoietic and nonhematopoietic cells confers natural killer cell resistance to murine cytomegalovirus. Proc Natl Acad Sci U S A. 2010;107:8754–8759. doi: 10.1073/pnas.0913126107. The transgenesis of the H2-Dk gene is shown to enhance MCMV resistance in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Fodil-Cornu N, Loredo-Osti JC, Vidal SM. NK cell receptor/H2-Dk-dependent host resistance to viral infection is quantitatively modulated by H2q inhibitory signals. PLoS Genet. 2011;7:e1001368. doi: 10.1371/journal.pgen.1001368. Shows that double congenic mice carrying NKC and H2k genomic regions from MA/My mice acquire resistance to MCMV but not mice carrying a single region. MCMV resistance of H2-Dk transgenic mice is impaired by the presence of H2q encoded molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Pyzik M, Charbonneau B, Gendron-Pontbriand EM, Babic M, Krmpotic A, Jonjic S, Vidal SM. Distinct MHC class I-dependent NK cell-activating receptors control cytomegalovirus infection in different mouse strains. J Exp Med. 2011;208:1105–1117. doi: 10.1084/jem.20101831. Numerous activating Ly49 receptors are shown to recognize MCMV infected cells in a similar manner that is MHC1 and gp34/m04 dependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Fang M, Orr MT, Spee P, Egebjerg T, Lanier LL, Sigal LJ. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity. 2011;34:579–589. doi: 10.1016/j.immuni.2011.02.015. The activating heterodimer NKG2E/CD94 is shown to recognize ectromelia infected cells expressing the non-classical MHC1 protein, Qa1b. Mice lacking either CD94 or Qa1b are unable to control ectromelia virus replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lisnic VJ, Krmpotic A, Jonjic S. Modulation of natural killer cell activity by viruses. Curr Opin Microbiol. 2010;13:530–539. doi: 10.1016/j.mib.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Babic M, Pyzik M, Zafirova B, Mitrovic M, Butorac V, Lanier LL, Krmpotic A, Vidal SM, Jonjic S. Cytomegalovirus immunoevasin reveals the physiological role of “missing self” recognition in natural killer cell dependent virus control in vivo. J Exp Med. 2010;207:2663–2673. doi: 10.1084/jem.20100921. This study shows that an MCMV mutant virus lacking gp34/m04, a protein that escorts MHC1 molecules to the surface of infected cells, is attenuated in mouse strains naturally susceptible to MCMV. This is the first study to demonstrate that missing self-dependent NK cell activation is biologically relevant for protection against a viral infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11:321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie X, Stadnisky MD, Brown MG. MHC class I Dk locus and Ly49G2+ NK cells confer H-2k resistance to murine cytomegalovirus. J Immunol. 2009;182:7163–7171. doi: 10.4049/jimmunol.0803933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barao I, Alvarez M, Ames E, Orr MT, Stefanski HE, Blazar BR, Lanier LL, Anderson SK, Redelman D, Murphy WJ. Mouse Ly49G2+ NK cells dominate early responses during both immune reconstitution and activation independently of MHC. Blood. 2011;117:7032–7041. doi: 10.1182/blood-2010-11-316653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X, Chen Y, Li X, Kong H, Yang S, Ye B, Cui D, Wu W, Li L. Dynamic variations in the peripheral blood lymphocyte subgroups of patients with 2009 pandemic H1N1 swine-origin influenza A virus infection. Virol J. 2011;8:215. doi: 10.1186/1743-422X-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Amadei B, Urbani S, Cazaly A, Fisicaro P, Zerbini A, Ahmed P, Missale G, Ferrari C, Khakoo SI. Activation of natural killer cells during acute infection with hepatitis C virus. Gastroenterology. 