Abstract
The epidermal growth factor receptor (EGFR) is upregulated within a high percentage of solid tumors and hence is an attractive target for tumor-targeted therapies including gene therapy. The natural EGFR ligand epidermal growth factor (EGF) has been used for this purpose, despite the risk of mitogenic effects due to EGFR activation. We have developed a fully synthetic, EGFR-targeted gene delivery system based on PEGylated linear polyethylenimine (LPEI), allowing evaluation of different EGFR-binding peptides in terms of transfection efficiency and EGFR activation. Peptide sequences directly derived from the human EGF molecule enhanced transfection efficiency with concomitant EGFR activation. Only the EGFR-binding peptide GE11, which has been identified by phage display technique, showed specific enhancement of transfection on EGFR-overexpressing tumor cells including glioblastoma and hepatoma, but without EGFR activation. EGFR targeting led to high levels of cell association of fluorescently labeled polyplexes after only 30 min of incubation. EGF pretreatment of cells induced enhanced cellular internalization of all polyplex types tested, pointing at generally enhanced macropinocytosis. EGF polyplexes diminished cell surface expression of EGFR for up to 4 hr, whereas GE11 polyplexes did not. In a clinically relevant orthotopic prostate cancer model, intratumorally injected GE11 polyplexes were superior in inducing transgene expression when compared with untargeted polyplexes.
In this paper, Schäfer and colleagues develop a fully synthetic EGFR-targeted gene delivery system based on PEGylated linear polyethylenimine and use it to investigate whether the efficiency of transduction and activation of EGFR can be uncoupled. Various EGFR-binding peptides for targeted gene delivery to glioblastoma, hepatoma, and prostate carcinoma were tested. The peptide GE11 was able to enhance transfection of EGFR-overexpressing tumor cells while avoiding undesirable EGFR activation.
Introduction
The epidermal growth factor receptor (EGFR), also known as the ERBB1 or HER1 receptor, belongs to the Erb receptor family, a group of four transmembrane receptors with intrinsic tyrosine kinase activity. Ligand binding to these receptors activates the kinase moiety and leads to autophosphorylation and downstream signaling (Schlessinger, 2002), which may result in proliferation, differentiation, enhanced cell migration and adhesion, or inhibition of apoptosis. EGFR is present on all epithelial and stromal cells as well as on several glial and smooth muscle cell types at a density of 4×104–1×105 molecules per cell (Wells, 1999). Up to 2×106 EGF receptors per cell, as well as receptor mutations associated with constitutive tyrosine kinase activity, have been described in numerous solid tumors including lung, liver, breast, and bladder cancer as well as in hepatocellular carcinoma and glioblastoma (Kim and Muller, 1999). This makes EGFR a suitable marker for targeted delivery of anticancer drugs (Khalil et al., 2003). Several EGFR-targeted macromolecular carriers, such as liposomes loaded with chemotherapeutics (Kim et al., 2009), nanocrystals (Diagaradjane et al., 2008), and also nucleic acids (Shir et al., 2006), have been successfully delivered in cell culture and in experimental animal models. EGF protein, either from natural sources or recombinant production, can be easily covalently coupled to gene carriers. Its high binding affinity for EGFR leads to highly specific cell binding and subsequent internalization (de Bruin et al., 2007) and allows systemic targeting of EGFR-overexpressing tumors (Wolschek et al., 2002). Binding and internalization of EGF, however, generally result in EGFR activation, which is a significant disadvantage in the case of antitumoral therapies. Hence, alternatives for efficient targeting in the absence of activation are needed. Also, the use of recombinant protein for gene delivery vectors can be expensive when moving toward preclinical and clinical development of such a synthetic gene vector. In this study, we evaluated various fully synthetic EGFR-binding peptides for targeted gene delivery to glioblastoma, hepatoma, and prostate carcinoma and studied the effect of such carriers on internalization and activation of EGFR. Activation of EGFR was observed not only with EGF protein bound to the gene carrier, but also with peptide sequences directly derived from EGF. This led to rapid internalization of the EGFR without receptor recycling for at least 4 hr. In sharp contrast, the GE11 peptide sequence identified by the phage display technique was able to allow highly efficient gene delivery to a broad range of tumor cells in vitro, but also in a clinically relevant, orthotopic model of prostate cancer without EGFR activation.
