SUMMARY
Cyclin D-dependent kinases (CDK4 and CDK6) are positive regulators of cell cycle entry, and they are overactive in the majority of human cancers. However, it is currently not completely understood by which cellular mechanisms CDK4/6 promote tumorigenesis, largely due to the limited number of identified substrates. Here we performed a systematic screen for substrates of cyclin D1-CDK4 and cyclin D3-CDK6. We identified the Forkhead Box M1 (FOXM1) transcription factor as a common critical phosphorylation target. CDK4/6 stabilize and activate FOXM1, thereby maintain expression of G1/S phase genes, suppress the levels of reactive oxygen species (ROS), and protect cancer cells from senescence. Melanoma cells, unlike melanocytes, are highly reliant on CDK4/6-mediated senescence suppression, which makes them particularly susceptible to CDK4/6 inhibition.
INTRODUCTION
Excessive cell proliferation induced by aberrant entry into the cell cycle is considered a hallmark of cancer. Commitment to cell cycle entry occurs during the G1 phase, when CDK4 and CDK6 form active complexes with one of the three D-type cyclins (D1, D2 or D3). These complexes promote G1-S transition in cancer cells by phosphorylating critical substrates, of which the Retinoblastoma tumor suppressor protein, RB1, as well as the related family members, RBL1 (p107) and RBL2 (p130), remain best characterized. Mechanistically, phosphorylation of RB proteins disables their function as transcriptional repressors to allow activation of the E2F-dependent transcriptional program, an important mediator of S-phase entry and initiation of DNA synthesis (Ortega et al., 2002; Sherr and Roberts, 1999). These processes are negatively regulated by INK4 proteins (including p15INK4 and p16INK4), which specifically inhibit the assembly and activation of cyclin D-CDK4/6 complexes.
It is therefore not surprising that CDK4 and its regulatory subunit, cyclin D1, are oncogenes; and recent findings have revealed that both are embedded in the ten most frequently amplified genomic loci in a diverse set of human cancers (Beroukhim et al., 2010). Conversely, the gene encoding p16INK4 exhibits more deletions than any other recessive cancer gene (Bignell et al., 2010). Moreover, cyclin D1-CDK4 is required for the formation of several tumor types, including breast and lung cancer, with the catalytic function of the CDK4 subunit being critically important (Yu et al., 2006; Landis et al., 2006; Puyol et al., 2010).
Despite of this, the full spectrum of the substrates phosphorylated by CDK4/6 remains unknown, although this information is crucial for our understanding of kinase function in human cancer. It is also unclear whether individual cyclin D1/D2/D3-CDK4/6 complexes target the same subset of proteins for phosphorylation, or whether they possess distinct substrate specificities. Yet, linking CDK4/6 to their substrates is particularly challenging; unlike other CDKs, CDK4 and CDK6 are not readily susceptible to chemical genetics approaches, using ‘bulky’ ATP. Classical substrate-trapping methods also pose inherent limitations, such as the transient nature of physical kinase-substrate interactions, the general difficulty to detect low-abundance proteins, and the experimental restriction of the analysis to certain cell or tissue types.
Here we sought to overcome these limitations, and to uncover genuine substrates of CDK4/6 across the human proteome. Through functional analysis of substrate phosphorylation we aim to define mechanisms by which CDK4/6 promote tumorigenesis in order to maximize the merits of CDK4/6 small molecule inhibitors for targeted therapy.
RESULTS
Cyclin D-CDK4/6 Substrate Phosphorylation Profiling in vitro and in Cells
Our strategy for substrate identification was to use computational tools to enrich for candidate substrates from the entire human proteome, and then to experimentally test the enriched proteins for phosphorylation in high-throughput kinase reactions, using individual cyclin D-CDK4/6 complexes. CDKs possess exquisite phosphorylation site selectivity, with the phosphate-acceptor residue preceding a proline. Moreover, the so-called ‘full’ CDK consensus site typically contains one or more basic residues downstream of the critical proline (Songyang et al., 1994). In keeping with this, we searched the SWISSPROT protein database with the Web-based program Scansite (Obenauer et al., 2003) for human nuclear proteins containing at least two CDK consensus sites. We did not include proteins with only one site, due to previous observations that CDKs phosphorylate their substrates preferentially on multiple sites (Ubersax et al., 2003). This in silico analysis led to the identification of 445 candidate protein-coding genes (Figure 1A).
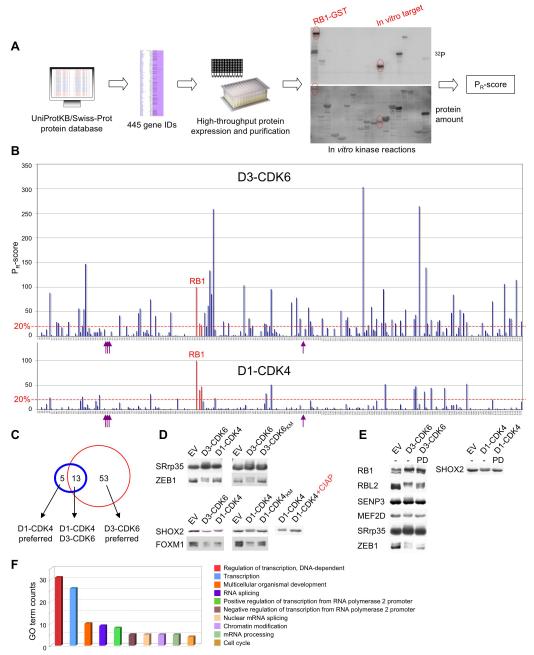
Figure 1. Proteome-Wide Identification of CDK4/6 Substrates.
(A) Schematic for the approach to substrate identification. All nuclear proteins with at least two potential CDK phosphorylation sites were selected from the protein database SWISS-PROT. Candidate proteins were expressed in E. coli, purified and tested for phosphorylation by recombinant cyclin D1-CDK4 and cyclin D3-CDK6. Reaction products were analyzed by SDS-PAGE (lower panel) and autoradiography (upper panel), and relative rates of phosphorylation (PR-scores) were calculated.
(B) Side-by-side comparison of phosphorylation of 285 enriched proteins by cyclin D3-CDK6 (upper panel) versus cyclin D1-CDK4 (lower panel). PR-scores are the rates of phosphorylation normalized to RB1, which was set to 100%. Proteins with PR-scores above 20% are referred to as in vitro substrates. Arrows indicate histone proteins; the three RB family proteins are highlighted in red.
(C) Overlap between cyclin D1-CDK4 (blue circle) and cyclin D3-CDK6 (red circle) in vitro substrates.
(D) In vitro substrates were co-expressed in HEK293 cells with either empty vector (EV) or wild-type cyclin-CDK complexes (left panel). Co-expression of wild type versus kinase-dead (KM) CDK versions is shown in the middle panel. Phosphorylated SHOX2 was treated with calf intestinal phosphatase (CIAP, right panel).
(E) Collapse in the mobility shift of seven CDK4/6 substrates upon short-term treatment (40 min) with the CDK4/6 specific inhibitor, PD0332991. GST-tagged substrates plus cyclin D-CDK complexes were expressed in HEK293 cells exposed to either 1 μM of the inhibitor (PD) or DMSO (−) 30 h post-transfection.
(F) ‘Biological process’ gene ontology (GO) terms were assigned to all 71 in vitro substrates. See also Figure S1 and Table S1.