2010;138:1536–1545. doi: 10.1053/j.gastro.2010.01.006. The first study to document acitvation of NK cells in the acute phase of hepatitis C virus infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Bjorkstrom NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, Michaelsson J, Malmberg KJ, Klingstrom J, Ahlm C, et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011;208:13–21. doi: 10.1084/jem.20100762. A comprehensive longitudinal analysis of NK cells in an acute human viral infection. The expansion and persisitence of an NKG2C-positive NK cell subset in repsonse to hantavirus infection is documented and suggested to be an HLA-E driven process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 44.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GW. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 45.Oliviero B, Varchetta S, Paudice E, Michelone G, Zaramella M, Mavilio D, De Filippi F, Bruno S, Mondelli MU. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology. 2009;137:1151–1160. 1160e1151–1157. doi: 10.1053/j.gastro.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 46*.Ahlenstiel G, Titerence RH, Koh C, Edlich B, Feld JJ, Rotman Y, Ghany MG, Hoofnagle JH, Liang TJ, Heller T, et al. Natural killer cells are polarized toward cytotoxicity in chronic hepatitis C in an interferon-alfa-dependent manner. Gastroenterology. 2010;138:325–335. e321–322. doi: 10.1053/j.gastro.2009.08.066. Changes in receptor expression and the phenotype of type 1 IFN reswponses are shown in NK cells from individuals chronically infected with HCV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulet S, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, Bernard NF. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS. 2008;22:595–599. doi: 10.1097/QAD.0b013e3282f56b23. [DOI] [PubMed] [Google Scholar]
- 48.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 49.Qi Y, Martin MP, Gao X, Jacobson L, Goedert JJ, Buchbinder S, Kirk GD, O'Brien SJ, Trowsdale J, Carrington M. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006;2:e79. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31:2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 52.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 53.Fuller CL, Ruthel G, Warfield KL, Swenson DL, Bosio CM, Aman MJ, Bavari S. NKp30-dependent cytolysis of filovirus-infected human dendritic cells. Cell Microbiol. 2007;9:962–976. doi: 10.1111/j.1462-5822.2006.00844.x. [DOI] [PubMed] [Google Scholar]
- 54.Chisholm SE, Howard K, Gomez MV, Reyburn HT. Expression of ICP0 is sufficient to trigger natural killer cell recognition of herpes simplex virus-infected cells by natural cytotoxicity receptors. J Infect Dis. 2007;195:1160–1168. doi: 10.1086/512862. [DOI] [PubMed] [Google Scholar]
- 55.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagne F, Ferracci G, Arosa FA, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chisholm SE, Reyburn HT. Recognition of vaccinia virus-infected cells by human natural killer cells depends on natural cytotoxicity receptors. J Virol. 2006;80:2225–2233. doi: 10.1128/JVI.80.5.2225-2233.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, Wills M, Lehner PJ. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2008;105:1656–1661. doi: 10.1073/pnas.0707883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Llano M, Guma M, Ortega M, Angulo A, Lopez-Botet M. Differential effects of US2, US6 and US11 human cytomegalovirus proteins on HLA class Ia and HLA-E expression: impact on target susceptibility to NK cell subsets. Eur J Immunol. 2003;33:2744–2754. doi: 10.1002/eji.200324182. [DOI] [PubMed] [Google Scholar]
- 59.Brooks CR, Elliott T, Parham P, Khakoo SI. The inhibitory receptor NKG2A determines lysis of vaccinia virus-infected autologous targets by NK cells. J Immunol. 2006;176:1141–1147. doi: 10.4049/jimmunol.176.2.1141. [DOI] [PubMed] [Google Scholar]
- 60.Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM, Bernard NF. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J Immunol. 2010;184:2057–2064. doi: 10.4049/jimmunol.0902621. [DOI] [PubMed] [Google Scholar]