Materials and Methods
Conjugate synthesis, plasmid DNA, polyplexes
Coupling of 2-kDa heterobifunctional polyethylene glycol (NHS-PEG-OPSS: (ortho-pyridyl)disulfide-PEG-succinimidyl ester; Rapp Polymere, Tübingen, Germany) to linear polyethylenimine (22 kDa; LPEI) was performed under water-free conditions in ethanol as described (Schaffert et al., 2011). In brief, LPEI dissolved in ethanol was reacted with a 3-fold molar excess of NHS-PEG-OPSS dissolved in dimethyl sulfoxide (DMSO). After 3 hr at room temperature the product, LPEI-PEG-OPSS, was purified by cation-exchange chromatography (Macro-Prep High S, HR10/10; Bio-Rad, Munich, Germany) with a salt gradient from 0.5 to 3 M NaCl in 20 mM HEPES, pH 7.4. The product eluted between 2.0 and 2.8 M NaCl, and was subsequently dialyzed against HBS (20 mM HEPES [pH 7.4], 150 mM NaCl). LPEI concentration was determined by copper assay (Ungaro et al., 2003), and the degree of PEGylation by proton nuclear magnetic resonance (1H NMR). For subsequent peptide coupling, LPEI-PEG-OPSS conjugates with molar ratios of PEG/LPEI between 0.8:1 and 1.5:1 were used. Peptides were produced by standard solid-phase synthesis with a purity greater than 95%. CMY (CMYIEALDKYA) and MYI (MYIEALDKYAC) were produced by IRIS Biotech (Marktredwitz, Germany), and GE11 (CYHWYGYTPQNVI) either by IRIS Biotech or Biosyntan (Berlin, Germany). For LPEI-PEG-GE11 synthesis, GE11 solution (0.27 μmol in 30% acetonitrile, 70% H2O, 0.1% trifluoroacetic acid [TFA]; total volume, 100 μl) was mixed with LPEI-PEG-OPSS (0.18 μmol of LPEI, 0.27 μmol of PEG-OPSS; total volume, 1000 μl) in 150 mM NaCl, 20 mM HEPES (pH 7.4). The pH of the reaction mixture was approximately pH 7.2. Reaction was completed after 2–4 hr at room temperature, when measuring the release of the dithiopyridone group at 343 nm. The mixture was purified by cation-exchange chromatography as described for LPEI-PEG-OPSS (see above), and the product LPEI-PEG-GE11 was dialyzed against HBS and stored frozen in aliquots at –80°C. LPEI-PEG-CMY and LPEI-PEG-MYI were produced in an analogous manner. Control conjugate LPEI-PEG was either used in the precursor form (LPEI-PEG-OPSS) or the terminal OPSS group was reacted with cysteine and purified by size-exclusion chromatography (SEC) on a Sephadex G-25 column (GE Healthcare Life Sciences, Freiburg, Germany), using 20 mM HEPES, pH 7.4. The amount of targeting peptides present in newly synthesized conjugates was quantified through reaction with 5,5′-dithiobis-nitrobenzoate (DTNB; Ellman's assay). DTNB converts thiols to a mixed disulfide while releasing 2-nitro-5-thiobenzoate (TNB), which is measured at 412 nm. Cysteine was used as standard. The synthesis of LPEI-PEG-EGF (containing recombinant murine EGF as targeting ligand) was carried out as described (Schaffert et al., 2011).
Cyanine 5 (Cy5) labeling of LPEI and conjugates was achieved with amine-reactive FluoroLink Cy5 monofunctional dye (GE Healthcare Life Sciences) as described (Boeckle et al., 2004); unreacted dye was removed by SEC.
Cytomegalovirus (CMV)-driven plasmids pEGFP-Luc (Promega, Mannheim, Germany), pCDNA3Luc (kindly provided by W.G. Kaelin, Dana-Farber Cancer Institute, Birmingham, AL), and pCMVLuc were propagated in Escherichia coli DH5α and purified endotoxin-free with an EndoFree plasmid Giga kit (Qiagen, Hilden, Germany) or by PlasmidFactory (Bielefeld, Germany). Plasmid pCpG-hCMV-Luc (human CMV enhancer and elongation factor 1α promoter driven; Navarro et al., 2010) was propagated in E. coli GT115 (Cayla-InvivoGen, Toulouse, France) under Zeocin selection pressure and purified by PlasmidFactory. Plasmid was covalently labeled with Cy5, using a Label IT kit (Mirus, Madison, WI) according to the manufacturer's instructions. For in vitro studies, polyplexes were generated in HEPES-buffered glucose (HBG; 20 mM HEPES [pH 7.1], 5% glucose [w/v]) or (for U87wtEGFR and U-87 MG cells) in HBS ½ (20 mM HEPES [pH 7.1], 2.5% glucose [w/v], 75 mM NaCl) at an N/P ratio (molar ratio of nitrogen in LPEI to phosphate in pDNA [plasmid DNA]) of 6 (corresponding to an LPEI/pDNA ratio [w/w] of 0.78/1) and a final pDNA concentration of 20 μg/ml. For in vivo studies, polyplexes were generated at 200 μg of pDNA per milliliter. Size and surface charge was determined with a Malvern Zetasizer (Malvern Instruments, Worcestershire, UK) as described (Schaffert et al., 2011).
Cell culture and in vitro transfections
HuH-7 (JCRB0403; Japanese Cancer Research Resources Bank, Tokyo, Japan) and HepG2 (HB-8065; American Type Culture Collection [ATCC], Manassas, VA) human hepatocellular carcinoma cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–F12 (1:1) supplemented with 10% fetal bovine serum (FBS) and antibiotics (penicillin, streptomycin). Human glioblastoma cell lines U-87 MG (HTB-14; ATCC) and U87MGwtEGFR overexpressing epidermal growth factor (kindly provided by A. Levitzki, Hebrew University, Jerusalem, Israel) were cultured in DMEM supplemented with 10% FBS and antibiotics (penicillin, streptomycin). U87MGwtEGFR cells were additionally kept under selection pressure by adding geneticin (G418; 0.4 mg/ml) to maintain EGFR overexpression.
For in vitro transfections followed by luciferase quantification, cells (U-87 MG, U87MGwtEGFR, and HepG2, 10,000 cells; HuH7, 5000 cells) were seeded in 200 μl of medium in 96-well plates (TPP, Trasadingen, Switzerland) 24 hr before transfection. For transfection, medium was replaced with 100 μl of fresh, FBS-containing medium and polyplexes containing the indicated amounts of plasmid were added. After 4 hr medium was replaced again and after 24 hr of transfection cells were lysed in cell lysis buffer (Promega) and luciferase activity was quantified with a luminometer (Berthold Lumat LB 9507; Berthold Technologies, Bad Wildbad, Germany) as described (Navarro et al., 2010). Two nanograms of recombinant luciferase (Promega) correspond to 107 relative light units (RLU). Under all transfection conditions applied, no toxicity was observed when measuring total protein or mitochondrial activity (MTT assay) 24 hr after transfection.