We reasoned that like RB proteins, purified putative substrates would become phosphorylated by recombinant cyclin D-CDK4/6 in in vitro assays. This approach excludes indirect phosphorylation events and makes use of the firmly established in vitro substrate selectivity of CDK4 in particular (Sherr and Roberts, 1999). We successfully expressed and purified 285 candidate proteins (Table S1) - corresponding to 231 genes - from E. coli and challenged them individually with either cyclin D1-CDK4 or cyclin D3-CDK6 complex, as these are the most characterized. All obtained relative protein phosphorylation scores (PR-scores) were plotted on a scale normalized to phosphorylation of RB1, the PR-score of which was set to 100% (Figure 1B). For detailed information of the PR-scores of all tested proteins see Table S1. To define in vitro substrates, a cut-off of 20% was applied; this threshold can be considered conservative, as it does not take into account RB1’s many phosphorylation sites (substrates phosphorylated at only two sites might be expected to have lower PR-scores than those phosphorylated at 10 sites, for example). However, we did not include such calculation in our analysis, as it assumes that phosphorylation of each site is equally efficient, but this scenario is unlikely and difficult to assess experimentally. Importantly, all three RB family members scored as substrates in both screens (Figure 1B). By contrast, histone proteins, which represent substrates of all CDKs, except cyclin D-CDK4/6 (Sherr and Roberts, 1999), were not phosphorylated, further confirming the specificity of our analysis. Intriguingly, we discovered major differences in substrate phosphorylation between the two kinases, with cyclin D3-CDK6 possessing a much broader substrate specificity than cyclin D1-CDK4 (Figure 1B). In total, our screen revealed thirteen common in vitro substrates, including RB1, RBL1 and RBL2, fifty three CDK6-D3 ‘preferred’ and only five CDK4-D1 ‘preferred’ in vitro substrates (Figure 1C; see Figure S1A for all substrate PR-scores).
To validate at least some of the proteins in human cells, all 71 in vitro substrates were expressed in HEK293 cells. Although, in many cases, phosphorylation of proteins does not result in changes in their electrophoretic migration on SDS gels (CDK4/6-mediated phosphorylation of RBL1 is an example), 8 proteins (RB1, RBL2, SENP3, MEF2D, SRrp35, ZEB1, SHOX2, FOXM1) showed reproducible mobility shifts with co-expression of the kinases, but not with kinase-inactive CDK versions (Figure 1D, Figure S1B). Concordantly, short-term treatment (40 min) with a CDK4/6 specific inhibitor, PD0332991 (Fry et al., 2004), abrogated the CDK-induced mobility shifts (Figure 1E). Further, the differences in in vitro substrate phosphorylation specificity of cyclin D1-CDK4 versus cyclin D3-CDK6 were largely recapitulated in cells (Figure 1D, Figure S1B). Together, this underscores the feasibility of our in vitro screening approach.
Cyclin D-CDK4/6 Activate FOXM1 Transcriptional Function
We noted with interest that many of the 71 in vitro substrates share a transcription regulatory function (Figure 1F). Such proteins are often underrepresented in traditional proteomic approaches that rely on mass spectrometry, due to their low abundance in cells. The FOXM1 transcription factor was chosen for further investigation for several reasons. First, FOXM1 was in vitro phosphorylated by cyclin D1-CDK4 and cyclin D3-CDK6 equally well, thus defining it as a universal CDK4/6 substrate (Figure S1A). Second, FOXM1 was among the in vivo validated phosphorylation targets as assessed by mobility shift analysis (Figure 1D). Third, some of its biological functions overlap with those regulated by CDK4/6 activity, such as the positive regulation of the cell cycle transition from G1 to S phase (Wang et al., 2002; Wang et al., 2008), suppression of cellular senescence (Wang et al., 2005; Park et al., 2009), and maintenance of pancreatic β-cell mass in vivo (Ackermann and Gannon, 2007). Fourth, like CDK4/6, FOXM1 is critically required for tumorigenesis, as observed for several cancer types (Kalinichenko et al., 2004; Liu et al., 2006).
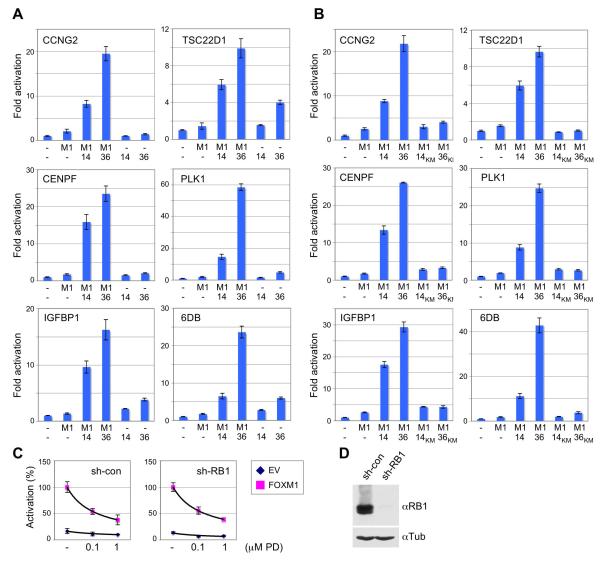
We first examined whether CDK4/6 alters the ability of FOXM1 to activate transcription. To this end, promoters derived from several FOXM1 target genes, the common Forkhead-responsive IGF-binding protein 1 (IGFBP1) promoter, and the 6DB sequence containing 6 tandem Forkhead-response elements, were transfected with or without FOXM1, and in the presence or absence of cyclin D1-CDK4 or cyclin D3-CDK6. Figure 2A illustrates that co-expression of the kinases in RB protein-deficient HeLa cells markedly potentiated FOXM1-mediated transcription from all tested promoters, whereas expression of the kinases without FOXM1 had much less effect. Moreover, activation of transcription required CDK4 and CDK6 catalytic activity (Figure 2B). Conversely, inhibition of endogenous CDK4/6 activity with the specific inhibitor, PD0332991, reduced FOXM1-mediated transcription in both, control shRNA and RB1 shRNA expressing U2OS cells (Figure 2C), in which the RB1 protein was reduced to undetectable levels (Figure 2D). Thus, CDK4/6 can drive FOXM1-mediated transcription in the absence of RB1.
Figure 2. Cyclin D-CDK4/6 Activate FOXM1 Transcriptional Function.
(A) Effects of individual and combined expression of FOXM1 and cyclin D-CDK4/6 complexes on reporter gene activation (+/−SD). Hela cells were transfected with the CCNG2, TSC22D1, IGFBP-1, 6DB, CENPF and PLK1 promoter-luciferase constructs, FOXM1 (M1), empty vector (-), cyclin D1-CDK4 (14) or cyclin D3-CDK6 (36).
(B) Reporter activity (+/−SD) in Hela cells after transfection of FOXM1 (M1) and cyclin D1-CDK4 (14), or cyclin D3-CDK6 (36), versus kinase-dead (KM) versions.
(C) U2OS cells stably expressing either control (sh-con) or RB1 targeting shRNA (sh-RB1) were transfected with the CCNG2 promoter-reporter, empty vector (EV, blue) or FOXM1 (pink), and treated with DMSO, 0.1 μM or 1 μM PD0332991 for 16 h. Values are mean +/−SD.
(D) RB1 protein expression in control (sh-con) and RB1 knockdown (sh-RB1) cells.