- 61.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 63.Knapp S, Warshow U, Hegazy D, Brackenbury L, Guha IN, Fowell A, Little AM, Alexander GJ, Rosenberg WM, Cramp ME, et al. Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology. 2010;51:1168–1175. doi: 10.1002/hep.23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romero V, Azocar J, Zuniga J, Clavijo OP, Terreros D, Gu X, Husain Z, Chung RT, Amos C, Yunis EJ. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008;45:2429–2436. doi: 10.1016/j.molimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vidal-Castineira JR, Lopez-Vazquez A, Diaz-Pena R, Alonso-Arias R, Martinez-Borra J, Perez R, Fernandez-Suarez J, Melon S, Prieto J, Rodrigo L, et al. Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol. 2010;84:475–481. doi: 10.1128/JVI.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A, Stathopoulos S, Middleton D, Mulder A, Claas FH, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. A novel mechanism of changes in the peptide repertoire presented by the MHC class 1 ligand of KIR efficiently remove the inhibitory effects by which NK cell inhibition can be overcome, is shown. Thus, viruses do not need to specifically down-regualte MHC class I in order to overcome inhibitory signals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dolan BP, Bennink JR, Yewdell JW. Translating DRiPs: progress in understanding viral and cellular sources of MHC class I peptide ligands. Cell Mol Life Sci. 2011;68:1481–1489. doi: 10.1007/s00018-011-0656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fadda L, O'Connor GM, Kumar S, Piechocka-Trocha A, Gardiner CM, Carrington M, McVicar DW, Altfeld M. Common HIV-1 peptide variants mediate differential binding of KIR3DL1 to HLA-Bw4 molecules. J Virol. 2011;85:5970–5974. doi: 10.1128/JVI.00412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69**.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. A large epidemiological study showing the association of a mutant peptide with the KIR2DL2 gene in HIV infection. Functional experiments show that the described mutation leads to inhibition of NK cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. NK cell activation is studied in fine detail and the activating potential of key NK cell receptors is dissected. Most receptors function as co-receptors, implying that stimulation of NK cells by one receptor is rarely adequate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horng T, Bezbradica JS, Medzhitov R. NKG2D signaling is coupled to the interleukin 15 receptor signaling pathway. Nat Immunol. 2007;8:1345–1352. doi: 10.1038/ni1524. [DOI] [PubMed] [Google Scholar]
- 72.Biron CA, Sen G. Innate immune responses to viral infection. In: Knipe DM, Howley PM, Walter Kluwer, editors. Fields Virology. Lippincott: Williams & Williams; 2007. pp. 249–278. [Google Scholar]
- 73**.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. Identification of the IL-28B gene as a key determinant in clearance of HCV infection during anti-viral therapy using a genome wide scan (GWS). This is shown to be by far the most important host-genetic determinant of the outcome of treatment. [DOI] [PubMed] [Google Scholar]
- 74*.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. The IL-28B gene is identified as also beneficial in the outcome of HCV infection. [DOI] [PubMed] [Google Scholar]
- 75*.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. A GWAS is used to determine the genes associated with a beneficial response to therapy in chronic hepatitis C virus infection. The IL-28B gene is found to also be associated with treatment outcome in the Japanese population. [DOI] [PubMed] [Google Scholar]
- 76*.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. The protective effect of the IL-28B against chronic HCV infection is documented. Individuals who spontaneously resolve acute HCV infection are shown to have an over-representation of the same IL-28B polymorphism responsible for protection in the treatment setting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77*.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. 1345e1331–1337. doi: 10.1053/j.gastro.2009.12.056. The IL-28B gene is identified as important in both spontaneously resolving HCV infection and treatment induced resolution. [DOI] [PubMed] [Google Scholar]