Western blot analysis
Cells (200,000) were seeded in 6-well plates 24 hr before transfection. Medium was replaced with 2 ml of fresh medium and polyplexes containing 2 μg of pDNA were added for 45 min. As a positive control, 108 pmol of free EGF (recombinant murine EGF; Peprotech, Hamburg, Germany) was added, corresponding to the amount of EGF present in LPEI-PEG-EGF polyplexes; untreated cells served as negative control. Thereafter, cells were washed with phosphate-buffered saline (PBS), harvested with trypsin–EDTA, resuspended in fresh medium, and pelleted for 5 min at 200×g. The pellet was washed twice with PBS and incubated on ice for 20 min with 50 μl of cell lysis buffer (Promega) supplemented with protease inhibitor cocktail (Roche, Mannheim, Germany) as well as phosphatase inhibitor mixtures I and II (Sigma-Aldrich, Munich, Germany), 10 μl of each per 1 ml. Undissolved cell debris was removed by centrifugation (15 min, 1000×g) and 40 μg (U87MGwtEGFR) or 10 μg (HuH7) of soluble protein (determined with a Pierce bicinchoninic acid [BCA] kit; Pierce Biotechnology, Rockford, IL) was separated by polyacrylamide gel electrophoresis and blotted onto a polyvinylidene difluoride membrane. Before antibody incubation, unspecific binding sites were blocked for 1 hr with 5% nonfat milk. Immunostaining was performed overnight at 4°C with monoclonal mouse anti-phospho-Erk (mitogen-activated protein kinase [MAPK]), polyclonal rabbit anti-Erk, monoclonal mouse anti-phospho-Akt (protein kinase B [PKB], Rac), and polyclonal rabbit anti-Akt antibodies (U87MGwtEGFR: New England BioLabs [Beverly, MA]; diluted 1:1000; HuH7: Cell Signaling Technologies [Danvers, MA], diluted 1:5000 [MAPK/Akt] or 1:2000 [pMAPK, pAkt]). Horseradish peroxidase-conjugated secondary anti-mouse and anti-rabbit antibodies (U87MGwtEGFR: Vector Laboratories [Burlingame, CA], diluted 1:2000; HuH7: Cell Signaling Technologies, diluted 1:10,000) and an enhanced chemiluminescence detection kit (ECL; Amersham, Arlington Heights, IL) were used for visualization; α-tubulin served as additional loading control.
Confocal laser scanning microscopy and flow cytometry
For laser scanning microscopy (LSM) studies, 5000 HuH7 cells were seeded per well in collagen A-coated Lab-Tek 8 chambered coverglasses (Nalge Nunc, Naperville, IL) 24 hr before transfection. Cells were incubated with Cy5-labeled polyplexes containing 500 ng of plasmid DNA in 400 μl of medium for up to 4 hr at 37°C, and thereafter were washed with PBS and fixed with 200 μl of 4% paraformaldehyde (PFA). Polyplexes contained either Cy5-labeled plasmid (20% of total plasmid) or Cy5-labeled conjugate, as indicated.
For antibody staining the cells were incubated successively for 1 hr with mouse anti-human EGFR (diluted 1:200, clone M3563; Dako, Hamburg, Germany) and goat anti-mouse IgG1 linked to Alexa Fluor 488 (diluted 1:400; Invitrogen, Karlsruhe, Germany) in 10% FBS in PBS, and thereafter were washed twice with 10% FBS in PBS and stored in 300 μl of PBS. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/ml in PBS). Cell imaging was performed with a confocal laser scanning microscope (LSM 510 META; Carl Zeiss, Jena, Germany) equipped with an argon and two helium/neon lasers delivering light at 488, 543, and 633 nm, respectively, as well as an argon laser emitting ultraviolet light at 364 nm. Light was collected through a 63×1.4 NA oil immersion objective (Carl Zeiss). Data acquisition was performed in multitrack mode (Cy5: excitation at 633 nm, emission at 650 nm longpass; Alexa 488: excitation at 488 nm, emission at 505–530 nm bandpass; DAPI: excitation at 364 nm, emission at 385–470 nm bandpass). Data were analyzed with AxioVision 4.6 software (Carl Zeiss).
For flow cytometry analyses, 100,000 HuH7 cells were seeded in 12-well plates (TPP) 24 hr before transfection. Thereafter medium was replaced with 1 ml of fresh culture medium and Cy5-labeled polyplexes (2 μg of plasmid per well; 20% of total plasmid Cy5 labeled) added. After 0.5 hr of incubation at 37°C cells were harvested by treatment with trypsin–EDTA solution and incubated for 1 hr with mouse anti-human EGFR (diluted 1:200, M3563; Dako, Hamburg, Germany) or mouse IgG1 control antibody (diluted 1:200, X0931; Dako) in 10% FBS in PBS, washed and stained with Alexa 488-conjugated goat anti-mouse antibody (diluted 1:400; Invitrogen). Thereafter cells were washed and DAPI (final concentration, 1 μg/ml) added to distinguish between live and dead cells. Cells were analyzed with a DakoCytomation ADP flow cytometer (Dako, Glostrup, Denmark) with Alexa 488 fluorescence excited at 488 nm and detected with a 530/40 nm bandpass filter. DAPI fluorescence was excited at 405 nm and detected with a 450/40 bandpass filter; Cy5 fluorescence was excited at 635 nm and emission was detected with a 665/20 nm bandpass filter. Cells were gated appropriately by forward versus sideward scatter and forward scatter versus DAPI to exclude cell debris and dead cells.