Cyclin D-CDK4/6 Activate FOXM1 Function by Multisite Phosphorylation
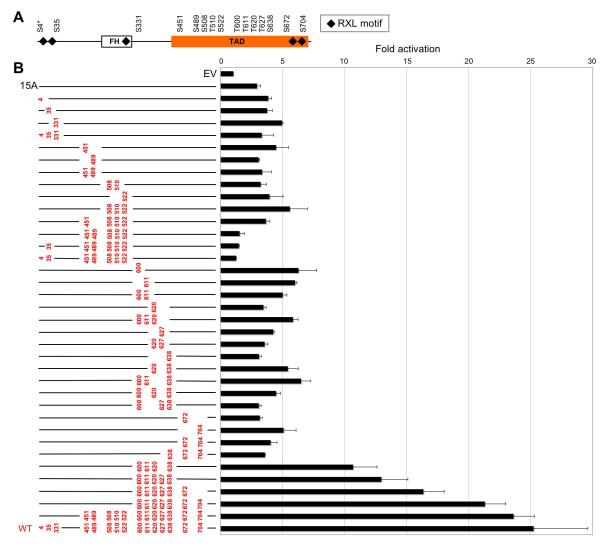
The data presented in Figures 1 and 2 indicate that CDK4/6 positively regulate FOXM1 activity by direct phosphorylation. FOXM1 harbors 5 RXL (cyclin binding) motifs and 15 CDK consensus sites, 12 of which are localized in its C-terminal transactivation domain (TAD) (Figure 3A). We first systematically determined the relative contribution of each of the 15 consensus sites, as well as combinations thereof, to FOXM1 transcriptional acitivity (Figure 3B). We then asked which of the sites are targeted by CDK4/6 (Figure 4). Compound mutation of all 15 sites to alanines led - like CDK4/6 inhibitor treatment (Figure 2C) - to greatly diminished FOXM1 transcriptional activity in U2OS cells (Figure 3B). We then introduced a series of ‘add-back’ (A → S/T) mutations into this template. As illustrated in Figure 3B, no single phosphorylation site was sufficient to mediate transactivation, nor was any tested combination of two or three sites. However, serial reconstitution of 5 to 7 sites in the FOXM1 C-terminus, comprising T600, T611, T620, T627, S638, S672 and S704, progressively increased transcription, consistent with the observation that T600, T611 and T638 critically contribute to FOXM1 function (Laoukili et al., 2008). By contrast, A → S/T replacements of an equal number of residues localized in the FOXM1 N-terminal and middle region, spanning S4, S35, S451, S489, S508, T510 and S522, did not lead to transactivation. Nevertheless, restoration of all 12 TAD sites (from S451 to S704) resulted in full activation. Thus, phosphorylation of the C-terminal region of the TAD domain is pivotal for driving transcription.
Figure 3. Contribution of CDK Consensus Sites to FOXM1 Transcriptional Function.
(A) Arrangement of Scansite CDK motifs in FOXM1. S4 (asterisks) is a typical S/T-P CDK site, but is not detected by Scansite, due to its close proximity to the N-terminus. FH, Forkhead domain; TAD, transactivation domain.
(B) Reporter gene activation (+/−SD) in U2OS cells transfected with the 6DB luciferase plasmid plus either empty vector (EV), the FOXM1 15 site to alanine mutant (‘15A’), FOXM1 ‘add-back’ mutants or wild-type FOXM1 (WT). All ‘add-back’ (A → S/T) mutated residues are highlighted in red; mutant alanines are not shown.
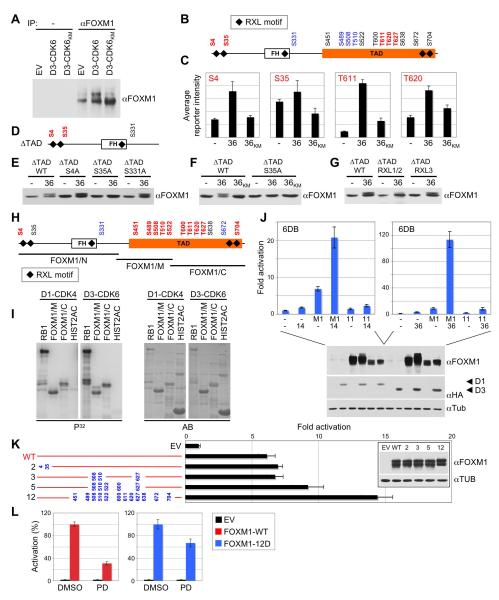
Figure 4. CDK4/6 Activate FOXM1 by Multisite Phosphorylation.
(A) Immunoprecipitation (IP) of endogenous FOXM1 from U2OS cells expressing either empty vector (EV), cyclin D3-CDK6, or cyclin D3 and kinase-inactive CDK6 (CDK6KM).
(B and C) FOXM1 was co-expressed with either empty vector (−), or cyclin D3 and CDK6 (36), or cyclin D3 and kinase-inactive CDK6KM (36KM) in HeLa cells, immunoprecipitated and subjected to quantitative mass spectrometry. In vivo phosphorylated sites are depicted in red; black sites were not accessible to analysis; no phosphorylation was detected on blue sites. Site-specific in vivo phosphorylation quantification (+/−SD) is shown in C.
(D) FOXM1 TAD deletion mutant (ΔTAD) with N-terminal CDK4/6 sites (red).
(E) HeLa cells were co-transfected with either empty vector (−), or cyclin D3 and CDK6 (36), plus FOXM1ΔTAD wild-type (WT) or phosphorylation-site mutants (S4A, S35A, S331A).
(F) FOXM1ΔTAD co-transfections with either empty vector (−), or cyclin D3 and CDK6 (36), or cyclin D3 and kinase-inactive CDK6KM (36KM).
(G) Co-transfections with either empty vector (−), or cyclin D3 and CDK6 (36), and FOXM1ΔTAD wild-type (WT) or RXL mutant versions, in which the RXL motif was replaced by AXA.
(H) FOXM1 fragments (black bars) were in vitro phosphorylated by cyclin D3-CDK6 and analyzed by mass spectrometry. Phosphorylated residues are depicted in red; unphosphorylated sites are blue; black sites were not accessible to analysis.
(I) In vitro phosphorylation assay performed using recombinant cyclin D1-CDK4 or cyclin D3-CDK6 with RB1, FOXM1 middle (M) or C-terminal fragment (C), and histone H2A. P32, autoradiogram; AB, Amidoblack protein staining for total protein amounts.
(J) Activation of the 6DB promoter (+/−SD) by wild-type FOXM1 (M1) versus the ‘11A’ CDK site to alanine mutant plus cyclin D1-CDK4 (14, left panel) or cyclin D3-CDK6 (36, right panel). Protein expression is shown (lower panel).
(K) Effect of phosphomimetic mutations on FOXM1 transcriptional activity (+/−SD). Hela cells were transfected with the 6DB promoter and either empty vector (EV), wild-type FOXM1 (WT), or phosphomimetic mutants (S/T residue numbers replaced by aspartic acid, blue). Expression of the mutant proteins is shown in the inlet.
(L) U2OS cells transfected with the 6DB promoter and either empty vector (EV), wild-type FOXM1 (WT) or the FOXM1 ‘12D’ mutant (12D) were treated with DMSO or 1μM PD0332991 (PD) for 16 h. Values are mean +/−SD. See also Figure S2.
Endogenous FOXM1 was phosphorylated in U2OS cells by cyclin D3-CDK6 in a kinase-dependent fashion as judged by mobility shift experiments (Figure 4A). Of note, previous studies have shown that FOXM1 is phosphorylated in vivo on at least nine S/T-P CDK consensus sites (Laoukili et al., 2008). To ascertain the phosphorylation state of individual CDK4/6-targeted sites in vivo, we turned to quantitative mass spectrometry, and sought to obtain site-specific phosphorylation quantification rather than absolute FOXM1 sequence coverage. As shown in Figures 4B and S2A, there were two in vivo CDK4/6 phosphorylation sites in the FOXM1 N-terminus, and at least three in the C-terminal region of the TAD, which is consistent with the finding that phosphorylation of this region is critical for FOXM1 function (Figure 3B). Importantly, phosphorylation at S4, S35, T611 and T620 was specifically induced by exogenous cyclin D3-CDK6 catalytic activity in HeLa cells (Figure 4C), although this analysis is limited by the basal phosphorylation state of individual sites in vivo, as some may be inherently phosphorylated by endogenous CDK activity. Phosphorylation quantification of T627 did not reach statistical significance, but in vivo phosphorylation of S35 was further confirmed by mobility shift analysis of a truncated FOXM1 protein (ΔTAD), which only contains the N-terminal CDK4/6 target sites and three RXL motifs (Figure 4D). As illustrated in Figure 4E, co-expression of cyclin D3-CDK6 led to decreased ΔTAD electrophoretic mobility, which specifically required S35 and CDK6 catalytic activity (Figure 4F). Moreover, FOXM1’s N-terminal RXL motifs 1 and 2, but not motif 3 in the FH domain (Figure 4G), are involved in CDK6-mediated phosphorylation of S35, which fits the observation that RXL sequences may allow for efficient phosphorylation of CDK sites lacking basic residues (Figure S2C; Leng et al., 2002).