- 78*.Dring MM, Morrison MH, McSharry BP, Guinan KJ, Hagan R, O'Farrelly C, Gardiner CM. Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc Natl Acad Sci U S A. 2011;108:5736–5741. doi: 10.1073/pnas.1016358108. A unique cohort of HCV exposed individuals is studied and a model of KIR and susceptibility to chronic HCV infection is suggested. Susceptibility is synergicstic in combination with the susceptible IL-28B genotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knapp S, Warshow U, Ho KM, Hegazy D, Little AM, Fowell A, Alexander G, Thursz M, Cramp M, Khakoo SI. A polymorphism in IL28B distinguishes exposed, uninfected individuals from spontaneous resolvers of HCV infection. Gastroenterology. 2011;141:320–325. doi: 10.1053/j.gastro.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lange CM, Moradpour D, Doehring A, Lehr HA, Mullhaupt B, Bibert S, Bochud PY, Antonino AT, Pascual M, Farnik H, et al. Impact of donor and recipient IL28B rs12979860 genotypes on hepatitis C virus liver graft reinfection. J Hepatol. 2011;55:322–327. doi: 10.1016/j.jhep.2010.10.037. [DOI] [PubMed] [Google Scholar]
- 82.Williams H, McAulay K, Macsween KF, Gallacher NJ, Higgins CD, Harrison N, Swerdlow AJ, Crawford DH. The immune response to primary EBV infection: a role for natural killer cells. Br J Haematol. 2005;129:266–274. doi: 10.1111/j.1365-2141.2005.05452.x. [DOI] [PubMed] [Google Scholar]
- 83.Meier UC, Owen RE, Taylor E, Worth A, Naoumov N, Willberg C, Tang K, Newton P, Pellegrino P, Williams I, et al. Shared alterations in NK cell frequency, phenotype, and function in chronic human immunodeficiency virus and hepatitis C virus infections. J Virol. 2005;79:12365–12374. doi: 10.1128/JVI.79.19.12365-12374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hong HS, Eberhard JM, Keudel P, Bollmann BA, Ballmaier M, Bhatnagar N, Zielinska-Skowronek M, Schmidt RE, Meyer-Olson D. HIV infection is associated with a preferential decline in less-differentiated CD56dim CD16+ NK cells. J Virol. 2010;84:1183–1188. doi: 10.1128/JVI.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morishima C, Paschal DM, Wang CC, Yoshihara CS, Wood BL, Yeo AE, Emerson SS, Shuhart MC, Gretch DR. Decreased NK cell frequency in chronic hepatitis C does not affect ex vivo cytolytic killing. Hepatology. 2006;43:573–580. doi: 10.1002/hep.21073. [DOI] [PubMed] [Google Scholar]
- 86.Golden-Mason L, Madrigal-Estebas L, McGrath E, Conroy MJ, Ryan EJ, Hegarty JE, O'Farrelly C, Doherty DG. Altered natural killer cell subset distributions in resolved and persistent hepatitis C virus infection following single source exposure. Gut. 2008;57:1121–1128. doi: 10.1136/gut.2007.130963. [DOI] [PubMed] [Google Scholar]
- 87.Tarazona R, Casado JG, Delarosa O, Torre-Cisneros J, Villanueva JL, Sanchez B, Galiani MD, Gonzalez R, Solana R, Pena J. Selective depletion of CD56(dim) NK cell subsets and maintenance of CD56(bright) NK cells in treatment-naive HIV-1-seropositive individuals. J Clin Immunol. 2002;22:176–183. doi: 10.1023/a:1015476114409. [DOI] [PubMed] [Google Scholar]
- 88.Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES, Miller JS, Carrington M, Altfeld M. HLA class I subtype-dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J Virol. 2009;83:6798–6805. doi: 10.1128/JVI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89**.Lee SH, Kim KS, Fodil-Cornu N, Vidal SM, Biron CA. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206:2235–2251. doi: 10.1084/jem.20082387. Demonstrates the importance of having an activating receptor-ligand pair in sustaining NK cells during profound MCMV infection of mice, and the IL-10 production by NK cells for immunoregulatory effects under these conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crawford A, Wherry EJ. The diversity of costimulatory and inhibitory receptor pathways and the regulation of antiviral T cell responses. Curr Opin Immunol. 