In vivo model
Human PC346C cells (van Weerden et al., 1996; Marques et al., 2006) were cultured in DMEM–F12 medium (Cambrex BioWhittaker, Verviers, Belgium) supplemented with 2% FBS (PAN Biotech, Aidenbach, Germany), 1% insulin–transferrin–selenium (GIBCO-BRL/Invitrogen), 0.01% bovine serum albumin (Boehringer Mannheim, Mannheim, Germany), epidermal growth factor (10 ng/ml; Sigma-Aldrich), penicillin–streptomycin antibiotics (penicillin, 100 U/ml; streptomycin, 100 mg/ml; Cambrex BioWhittaker) plus the following additions: fibronectin (100 ng/ml; Harbor Bio-Products, Tebu-bio, Boechout, Belgium), fetuin (20 ng/ml; ICN Biomedicals, Zoetermeer, The Netherlands), cholera toxin (50 ng/ml), 0.1 mM phosphoethanolamine, triiodothyronine (0.6 ng/ml), dexamethasone (500 ng/ml) (all from Sigma-Aldrich), and 0.1 nM 17-methyltrienolone (R1881; New England Nuclear/PerkinElmer Life Sciences, Boston MA). Cells were grown in T25 Primaria tissue culture flasks (BD Biosciences, Benelux, The Netherlands) at 37°C under a 5% CO2 humidified atmosphere. When near confluency was reached, cells were harvested with a trypsin–EDTA solution (Lonza, Verviers, Belgium) consisting of trypsin–Versene (170,000 U/liter) and EDTA (200 mg/liter). Suspensions of cells were dissolved in PBS to yield approximately 1×106 cells/20 μl for orthotopic injection.
Six- to 7-week-old male NMRI nu/nu mice (Taconic, Ry, Denmark) were inoculated with PC346C cells via the dorsolateral prostate. Mice were kept in individually ventilated cages (Tecniplast, Buguggiate, Italy) under a 12-hr light/dark cycle and received irradiated chow and acidified drinking water ad libitum. Orthotopic tumor growth was monitored over time by transrectal ultrasonography of the mouse prostate (Kraaij et al., 2002). When tumors reached a size of 50–100 mm3 (15–20 days after inoculation), polyplexes formed with the luciferase reporter plasmid pCpG-hCMV-Luc were injected directly into the prostate tumor tissue at a concentration of 200 μg/ml in a total volume of 50 μl. Luciferase expression was detected by in vivo imaging. Luciferin (Xenogen/Caliper Life Sciences, Hopkinton, MA) in saline (B. Braun Melsungen, Melsungen, Germany) was administered intraperitoneally at a dose of 150 mg/kg. Imaging was performed with an IVIS imaging system 200 series with Living Image software version 2.5 (Xenogen/Caliper Life Sciences). Luciferase expression levels were quantified through region of interest measurements. Animal experiments were approved by the Dutch Animal Experimental Committee (DEC) and performed in agreement with The Netherlands Experiments on Animals Act (1977) and the European Convention for Protection of Vertebrate Animals Used for Experimental Purposes (Strasbourg, March 18, 1986). Tumors were harvested after sacrifice and lysed in 500 μl of lysis buffer (1 mM dithiothreitol [DTT], 1% Triton X-100, 15% glycerol, 8 mM MgCl2, and 25 mM Tris phosphate [pH 7.8]). Next, 100 μl of luciferin (0.25 μM) and 0.25 μM ATP (both from AppliChem, Darmstadt, Germany) in lysis buffer were added to 100 μl of each extract, and luciferase activity was measured with a GloMax luminometer (Promega, Leiden, The Netherlands). After a delay of 2 sec, light emission was recorded for 5 sec. Luciferase expression was normalized for total protein content (Lowry assay; Bio-Rad).
Results
Conjugate synthesis and biophysics of polyplexes
EGFR-binding peptides and recombinant, murine EGF (EGF) have been coupled to LPEI via a heterobifunctional PEG linker as described (Schaffert et al., 2011). Coupling of NHS-PEG-OPSS to LPEI was carried out in ethanolic solution. PEG linker not coupled to LPEI was removed by cation-exchange chromatography. The uncharged PEG was found in the flowthough (0.5 M NaCl, 20 mM HEPES), whereas LPEI-PEG-OPSS containing conjugate eluted at NaCl concentrations between 2.0 and 2.8 M. When reacting LPEI with a 3-fold molar excess of NHS-PEG-OPSS, LPEI-PEG-OPSS with an LPEI/PEG molar ratio of 1:1.5 (M/M) was obtained as determined by 1H NMR. In addition, the OPSS content was quantified by adding DTT to an aliquot of LPEI-PEG-OPSS. The absorption of the thiopyridone group released was measured at 343 nm (ɛ=8080 M–1), and the LPEI/OPSS ratio was determined as 1:1.5 (M/M). To a peptide sequence derived from the middle loop of human EGF (MYIEALDKYA; Komoriya et al., 1984) a terminal cysteine was added, either at the N terminus (CMYIEALDKYA, termed CMY) or the C terminus (MYIEALDKYAC, termed MYI); the peptide sequence GE11 (YHWYGYFPQNVI; Li et al., 2005) was synthesized with an N-terminal cysteine (CYHWYGYTPQNVI, termed GE11). Cysteine or cysteine-modified peptides were added to LPEI-PEG-OPSS and the reaction of peptides or cysteine with the OPSS group was quantitative as measured by thiopyridone release at 343 nm. After a 1-hr reaction time at room temperature no further increase in absorption at 343 nm was observed. The resulting conjugates were purified by cation-exchange chromatography to remove unbound peptide or cysteine and by-products (i.e., thiopyridone). The isolated product LPEI-PEG-peptide or LPEI-PEG-Cys was further analyzed for LPEI content (copper assay) and residual OPSS content. As no residual OPSS was detected, complete reaction of peptide with LPEI-PEG-OPSS was assumed, giving an LPEI/PEG/peptide ratio of 1:1.5:1.5. Also, no free thiol groups were present in the conjugates, indicating the absence of free cysteine or peptide. All conjugates were able to condense plasmid DNA in a similar way as unmodified LPEI (data not shown), and particle sizes of polyplexes generated in HBG buffer at an N/P ratio of 6 ranged between 90 and 120 nm (Supplementary Table S1; supplementary data are available online at www.liebertonline.com/hum). For EGF-based polyplexes, 50% of LPEI-PEG-EGF conjugate was replaced by LPEI-PEG-Cys conjugate, as otherwise polyplexes tended to aggregate, an observation already made with similar, but branched polyethylenimine-based polyplexes (Ogris et al., 2003). Compared with LPEI polyplexes, all PEG-containing polyplexes exhibited a reduced ζ potential (+25 mV for LPEI vs. +11–19 mV for all other PEG-containing conjugates). Apparently, the 2-kDa PEG reduces the surface charge, but did not lead to completely charge neutral particles. Because of its size of 2 kDa, it also did not negatively influence DNA condensation, as it has been observed with larger (3400-Da) PEG molecules (Erbacher et al., 1999).