Direct FOXM1 multisite phosphorylation by CDK4/6 was further confirmed in in vitro assays. When three FOXM1 fragments covering the entire protein sequence were incubated with recombinant cyclin D3-CDK6 complex and subjected to mass spectrometry, there were 10 in vitro-phosphorylated sites, although S35 and S638 were not susceptible to analysis (Figure 4H, Figure S2B). Notably, most in vitro sites are localized in the TAD (Figure 4H), and the FOXM1 fragments comprising the TAD were phosphorylated by cyclin D1-CDK4 and cyclin D3-CDK6 in in vitro assays (Figure 4I). Accordingly, mutation of all 10 phosphorylation sites within the TAD plus S638 to alanine (the ‘11A’ mutant) greatly diminished FOXM1 activation by CDK4 and CDK6 (Figure 4J). Finally, compound mutation of all RXL motifs in the N-terminus and the TAD did not affect CDK4/6-mediated activation of FOXM1 (Figures S2D, S2E). Thus, although RXL sequences aid in phosphorylation of certain sites, such as S35, they are not required for FOXM1 activation. In sum, these data identify five overlapping in vivo and in vitro CDK4/6 target sites in FOXM1 (S4, S35, T611, T620 and T627), although the phosphorylation site coverage of our in vivo analysis was only fragmentary. We conclude that multisite phosphorylation of the FOXM1 C-terminus by CDK4/6 is required for full activation of FOXM1 transcriptional function.
We next sought to obtain a FOXM1 phosphomimetic mutant, which should remain active under conditions of acute CDK4/6 inhibition. When we progressively replaced all S/T-P CDK consensus sites in the TAD with aspartic acid residues, there was an increase in overall FOXM1-mediated transcription (Figure 4K), although to a much lesser extent than was the case upon co-expression of wild-type FOXM1 with cyclin D-CDK4/6 (Figure 2A). Notably, aspartic acid has only a single negative charge, so more aspartic acid residues may be needed than phosphates to efficiently activate FOXM1. Most importantly, however, while PD0332991 treatment substantially diminished the function of wild-type FOXM1 (by 70%), the reduction was much less when transcription was driven by the FOXM1 ‘12D’ mutant (Figure 4L). Thus, a multisite phosphorylation mechanism, which activates FOXM1 by converting its C-terminal domain into an acidic-type TAD, is highly plausible.
Cyclin D-CDK4/6 Stabilize the FOXM1 Protein by Multisite Phosphorylation
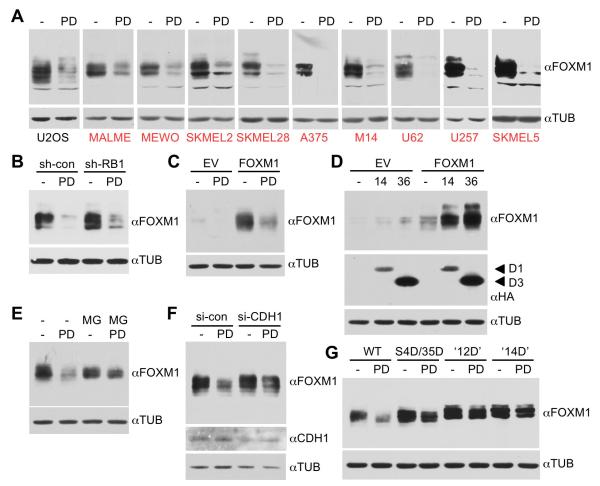
We next asked whether cyclin D-CDK4/6 activity also regulates the expression of FOXM1 itself. To this end, we treated a large panel of human cancer cell lines with the specific CDK4/6 inhibitor, PD0332991. We noted with interest that CDK4/6 inhibition partially to completely ablated expression of the FOXM1 protein, with the most dramatic effects seen in human melanoma cell lines (Figure 5A). By contrast, inhibition of CDK2 had no such effect (Figure S3A). We next explored the mechanism by which PD0332991 decreased FOXM1 levels. Importantly, this mechanism was independent of RB1, as FOXM1 expression was still efficiently reduced in RB1 knockdown cells (Figure 5B, Figure 2D). Moreover, when FOXM1 was expressed from a constitutive, but weak promoter (which is used here throughout Figure 5) in U2OS cells, it became apparent that PD0332991 treatment reduced endogenously as well as exogenously expressed FOXM1 protein to a comparable extent (left panel of Figure 5A vs. Figure 5C). Conversely, when cyclin D-CDK complexes were ectopically expressed in HeLa cells with or without FOXM1, they upregulated both exogenously as well as endogenously expressed FOXM1 protein (Figure 5D). By contrast, when FOXM1 was expressed at much higher levels using CMV-based plasmids, the total protein amount barely changed in response to either CDK4/6 inhibition or ectopic expression (Figure S3B, see also Figures 3, 4). This is understandable, as the relative reduction of the protein by PD0332991 becomes negligible under these conditions.
Figure 5. CDK4/6 Stabilize FOXM1 by Multisite Phosphorylation.
(A) U2OS and melanoma cells (red) were incubated with DMSO or 1 μM PD0332991 (PD) for 16 h, and endogenous FOXM1 protein levels were analyzed.
(B) U2OS cells stably expressing either control (sh-con) or RB1 targeting shRNA (sh-RB1) were treated with PD0332991 and endogenous FOXM1 was analyzed.
(C) U2OS cells were transfected with pBABE empty vector (EV) or pBABE-FOXM1 and treated with PD0332991.
(D) Cells were co-transfected with either pBABE empty vector (EV) or pBABE-FOXM1, and cyclin D1-CDK4 (14) or cyclin D3-CDK6 (36). Expression of the HA-tagged cyclins is shown below.
(E) U2OS cells transfected with pBABE-FOXM1 were treated with 5 μM MG132 (MG) 4 h prior to cell lysis.
(F) U2OS cells were transfected with pBABE-FOXM1 and either control (si-con) or CDH1 siRNA (si-CDH1).
(G) Effect of replacing the FOXM1 N-terminal (S4/S35), 12 TAD or all 14 CDK (12 TAD plus N-terminal) sites with aspartic acid residues on FOXM1 protein levels. U2OS cells were transfected with either pBABE wild-type FOXM1 (WT), FOXM1 S4/S35 to D (S4D/S35D), ‘12D’ or ‘14D’ mutant. Cells were treated with DMSO (−) or 1μM PD0332991 (PD) for 16 h. See also Figure S3.
Since these results suggested a post-transcriptional mechanism of regulation, we next asked whether cyclin D-CDK4/6 activity interferes with FOXM1 protein degradation. Indeed, treatment of U2OS cells with a proteasomal inhibitor, MG132, blocked PD0332991-induced FOXM1 turnover (Figure 5E). Likewise, ablation of the anaphase-promoting complex (APC) subunit CDH1, a mediator of FOXM1 proteasomal degradation (Laoukili et al., 2008), partially protected this transcription factor from proteolysis (Figure 5F).
We finally examined if FOXM1 protein turnover is regulated by phosphorylation of FOXM1 itself. To address this, we took advantage of FOXM1 phosphomimetic mutants, harboring replacements of either the N-terminal CDK4/6 target sites (S4D/S35D), the 12 TAD CDK sites (‘12D’), or all 14 CDK sites, including S4/S35 (the ‘14D mutant’). As illustrated in Figure 5G, ‘simulated’ multisite phosphorylation of the 12 TAD or all 14 sites partially protected FOXM1 from drug-induced degradation. Degradation susceptibility of FOXM1 S4D/S35D was less reduced, with a rank order of wild-type FOXM1 > S4D/S35D > ‘12D’ = ‘14D’. This suggests that multisite phosphorylation converts FOXM1 to a more degradation-resistant state, in addition to activating its transcriptional function. Such a mechanism is indirectly supported by the observation that FOXM1 protein expression is induced at the end of G1 (Major et al., 2007), a cell cycle phase where cyclin D-CDK4/6 are thought to be maximally active.