2009;21:179–186. doi: 10.1016/j.coi.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pelletier S, Drouin C, Bedard N, Khakoo SI, Bruneau J, Shoukry NH. Increased degranulation of natural killer cells during acute HCV correlates with the magnitude of virus-specific T cell responses. J Hepatol. 2010;53:805–816. doi: 10.1016/j.jhep.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Maria A, Fogli M, Mazza S, Basso M, Picciotto A, Costa P, Congia S, Mingari MC, Moretta L. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol. 2007;37:445–455. doi: 10.1002/eji.200635989. [DOI] [PubMed] [Google Scholar]
- 93**.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. Demonstrates the infection-induced development of long-lived NK cells expressing the Ly49H activating receptor recognizing MCMV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94**.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. Demonstrates vaccination-induced long-lived NK cells to result in enhanced defense against viral challenges under immunodeficient conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun JC, Beilke JN, Bezman NA, Lanier LL. Homeostatic proliferation generates long-lived natural killer cells that respond against viral infection. J Exp Med. 2011;208:357–368. doi: 10.1084/jem.20100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Biron CA. More things in heaven and earth: defining innate and adaptive immunity. Nat Immunol. 2010;11:1080–1082. doi: 10.1038/ni1210-1080. [DOI] [PubMed] [Google Scholar]
- 97.Biron CA, Dalod M, Salazar-Mather TP. Innate immunity and viral infections. In: Kaufman SHE, Sherr A, Ahmed R, editors. Immunology of Infectious Diseases. American Society for Microbiology; 2002. pp. 139–160. [Google Scholar]
- 98.Orange JS, Biron CA. An absolute and restricted requirement for IL-12 in natural killer cell IFN-gamma production and antiviral defense. Studies of natural killer and T cell responses in contrasting viral infections. J Immunol. 1996;156:1138–1142. [PubMed] [Google Scholar]
- 99.Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, Caligiuri MA, Durbin JE, Biron CA. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol. 2002;169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- 100.Pien GC, Satoskar AR, Takeda K, Akira S, Biron CA. Cutting edge: selective IL-18 requirements for induction of compartmental IFN-gamma responses during viral infection. J Immunol. 2000;165:4787–4791. doi: 10.4049/jimmunol.165.9.4787. [DOI] [PubMed] [Google Scholar]
- 101.Asselin-Paturel C, Boonstra A, Dalod M, Durand I, Yessaad N, Dezutter-Dambuyant C, Vicari A, O'Garra A, Biron C, Briere F, et al. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat Immunol. 2001;2:1144–1150. doi: 10.1038/ni736. [DOI] [PubMed] [Google Scholar]
- 102.Delale T, Paquin A, Asselin-Paturel C, Dalod M, Brizard G, Bates EE, Kastner P, Chan S, Akira S, Vicari A, et al. MyD88-dependent and -independent murine cytomegalovirus sensing for IFN-alpha release and initiation of immune responses in vivo. J Immunol. 2005;175:6723–6732. doi: 10.4049/jimmunol.175.10.6723. [DOI] [PubMed] [Google Scholar]
- 103.Krug A, French AR, Barchet W, Fischer JA, Dzionek A, Pingel JT, Orihuela MM, Akira S, Yokoyama WM, Colonna M. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 104.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pien GC, Nguyen KB, Malmgaard L, Satoskar AR, Biron CA. A unique mechanism for innate cytokine promotion of T cell responses to viral infections. J Immunol. 2002;169:5827–5837. doi: 10.4049/jimmunol.169.10.5827. [DOI] [PubMed] [Google Scholar]
- 106.Louten J, van Rooijen N, Biron CA. Type 1 IFN deficiency in the absence of normal splenic architecture during lymphocytic choriomeningitis virus infection. J Immunol. 2006;177:3266–3272. doi: 10.4049/jimmunol.177.5.3266. [DOI] [PubMed] [Google Scholar]