Enhanced in vitro transfection with EGFR-targeted peptide polyplexes
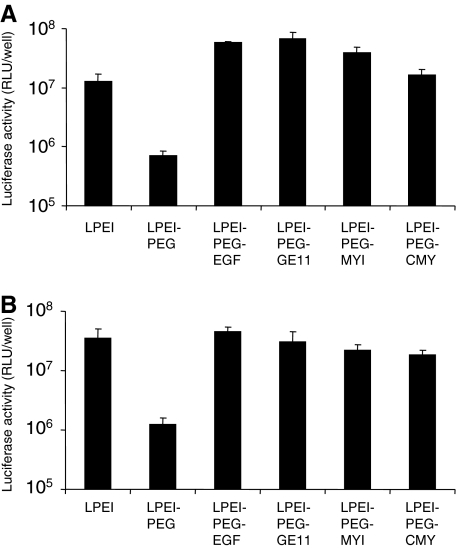
EGFR-targeted and untargeted polyplexes were used for transfection of human glioblastoma cell lines U87MGwtEGFR (high EGFR level after retroviral transduction to express additional EGFR) and U-87 MG (medium EGFR level; see Supplementary Fig. S1). On both cell lines, the modification of LPEI with 2-kDa PEG reduced transfection efficiency by about 90%; in sharp contrast, all EGFR-targeted polyplex formulations led to fully regained transfection efficiency on U-87 MG cells, whereas on U87MGwtEGFR cells a 5-fold increased transgene activity compared with LPEI polyplexes was observed (Fig. 1).
FIG. 1.
In vitro transfection of epidermal growth factor receptor (EGFR)-overexpressing glioma cells. (A) U87MGwtEGFR cells and (B) U-87 MG cells were transfected in 96-well plates with the indicated polyplexes containing luciferase-encoding plasmid (N/P ratio [(molar ratio of nitrogen in LPEI to phosphate in pDNA], 6; 200 ng of pDNA per well) in 100 μl of serum-containing medium. After 4 hr, medium was exchanged and luciferase activity was measured 24 hr after transfection as described in Materials and Methods. (n=3; error bars indicate the SD.)
EGFR activation
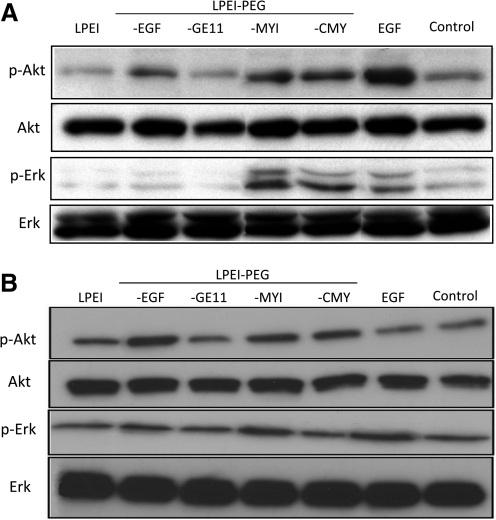
On ligand binding, activation of EGFR triggers, besides others, the ERK and PI3K/Akt pathways. Therefore, activation of these pathways through addition of complexes containing putative EGFR-targeting ligands was examined on U87MGwtEGFR and HuH7 cells. After 45 min of polyplex incubation, cells were harvested and phosphorylation of Erk and Akt was analyzed by Western blotting (Fig. 2). U87MGwtEGFR and HuH7 cells already showed basal, latent EGFR activation, as both p-Erk and p-Akt were detected. On incubation of U87MGwtEGFR cells with the EGF control or polyplexes containing EGF, CMY, or MYI, a strong increase in p-Akt was found, whereas p-Erk levels were elevated with CMY- and MYI-targeted polyplexes and the EGF (0.54 nmol/ml) control (Fig. 2A). In sharp contrast, GE11-targeted polyplexes did not activate Erk or Akt, as similar p-Erk and p-Akt levels were obtained with untreated cells or cells transfected with LPEI polyplexes. Similar observations were made on HuH7 cells (Fig. 2B), with the exception that the p-Akt increase was less pronounced and at the time point analyzed (45 min), no p-Erk was observed any longer when treating HuH7 cells with EGF alone. Apparently, enhanced transfection levels can be achieved with GE11-targeted polyplexes on EGFR-overexpressing U87MGwtEGFR cells without activation of EGFR signaling pathway. For this purpose, GE11-targeted polyplexes were further evaluated on other EGFR-overexpressing tumor cell lines, both in vitro and in vivo.