CDK4/6 → FOXM1 Signaling Suppresses Senescence in Cancer Cells
It is currently not completely understood how hyperactive cyclin D-CDK4/6 promotes tumorigenesis. One possible mechanism is to overcome cellular senescence, a permanent form of growth arrest that is normally enforced in cells by aberrant oncogenic signaling and that prevents them from progressing towards malignancy (Campisi and d’Adda di Fagagna, 2007).
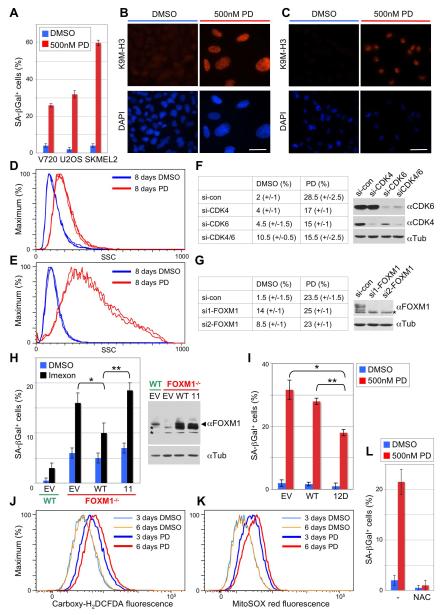
We have previously shown that cyclin D1-CDK4 catalytic activity is critically required in a murine model of HER2-driven breast cancer (Yu et al., 2006; Landis et al., 2006). We now asked if treatment of cells derived from these mouse tumors (V720 cells) with PD0332991 ‘reactivates’ the senescence program. We also examined the effects of treatment of human osteosarcoma (U2OS) and melanoma cells (SKMEL2). As shown in Figure 6A, acute inhibition of CDK4/6 increased the activity of SA-β-Gal, a marker for senescent cells, in all three cell lines, whereas inhibition of CDK2 had only marginal effects (Figure S4A). Other markers of senescence were also detectable, such as global chromatin modifications, including tri-methylated lysine 9 of histone H3 (K9M3-H3, Figures 6B, 6C), and a dramatic increase in overall cellular granularity (Figures 6D, 6E).
Figure 6. Inhibition of CDK4/6 Catalytic Activity Enforces a ROS Dependent Senescence Program by Disabling FOXM1.
(A) Percentage of SA-β-galactosidase positive cells (+/−SD) among breast cancer (V720), osteosarcoma (U2OS) and melanoma (SKMEL2) cell lines, each treated with either DMSO or 500 nM PD0332991 for 8 days.
(B and C) Immunofluorescence staining of U2OS cells (B) and SKMEL2 cells (C) using anti-trimethyl K9 histone H3 antibody (K9M-H3, upper panel) or DAPI (lower panel). Cells were treated with either DMSO or PD0332991 as above. Scale bar represents 10 μm.
(D and E) Quantification of cellular granularity by FACS. Side scatter (SSC) analysis of U2OS (D) and SKMEL2 cells (E), treated with either DMSO or 500 nM PD0332991 as above. Graphs are from triplicate experiments.
(F and G) Percentage of SA-β-galactosidase positive U2OS cells upon knockdown of CDK4/6 (F) and FOXM1 (G). Cells were transiently transfected with the indicated siRNAs, followed by 8-day PD0332991 treatment 48 h post-transfection. si-con, control siRNA; si-CDK4, CDK4-specific siRNA; si-CDK6, CDK6-specific siRNA; si1-FOXM1 and si2-FOXM1, two different siRNAs targeting FOXM1. The asterisk indicates an unspecific band.
(H) Wild type (WT) and FOXM1 knockout MEFs (FOXM1-/-) were transduced with either pBABE empty vector (EV), wild type FOXM1 (WT) or the 11-site phosphorylation mutant (11), and FOXM1 expression was analyzed (right panel; asterisks indicate unspecific bands). Cells were treated with either DMSO or 100 μM of the ROS inducing drug, Imexon. SA-β-galactosidase positive cells were counted after 4 days of treatment. Error bars, mean of SD; *p<0.05, **p<0.01.
(I) SA-β-galactosidase positive U2OS cells (+/−SD) transfected with either empty vector (EV), wild-type FOXM1 (WT), or the ‘12D’ mutant (expressed from the CMV promoter). Cells were treated as in F. Error bars, mean of SD; *p<0.05, **p<0.01.
(J) Measurement of cytoplasmic ROS levels with Carboxy-H2DCFDA by FACS in U2OS cells treated with either DMSO or 500 nM PD0332991 for 3 and 6 days.
(K) Quantification of mitochondrial ROS levels with MitoSOX red in U2OS cells treated as in J.
(L) SA-β-galactosidase positive U2OS cells (+/−SD) after simultaneous incubation with 1 mM N-acetyl-cysteine (NAC) and either DMSO or PD0332991. See also Figure S4.
Importantly, initiation of this senescence program was not merely a consequence of cell cycle inhibition by PD0332991 (Figures S4B, S4C, S4D). Moreover, like CDK4/6 inhibitor treatment, siRNA-mediated knockdown of CDK4 and CDK6 in combination led to a significant increase in basal senescence, although to a lesser extent (Figure 6F). These results are consistent with previous observations that genetic ablation of CDK4 leads to the induction of cellular senescence (Zou et al., 2002; Puyol et al., 2010). Surprisingly, however, PD0332991-triggered senescence is partially RB1-independent, as depletion of this bona fide substrate in U2OS cells decreased senescence by less than 50% (Figure S4E, S4F, S4G). This observation led us to examine whether FOXM1 is involved in CDK4/6-mediated senescence suppression. Of note, FOXM1 is a transcriptional activator, and the effects of its depletion on senescence are therefore opposite to those of ablation of RB1, which acts as transcriptional repressor. Thus, while RB1 ablation decreased basal senescence (Figure S4E), transfection of U2OS cells with two different FOXM1 siRNAs resulted in a substantial increase in basal senescence (from 1.5% to 14% and 9%, respectively). Importantly, FOXM1 knockdown did not further enhance PD0332991-induced SA-β-Gal activity (Figure 6G).
To further substantiate the involvement of FOXM1, we reconstituted FOXM1 knockout MEFs (Laoukili et al., 2005) with wild-type FOXM1 or the inactive ‘11’ site mutant (Figure 4J). As immortalized FOXM1 knockout MEFs show only a modest increase in senescence (approximately 5%), we challenged both cell populations with the reactive oxygene species (ROS)-inducing drug, Imexon (ROS are known to induce senescence). Although Imexon increased absolute SA-β-Gal activity in FOXM1 knockout cells expressing an empty vector (EV), it did barely so in the wild-type cells (Figure 6H). Importantly, re-expression of wild-type FOXM1, but not the inactive ‘11’ phosphorylation site mutant, significantly lowered Imexon-induced SA-β-Gal activity.
Conversely, when we expressed constitutively active FOXM1 (the ‘12D’ phosphomimetic mutant) in U2OS cells and quantified SA-β-Gal activity following treatment with PD0332991, the senescence response was reduced by 36% (Figure 6I). By contrast, there were no significant differences between cells transfected with empty vector or wild-type FOXM1 (Figure 6I). Taken together, these data strongly suggest that CDK4/6 phosphorylates FOXM1 to protect cells from senescence, consistent with previous findings of FOXM1 exhibiting senescence-suppressing activity (Wang et al., 2009), presumably by repressing the levels of ROS (Park et al., 2009). Notably, elevated ROS levels were detectable in the cytoplasm and mitochondria of U2OS cells as early as 3 days post-PD0332991 treatment, and they further increased with time (Figures 6J, 6K). Concordantly, when ROS were scavenged by treatment of U2OS cells with an antioxidant, N-acetyl-cysteine (NAC), PD0332991-induced senescence was abrogated (Figure 6L).