- 107.Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med. 2007;204:2383–2396. doi: 10.1084/jem.20070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108*.Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, Lebedeva M, DeCamp A, Li D, Grove D, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. An intersting comparison of cytokine responses in the acute phase of infection amongst three viruses associated with chronic diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109**.Dunn C, Peppa D, Khanna P, Nebbia G, Jones M, Brendish N, Lascar RM, Brown D, Gilson RJ, Tedder RJ, et al. Temporal analysis of early immune responses in patients with acute hepatitis B virus infection. Gastroenterology. 2009;137:1289–1300. doi: 10.1053/j.gastro.2009.06.054. A comprehensive longitudinal analysis of the early events in acute hepatitis B infection, documenting early cytokine profiles as well as NK cell response. [DOI] [PubMed] [Google Scholar]
- 110.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wieland S, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lanford RE, Feng Z, Chavez D, Guerra B, Brasky KM, Zhou Y, Yamane D, Perelson AS, Walker CM, Lemon SM. Acute hepatitis A virus infection is associated with a limited type 1 interferon response and persistence of intrahepatic viral RNA. Proc Natl Acad Sci U S A. 2011;108:11223–11228. doi: 10.1073/pnas.1101939108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 114.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wong LH, Sim H, Chatterjee-Kishore M, Hatzinisiriou I, Devenish RJ, Stark G, Ralph SJ. Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. J Biol Chem. 2002;277:19408–19417. doi: 10.1074/jbc.M111302200. [DOI] [PubMed] [Google Scholar]
- 117.Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gil MP, Salomon R, Louten J, Biron CA. Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood. 2006;107:987–993. doi: 10.1182/blood-2005-07-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Orange JS, Biron CA. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol. 1996;156:4746–4756. [PubMed] [Google Scholar]
- 120.French AR, Sjolin H, Kim S, Koka R, Yang L, Young DA, Cerboni C, Tomasello E, Ma A, Vivier E, et al. DAP12 signaling directly augments proproliferative cytokine stimulation of NK cells during viral infections. J Immunol. 2006;177:4981–4990. doi: 10.4049/jimmunol.177.8.4981. [DOI] [PubMed] [Google Scholar]
- 121.Sun JC, Ma A, Lanier LL. Cutting edge: IL-15-independent NK cell response to mouse cytomegalovirus infection. J Immunol. 2009;183:2911–2914. doi: 10.4049/jimmunol.0901872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122**.Miyagi T, Takehara T, Nishio K, Shimizu S, Kohga K, Li W, Tatsumi T, Hiramatsu N, Kanto T, Hayashi N. Altered interferon-alpha-signaling in natural killer cells from patients with chronic hepatitis C virus infection. J Hepatol. 2010;53:424–430. doi: 10.1016/j.jhep.2010.03.018. Demonstrates changes in type 1 IFN access to STAT4 in NK cells associated with levels of STAT1 in patients with chronic HCV. [DOI] [PubMed] [Google Scholar]
- 123**.Mack EA, Kallal LE, Demers DA, Biron CA. Type 1 Interferon Induction of Natural Killer Cell Gamma Interferon Production for Defense during Lymphocytic Choriomeningitis Virus Infection. MBio. 2011;2:e00169–11. doi: 10.1128/mBio.00169-11. Demonstrates early local induction of NK cell IFNγ by type 1 IFNs following LCMV infection of mice. The response is shown to be dependent on STAT4 and observed piror to the induction of elevated STAT1 in NK cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Farag SS, VanDeusen JB, Fehniger TA, Caligiuri MA. Biology and clinical impact of human natural killer cells. Int J Hematol. 2003;78:7–17. doi: 10.1007/BF02983234. [DOI] [PubMed] [Google Scholar]
- 125.Hayakawa Y, Huntington ND, Nutt SL, Smyth MJ. Functional subsets of mouse natural killer cells. Immunol Rev. 2006;214:47–55. doi: 10.1111/j.1600-065X.2006.00454.x. [DOI] [PubMed] [Google Scholar]