FIG. 2.
EGFR activation after polyplex treatment. (A) U87MGwtEGFR and (B) HuH7 cells were transfected for 45 min with the indicated polyplexes containing DNA (2 μg/ml) and EGFR activation was monitored in cell lysates, using antibodies directed against the EGFR downstream signals Akt and Erk and the respective phosphorylated forms (p-Akt and p-Erk). Lanes 1–5, after incubation with the indicated polyplex; lane 6, incubation with EGF (0.54 nmol/ml); lane 7, medium only.
EGFR targeting in hepatoma
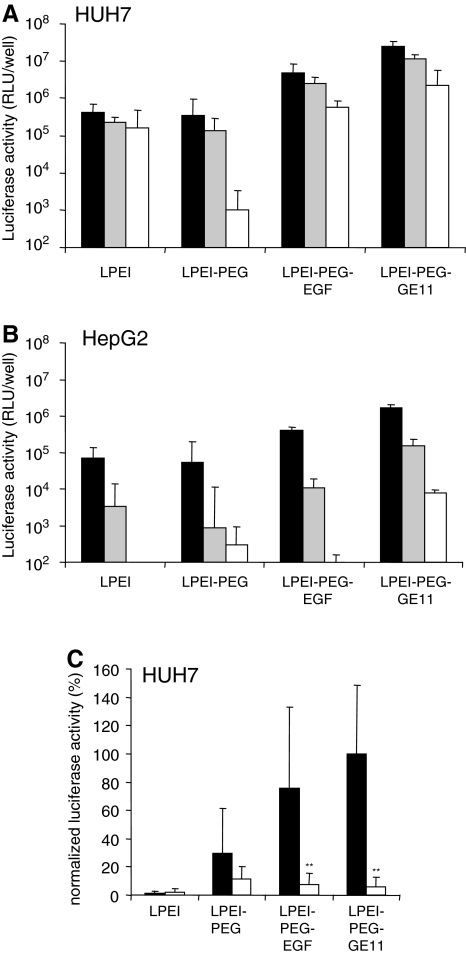
Similar to many glioblastoma-derived tumor cells, hepatocellular carcinoma often exhibits upregulated EGFR expression (Moon et al., 2006). Here we evaluated transfection performance on two hepatoma cell lines, namely HuH7 (high EGFR level) and HepG2 (low EGFR level; see Supplementary Fig. S1). GE11-targeted polyplexes were directly compared with EGF polyplexes and untargeted polyplexes (Fig. 3). In addition, the individual polyplexes were evaluated at various pDNA concentrations. Whereas the detargeting effect due to PEG modification was observed only on HuH7 cells at the lowest polyplex concentration, GE11 polyplexes were superior even to EGF-targeted polyplexes, resulting in a 25-fold (HepG2) or 60-fold (HuH7) increase in transfection efficiency compared with untargeted polyplexes. To ensure that the GE11-mediated increase in transfection efficiency was due to EGFR targeting, competitive inhibition experiments were carried out on HuH7 cells with a 1000-fold molar excess of free EGF present during transfection (Fig. 3C). Under these conditions only EGF- and GE11-containing polyplexes exhibited significantly reduced transfection efficiency. It must be noted that LPEI polyplexes in the presence of free EGF were, although not statistically significantly so, slightly more active in transfection, which could be due to EGF-induced macropinocytosis and mitogenicity (see below).
FIG. 3.
Transfection efficiency and specificity of GE11 polyplexes in hepatoma. (A and C) HuH7 and (B) HepG2 human hepatoma cells were seeded in 96-well plates (5000 cells per well) and transfected after 24 hr with the indicated polyplexes containing pEGFP-Luc plasmid for 4 hr. After a further 24 hr, cells were harvested and luciferase activity was quantified. (A and B) Cell lines were transfected with the indicated polyplexes with 500 ng (solid columns), 200 ng (shaded columns) or 50 ng (open columns) of plasmid per well. Relative light units (RLU) per well are shown. (n=3; error bars indicate the SD.) (C) HuH7 cells were transfected as described in (A) with 200 ng/well either in the absence (solid columns) or presence (open columns) of a 1000-fold molar excess of free EGF (based on the EGF amount present in LPEI-PEG-EGF polyplexes) for 4 hr and luciferase activity was quantified 24 hr thereafter. Normalized luciferase activity (n=6; error bars indicate the SD) is shown (mean LPEI-PEG-GE11, 100%). **p<0.01, standard medium versus a 1000-fold excess of free EGF, U test (Mann-Whitney).