CDK4/6 → FOXM1 Signaling Induces Expression of Critical G1/S Genes and Promotes S Phase Entry
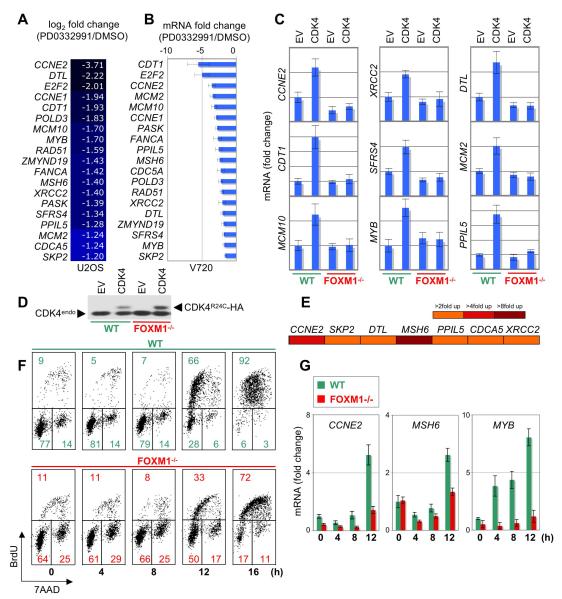
We also monitored the early transcriptional response to acute inhibition of CDK4/6 catalytic activity. As PD0332991 substantially induced senescence in U2OS cells and ErbB2-overexpressing mouse mammary tumor cells (V720, Figure 6A), we hypothesized that the identification of overlapping genes exhibiting differential expression in both cell lines (human osteosarcoma and mouse breast cancer) would uncover ‘common’ CDK4/6 regulated genes potentially contributing to senescence regulation, rather than cell type-specific transcriptional changes. We therefore performed gene expression arrays in combination with QPCR analysis from U2OS and V720 cells treated with or without PD0332991 for 4 hours. Although this did not result in significant changes in the cell cycle profile (Figure S5), it led to a significant decrease in the expression of 19 ‘common’ genes (Figures 7A, 7B).
Figure 7. FOXM1 Activates Key G1/S Genes Downstream of CDK4 Function and Promotes G1/S Transition.
(A and B) 19 ‘common’ cyclin D-CDK4/6-regulated genes obtained by microarray analysis from U2OS cells (A), and QPCR analysis (+/−SD) from V720 mouse mammary tumor cells (B). Each cell line was treated with either DMSO or PD0332991 for 4 h. For each cell line, genes are arranged from the highest to lowest change in expression.
(C) QPCR analysis of the indicated genes in wild-type (WT) and FOXM1-/- MEFs, each stably expressing either pBABE empty vector (EV) or CDK4R24C (CDK4). Values derived from WT-EV cells were set to 1-fold change in expression. Values are mean +/−SD.
(D) Protein expression of HA-tagged CDK4R24C and endogenous CDK4 (CDK4endo) in wild-type (WT) and FOXM1-/- MEFs.
(E) U2OS cells were transfected with the indicated promoter-luciferase constructs plus either empty vector or FOXM1. Reporter assay results are visualized as a heat map (scale, right panel).
(F and G) Wild-type (WT) and FOXM1-/- MEFs were serum starved for 48 h, followed by serum addition at the indicated time points. Cell cycle parameters (F) and mRNA expression (G) were subsequently assessed by FACS and QPCR analysis (+/−SD), respectively. See also Figure S5.
Expectedly, several of those genes, including cyclin E1 and E2, are known to be induced at the G1/S phase transition and have previously been identified as transcriptional targets of the canonical RB-E2F pathway (Bracken et al., 2004). To test the involvement of FOXM1 in the regulation of these 19 genes, we compared their expression in wild-type and FOXM1 knockout MEFs, each expressing either empty vector (EV) or constitutively active CDK4R24C. As illustrated in Figure 7C, CDK4 expression substantially increased (>1.75-fold) the mRNA levels of 9 out of 19 genes in the wild-type cells. However, none of the genes were induced in the absence of FOXM1, indicating that this transcription factor positively regulates expression of G1/S phase genes like cyclin E2 (CCNE2), MYB, MCM2, MCM10 and CDT1, which are critical for initiation of DNA replication, but also activates genes involved in the regulation of DNA repair (XRCC2) and mRNA splicing (SFRS4). Accordingly, ectopic expression of FOXM1 induced transcription of a representative panel of PD0332991-regulated genes, including cyclin E2, MSH6 and SKP2, an established FOXM1 target gene (Wang et al., 2005) (Figure 7E). Moreover, the induction of cyclin E, MSH6 and MYB was blunted during G1/S transition in FOXM1 knockout MEFs (Figure 7G). Concordantly, these cells had a delay in S phase entry (Figure 7F), which fits previous observations of FOXM1 promoting G1/S phase transition (Wang et al., 2002; Wang et al., 2008).
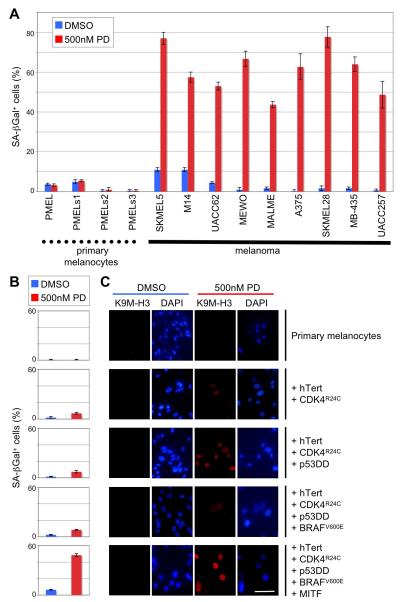
PD0332991 Triggers Massive Senescence in Malignant Melanoma Cells, but not in Normal Melanocytes
The strong reduction of the FOXM1 protein in malignant melanoma cell lines (Figure 5A) and the dramatic senescence response triggered by PD0332991 in SKMEL2 melanoma cells (Figures 6A, 6C, 6E) prompted us to investigate whether this senescence induction is common among cell lines derived from this tumor type, which is notoriously known for its chemoresistance. It was also unclear whether PD0332991 elicits similar effects in ‘normal’ melanocytes. As shown in Figure 8A, the drug triggered profound senescence in all tested human melanoma cell lines (in addition to SKMEL2, Figure 6A), but not in the primary human melanocytes derived from four different donors. Moreover, when these PD0332991-unresponsive melanocytes were transformed by serial introduction of common melanoma oncogenes, including CDK4R24C, BRAFV600E and MITF as previously described (Garraway et al., 2005), only the fully transformed cells showed a substantial drug-induced senescence response, as judged by the increase in SA-β-Gal activity (Figure 8B) and histone H3 lysine 9 trimethylation (Figure 8C). By comparison, transduction of constitutively active CDK4 (CDK4R24C) had only a moderate effect, indicating that hyperactivation of this kinase is a prerequisite but not sufficient event in rendering melanocytes fully susceptible to CDK4/6 inhibitor-induced senescence.
Figure 8. PD0332991-Induced Cellular Senescence is Linked to CDK4 Hyperactivation and the Transformed Phenotype.
(A) Percentage of SA-β-galactosidase positive (+/−SD) primary melanocytes versus malignant melanoma cells following treatment with PD0332991 or DMSO for 8 days.
(B and C) Successive transduction (top to bottom) of primary melanocytes with hTERT, CDK4R24C, dominant-negative p53 (p53DD), BRAFV600E and MITF renders these cells susceptible to PD0332991-induced senescence. Panel B shows the percentage of SA-β-galactosidase positive cells (+/−SD) following PD0332991 or DMSO treatment for 8 days. In panel C, trimethylation of histone H3 lysine 9 (K9M-H3) is visualized by immunofluorescence (red). Nuclear DNA was stained with DAPI (blue). Scale bar represents 10 μm.