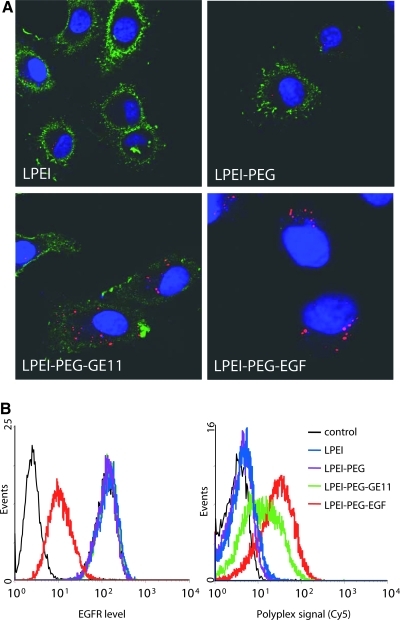
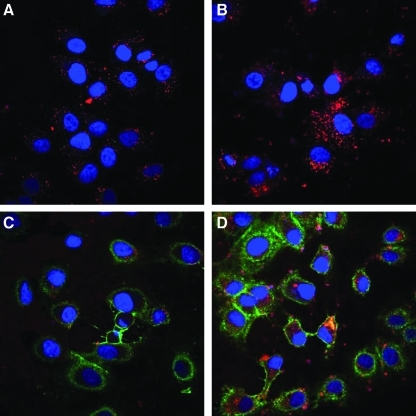
As binding and internalization of EGFR-targeted polyplexes has been reported to occur within less than 30 min of incubation (de Bruin et al., 2007), we also evaluated cellular binding and uptake of polyplexes generated with fluorescently labeled pDNA on HuH7 cells after only 30 min of incubation (Fig. 4). In addition, anti-EGFR staining after cell fixation allowed evaluation of EGFR levels on the cell surface. For LPEI and LPEI-PEG polyplexes, a low level of cell-associated polyplexes is seen (Fig. 4A, top and Fig. 4B, right), whereas with EGF and GE11 polyplexes cellular association is clearly higher (Fig. 4A, bottom and Fig. 4B, right). An even more striking difference can be observed for the surface-located EGFR: with untargeted and GE11-targeted polyplexes, cell surface EGFR levels are similar to that of untreated cells, whereas after incubation with EGF-targeted polyplexes almost no surface-located EGFR can be found (Fig. 4A, bottom; Fig. 4B, left). Apparently, LPEI-conjugated EGF triggered EGFR internalization without allowing for relocation of the receptor to the surface within 30 min. As more than 60% of polycation in PEI-based polyplexes with an N/P ratio of 6 can be free in solution (Boeckle et al., 2004), we also examined the uptake pattern of polyplexes formed with fluorescently labeled conjugates (Fig. 5). Already after 30 min of incubation with LPEI-PEG-EGF polyplexes, an intense fluorescent pattern of EGF conjugate can be observed, also in close vicinity to the nucleus (Fig. 5A), a result that is even more pronounced after 4 hr of incubation (Fig. 5B). Cy5-labeled LPEI-PEG-GE11 is already surface associated after 30 min (Fig. 5C), but to a clearly lower extent when compared with Cy5-labeled LPEI-PEG-EGF; after 4 hr of incubation the level of cell association with LPEI-PEG-GE11 is almost similar to that of labeled LPEI-PEG-EGF (Fig. 5D). Note also that after 4 hr of incubation with EGF polyplexes no surface-located EGFR can be found.
FIG. 4.
Influence of polyplex type on cellular EGFR level and uptake. HuH7 human hepatoma cells were seeded in chamber slides (A, 5000 cells) or 12-well plates (B, 100,000 cells) and 24 hr thereafter were incubated for 30 min at 37°C with the indicated polyplexes containing Cy5-labeled plasmid DNA (A, 500 ng; B, 2 μg). (A) Cells were fixed with 4% paraformaldehyde and stained for EGFR (Alexa Fluor 488-conjugated mouse anti-human EGFR, goat anti-mouse antibody), and nuclei were counterstained with DAPI. Pictures were taken with an LSM 510 META laser scanning microscope, and central sections are shown. Red, Cy5-labeled polyplexes; green, EGFR; blue, cell nuclei. (B) Cells were harvested by trypsinization, stained for EGFR (Alexa Fluor 488-conjugated mouse anti-human EGFR, goat anti-mouse antibody) and analyzed by flow cytometry. Left: EGFR level. Right: cellular level of Cy5-labeled polyplexes.
FIG. 5.
Intracellular uptake of labeled conjugate over time. HuH7 human hepatoma cells were seeded in chamber slides as described in Fig. 4A and thereafter incubated with (A and B) LPEI-PEG-EGF or (C and D) LPEI-PEG-GE11 polyplexes containing Cy5-labeled conjugate (50% of total conjugate Cy5-LPEI-PEG-EGF or Cy5-LPEI-PEG–GE11, 50% LPEI-PEG) for (A and C) 30 min or (B and D) 4 hr at 37°C. Cells were fixed with 4% paraformaldehyde and stained for EGFR (Alexa Fluor 488-conjugated mouse anti-human EGFR, goat anti-mouse antibody) and nuclei were counterstained with DAPI. Pictures were taken on a Zeiss LSM 510 META laser scanning microscope, and central sections are shown. Red, Cy5-labeled polyplexes; green, EGFR; blue, cell nuclei.