DISCUSSION
Identification of CDK4/6 Substrates by Combined Approaches
We performed here an unbiased systematic substrate screen for CDK4 and CDK6, and generated a list of 68 potential human phosphorylation targets, in addition to the three RB family proteins (Figure S1). This in vitro substrate resource is not confined to proteins expressed in certain cell, organ, tissue or tumor types, and can be used as a basis for future analyses of CDK4/6 molecular functions in a wide variety of developmental and disease processes, ranging from cancer to diabetes.
Several general findings emerged from our combinatorial screen. First, there is considerable difference in substrate specificity between the cyclin D1-CDK4 and cyclin D3-CDK6 complex, with the latter phosphorylating a much broader spectrum of substrates. A striking example is the potent in vitro phosphorylation of splicing regulatory proteins, such as SFRS16, TRA2A, SFRS10, by cyclin D3/CDK6, but not cyclin D1/CDK4 (Figure S1). Second, phosphorylation of substrates by CDK4/6 can induce protein stabilization, as shown here for FOXM1 (Figure 5). Notably, we did not observe such stabilization under conditions of strong (CMV promoter-driven) ectopic substrate expression, which overrides the subtle effects of CDK-induced protein stabilization (Figure S3B). Thus, from our validation screen, it is not possible at present to estimate the proportion of substrates stabilized by phosphorylation. Third, in addition to inactivating transcriptional repressors like RB1, CDK4 and CDK6 can directly activate transcription factors by phosphorylation, thereby driving specific gene expression.
Although our substrate screen is unbiased, it is clear that the number of in vitro substrates presented here is an underestimation of the total number of potential phosphorylation targets in the entire human proteome. Our computational substrate enrichment analysis was based on two assumptions: the potential substrates bear at least two CDK consensus sites, and they possess the annotation keyword ‘nuclear’ in their SWISSPROT protein entry. There will be cases where genuine substrates have only one CDK motif, and where protein database-listed annotations are not correctly defined (proteins can extensively shuttle between cytoplasm and nucleus, and might not be categorized as ‘nuclear’ in annotation analysis). Further, our current ORFeome library is not complete, and some proteins are not susceptible to expression in E. coli. Together, these limitations reduced the number of proteins subjected to the screen. In addition, our in vivo substrate validation method by mobility shift analysis is not applicable to all identified in vitro substrates. Thus, although our combined screening approach revealed eight confirmed substrates (SENP3, MEF2D, SRrp35, ZEB1, SHOX2, FOXM1, RB1, RBL2), the real number is likely to be substantially higher.
CDK4/6 Regulate FOXM1 Activity and Stability by Multisite Phosphorylation
FOXM1 is a master regulator of the cell cycle, being required for timely entry into both S phase and mitosis (Figure 7E) (Wang et al., 2002; Wang et al., 2008; Laoukili et al., 2005). Phosphorylation of FOXM1 starts in G1, and continues during S, G2 and M phases of the cell cycle (Major et al., 2007). The data presented here suggest that CDK4/6 initiate FOXM1 phosphorylation, thereby causing accumulation of this transcription factor in cells. At the same time, phosphorylation directly activates FOXM1 transcriptional function, consistent with previous observations that cyclin D1-CDK4 strongly activates FOXM1 in reporter gene assays (Wierstra and Alves, 2006), as does cyclin E-CDK2, although to a lesser extent (Luescher-Firzlaff et al., 2006). It is plausible that the coexistence of these two regulatory mechanisms (stabilization and activation) provides CDK4/6 with a more powerful and elaborate control over transcription factor function than would either mechanism alone.
How is FOXM1 activated in cancer cells by CDK4/6-mediated phosphorylation? It is clear from Figure 3B that seven C-terminal phosphorylation sites, spanning T600 to S704, critically contribute to FOXM1 function. Moreover, there is a direct correlation between the number of accessible CDK sites in this C-terminal region and FOXM1 transcriptional function, suggesting that CDKs activate this transcription factor according to a quantitative model, which would allow to ‘fine-tune’ FOXM1 function to spatial and temporal gradients of CDK activity. Based on the data presented in Figure 3, it is likely that phosphorylation simply incorporates negative charge into the C-terminal TAD, thereby progressively activating FOXM1, possibly by relieving autorepression by the N-terminal domain (Laoukili et al., 2008).
CDKs have not previously been implicated in FOXM1 protein stabilization, although this is an important aspect of FOXM1 regulation (Figure 5A). FOXM1 degradation is mediated through three degradation (D) boxes and one KEN box, all of which are localized in the FOXM1 N-terminus (Laoukili et al., 2008). Although CDK4/6 phosphorylate the N-terminal sites, S4 and S35 in vivo (Figure 4C), phosphomimetic mutations at these two sites were not sufficient to protect FOXM1 from PD0332991-induced degradation (Figure 5G). Indeed, the data suggest that phosphorylation of more sites in the TAD is required to efficiently stabilize FOXM1 (Figure 5G). Thus, we favor the idea that multisite phosphorylation, concomitantly with conferring transcriptional activation, converts FOXM1 to a more proteolysis-resistant conformation by making the degradation boxes less accessible to CDH1 (Figures 5F, 5G).
It has been shown that CDK2 phosphorylates FOXM1 (Laoukili et al., 2008), yet inhibition of CDK2 did not change the steady-state level of this transcription factor (Figure S3A) and did not cause senescence in U2OS and SKMEL2 cells (Figure S4A), which is in sharp contrast to the consequences of inhibition of CDK4/6 (Figure 6). Moreover, the critical requirement of CDK4/6 for tumor formation makes them the most likely candidate kinases to account for increased FOXM1 function in many tumors, such as HER2-positive breast cancers, in which expression of HER2 and FOXM1 are tightly correlated (Francis et al., 2009).
FOXM1 and RB1
Although RB1 is considered the major substrate of cyclin D-dependent kinases, emerging evidence indicates that CDK4/6 → RB1 signaling does not mediate all CDK4/6 catalytic functions (this study; Dean et al., 2010; Haferkamp et al., 2008). It is important to note that much of the experimental evidence describing RB1 as the central cyclin D-CDK4/6 substrate stems from the analysis of RB1-negative cell lines, in which CDK4/6 inhibition had no phenotypic consequences. However, RB1-negative cancer cell lines, such as HeLa, ‘overexpress’ p16 and therefore lack functional cyclin D-CDK4/6 complexes, as the CDK4/6 subunits form stable binary complexes with p16 INK4A (Ruas and Peters, 1998). This makes the interpretation of the data obtained from RB1-negative cells difficult.
To overcome this limitation by outcompeting p16, we overexpressed D-type cyclins together with the CDK4/6 subunit in HeLa cells (Figures 3A, 3B). Using this cell line, we demonstrated that CDK4/6 activate FOXM1 independently of RB proteins (Figure 2). It was recently reported that FOXM1 activation by CDK4 is lost in RB1-negative cells, but these findings rest on the expression of FOXM1 fragments lacking the N-terminal repression domain (Wierstra and Alves, 2006). Although we do not exclude the possibility that additional mechanisms contribute to the regulation of FOXM1 function, and that RB1 may play a role in this process, the data presented here strongly suggest that CDK4/6 activate and stabilize FOXM1 by direct phosphorylation.
CDK4/6 Substrate Phosphorylation and Senescence Suppression
Senescence is a tumor-suppressive mechanism, and it must be overcome during cell immortalization and transformation. Indeed, CDK4/6 overactivation is a common hallmark of human cancers, and ectopic expression of both kinases substantially extends the proliferative lifespan of primary cells, such as human fibroblasts (Ruas et al., 2007). Several of our identified in vitro substrates (Figure S1), including FOXM1, MYC and ZEB1, were shown to suppress cellular senescence (Wu et al., 2007; Liu et al., 2008). It is therefore likely that CDK4/6 maintain senescence suppression through phosphorylation of several transcriptional regulators, including RB1 and FOXM1. This might, in fact, explain why FOXM1 knockdown triggers senescence in only 9 to 14% of U2OS cells (Figure 6G), as compared to approximately 25-30% induced by acute inhibition of CDK4/6 with PD0332991 in the same cell type (Figure 6F). In melanoma cells, PD0332991-induced senescence was markedly higher, indicating that cells derived from this tumor type are particularly reliant on senescence suppression by CDK4/6 catalytic activity (Figure 8). Importantly, this reliance likely rests on a combination of signals emanating from several oncogenes, such as BRAF and MITF (in addition to CDK4). These findings provide a rationale for the treatment of melanoma with small molecule inhibitors of CDK4/6.
EXPERIMENTAL PROCEDURES
In Silico Screen of the Human Proteome
The UniProtKB/SWISS-PROT protein database was searched for CDK-motif containing protein sequences using Scansite 2.0 (Obenauer et al., 2003). Homo sapiens was chosen as “single species”, and the keyword “nucleus” was specified to enrich for nuclear proteins. Detailed information can be found in the Supplemental Experimental Procedures.
High-Throughput Protein Expression in E. coli and Purification
Bacterial protein expression and cell lysis was performed according to Braun et al. (2002). GST fusion proteins were enriched and purified with a ME200 12 channel electronic pipettor equipped with 20 μl Glutathione PhyTip columns (Phynexus).
Phosphorylation Quantification and PR-Score Calculation
Kinase reaction products were loaded on SDS gels and transferred to nitrocellulose membranes. Membranes were stained with Amido black 10B, dried and exposed to x-ray film. The relative phosphorylation score (PR-score) was calculated from ratio of the radioactive signal from the autoradiograph (32P) and the amount of respective fusion protein from the membrane (AB) according to the following formula: PR-score = 32PS/ABS × ABRB1/32PRB1 × 100%, were S is the substrate, and RB1 the positive control.
Quantification of In Vivo Phosphorylation Sites by Mass Spectrometry
FLAG-FOXM1 was immunoprecipitated from HeLa cells and digested with LysC. Resulting peptides were incubated with amino reactive tandem mass tag reagents (TMT) and assigned with different mass tags. Samples were analyzed by mass spectrometry, and TMT reporter ion intensities normalized to the accumulation time of each MS3 spectrum.
Gene Expression Analysis
Total RNA was isolated from cells using the RNeasy kit (Qiagen), and reverse-transcribed with the ABI reverse transcription kit. Biotin-labeled cDNA was hybridized to Affymetrix human 1.0 exon arrays, and derived CEL files were analyzed with Partek’s Genomics Suite software (Partek, St. Louis, MO).
Supplementary Material
Highlights.
Assembly of an in vitro CDK4/6 substrate resource across the human proteome
FOXM1 phosphorylation by CDK4/6 results in protein stabilization and activation
FOXM1 is critical for CDK4/6-mediated cell cycle entry and senescence suppression
CDK4/6 blockade in melanoma cells leads to FOXM1 degradation and massive senescence
SIGNIFICANCE.
CDK4/6 catalytic activity is thought to be critically required for tumorigenesis; and clinical trials with small molecule inhibitors, such as PD0332991, are currently under way. Even so, there is little information about the spectrum of substrates targeted by CDK4/6. This information is not only critical for our understanding of cell cycle control and its deregulation in cancer, but may also aid in optimizing current therapeutic strategies. In this study we have assembled a resource of CDK4/6 substrates across the human proteome. Our analyses suggest that therapeutic targeting of CDK4/6 → FOXM1 signaling might be an effective strategy to enforce senescence in cancer cells, and that small molecule inhibitors of CDK4/6 should be tested in clinical trials against malignant melanoma.
ACKNOWLEDGEMENTS
We thank Drs. Rene Medema for providing FOXM1 plasmids, IGFBP-1, 6DB, CENPF and PLK1 promoter sequences and FOXM1 knockout MEFs; David Fruman and Richard Assoian for CCNG2 and SKP2 promoters, Reuven Agami for pSUPER-CDH1, Sunkyu Kim and Nina Ilic for cell lines. We also thank Philippe Lamesch, Kourosh Salehi-Ashtiani and Josh LaBaer for help with the ORFeome libraries. P. H. was supported by a fellowship from the Swedish Wenner-Gren Foundations. This work was supported by NIH grants R01 CA083688 to P.S., and RL1 DE019022 to S.P.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS Exon array data were deposited in the NCBI Gene Expression Omnibus (GEO) database (GSE32182).
SUPPLEMENTAL INFORMATION Supplemental information includes five figures, one table as separate Excel file, Supplemental Experimental Procedures and Supplemental References, and can be found with this article online.
REFERENCES
- Ackermann AM, Gannon M. Molecular regulation of pancreatic beta-cell mass development, maintenance, and expansion. J Mol Endocrinol. 2007;38:193–206. doi: 10.1677/JME-06-0053. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Braun P, Hu Y, Shen B, Halleck A, Koundinya M, Harlow E, LaBaer J. High throughput protein production for functional proteomics. Proc Natl Acad Sci. 2002;5:2654–2659. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- Francis RE, Myatt SS, Krol J, Hartman J, Peck B, McGovern UB, Wang J, Guest SK, Filipovic A, Gojis O, et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol. 2009;35:57–68. doi: 10.3892/ijo_00000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK, Toogood PL. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analysis identify MITF as a lineage survival oncogene amplified in maligant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Haferkamp S, Becker TM, Scurr LL, Kefford RF, Rizos H. p16INK4a-induced senescence is disabled by melanoma-associated mutations. Aging Cell. 2008;7:733–745. doi: 10.1111/j.1474-9726.2008.00422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, Shin B, Datta A, Raychaudhuri P, Costa RH. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9:13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, Heck AJ, Medema RH. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol Cell Biol. 2008;28:3076–3087. doi: 10.1128/MCB.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008;7:2720–2726. doi: 10.4161/cc.7.17.6580. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Leng X, Noble M, Adams PD, Qin J, Harper JW. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol Cell Biol. 2002;22:2242–2254. doi: 10.1128/MCB.22.7.2242-2254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean D. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–588. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luescher-Firzlaff JM, Lilischkis R, Luescher B. Regulation of the transcription factor FOXM1c by Cyclin E/CDK2. FEBS Lett. 2006;580:1716–1722. doi: 10.1016/j.febslet.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- Park HJ, Carr JR, Wang Z, Nogueira V, Hay N, Tyner AL, Lau LF, Costa RH, Raychaudhuri P. FoxM1, a critical regulator of oxidative stress during oncogenesis. Embo J. 2009;28:2908–2918. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, Guerra C, Santamaria D, Barbacid M. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- Ruas M, Gregory F, Jones R, Poolman R, Starborg M, Rowe J, Brookes S, Peters G. CDK4 and CDK6 Delay Senescence by Kinase-Dependent and p16INK4a-Independent Mechanisms. Mol. Cell. Biol. 2007;27:4273–4282. doi: 10.1128/MCB.02286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes DE, Ackerson T, Major ML, Kalinichenko VV, Costa RH, Raychaudhuri P, Tyner AL, Lau LF. FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J Biol Chem. 2008;283:20770–20778. doi: 10.1074/jbc.M709892200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I, Alves J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biol Chem. 2006;387:949–962. doi: 10.1515/BC.2006.119. [DOI] [PubMed] [Google Scholar]
- Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl. Acad. Sci. USA. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Sicinska E, Geng Y, Ahnstrom M, Zagozdzon A, Kong Y, Gardner H, Kiyokawa H, Harris LN, Stal O, Sicinski P. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23–32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Zou X, Ray D, Aziyu A, Christov K, Boiko AD, Gudkov AV, Kiyokawa H. Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev. 2002;16:2923–2934. doi: 10.1101/gad.1033002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.