In vivo performance of GE11 polyplexes in a preclinical, orthotopic prostate cancer model
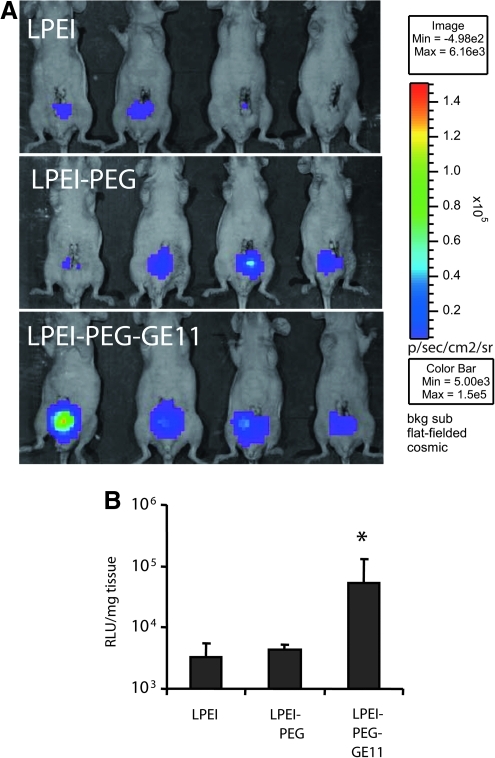
The human prostate cancer cell line PC346C was developed from the PC346 xenograft originating from transurethral resection material of the tumor of a patient with nonprogressive prostate cancer. Both xenograft and cell line are androgen responsive, secrete prostate-specific antigen (PSA), and harbor the wild-type androgen receptor (van Weerden et al., 1996; Marques et al., 2006). In addition, PC346 xenograft and cell line express EGFR (Supplementary Fig. S1). In vitro transfections revealed that this cell line is in principle transfectable with LPEI-based polyplexes under similar conditions as described for the other cell lines, although expression levels were low and variable (data not shown). Two to 3 weeks after intraprostatic injections of PC346C cells, prostate carcinomas developed. Polyplexes were generated with an in vivo-optimized luciferase reporter plasmid (Navarro et al., 2010) and directly injected into the prostate tumors. Bioluminescence imaging 48 hr after intratumoral injection clearly showed enhanced transgene expression with GE11-targeted polyplexes (Fig. 6). In tumor lysates, GE11 polyplexes led to 10-fold, significantly increased transgene expression. When applying polyplexes with fluorescently labeled pDNA (Cy5), we observed improved tissue distribution with the PEG-containing polyplex formulation when compared with LPEI polyplexes (data not shown).
FIG. 6.
Injection of polyplexes into orthotopic prostate tumors. Immune-deficient NMRI nu/nu mice carrying orthotopic PC346C prostate tumors were injected with 50 μl of polyplexes containing 10 μg of plasmid pCpG-hCMV-Luc. (A) Forty-eight hours after injection, bioluminescence imaging was performed (top, LPEI; middle, LPEI-PEG; bottom, LPEI-PEG-GE11). (B) Forty-eight hours after transfection mice were killed, tumors were resected and extracted, and luciferase activity was determined as relative light units per milligram of tissue (*p<0.05, LPEI vs. LPEI-PEG-GE11, U test).
Discussion
EGF has been intensively used for EGFR-targeted gene delivery approaches, both with viral and nonviral vectors (Peng et al., 2001; Wolschek et al., 2002; Ogris et al., 2003; Pereboeva et al., 2007). In our own work, we have most often used murine EGF, which, in contrast to human EGF, does not carry any lysines in its primary amino acid sequence (Blessing et al., 2001; Wolschek et al., 2002; Ogris et al., 2003). Hence, convenient coupling via the N-terminal primary amine to heterobifunctional cross-linkers, such as N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP), can occur without interfering with the binding domain of EGF. We have developed an improved method to couple EGF via a heterobifunctional PEG cross-linker to LPEI (Schaffert et al., 2011). To enhance the reaction efficiency between the amine-reactive N-hydroxy-succinimidyl ester (NHS) on OPSS-PEG-NHS with the secondary amines on LPEI, the reaction was carried out under water-free conditions in absolute ethanol. After purification and dialysis, thiolated EGF (after N-terminal modification with SPDP and subsequent reduction with DTT) could be specifically coupled to the distal OPSS group of PEG. In addition to recombinant EGF, we have chosen two peptide sequences known to interact with EGFR. One sequence was derived from the middle loop of human EGF (Komoriya et al., 1984) to which we added a cysteine either at the N or C terminus (CMY and MYI, respectively). The other peptide sequence, designated GE11, was described by Li and colleagues (2005) and was modified by the addition of an N-terminal cysteine. When directly comparing peptide polyplexes and EGF polyplexes in terms of transfection efficiency on EGFR-positive glioblastoma cells, all of them were at least as efficient as untargeted but strongly positively charged LPEI polyplexes and 10- to 100-fold more efficient than untargeted, PEG-containing control polyplexes. The EGFR-targeting effect was shown on both highly EGFR-overexpressing U87MGwtEGFR cells (106 EGFRs per cell; Shir et al., 2006) as well as the wild-type cell line U-87 MG (105 EGFRs per cell). Although only 2 kDa in size, the PEG molecule alone significantly reduced transgene expression in the glioblastoma cell lines. This detargeting can be explained by two effects: (1) PEG reduces the surface charge of polyplexes (see also Supplementary Table S1) and subsequently cellular polyplex association; and (2) after internalization, bulky molecules such as PEG or protein ligands can hamper endosomal release by negatively interfering with the intrinsic endosomolytic mechanisms of PEI, namely, the proton sponge effect and the rupture of the endosomal membrane (Walker et al., 2005; de Bruin et al., 2008). It is also important to note that the 2-kDa PEG spacer is sufficient to prevent aggregation-induced transfection of lung tissue after systemic application of polyplexes (Klutz et al., 2011), carrying the sodium iodide symporter (NIS) as a marker and therapeutic gene, in mice. LPEI polyplexes induced high levels of NIS mRNA expression in lung tissue. In sharp contrast, NIS mRNA was detected neither with LPEI-PEG-GE11- nor LPEI-PEG-Cys-based polyplexes.
Binding of EGF to EGFR rapidly induces receptor dimerization and internalization, which in turn activates the intracellular tyrosine kinase leading to auto- and transphosphorylation of the tyrosine residues. Activation of EGFR triggers, besides others, the Erk and PI3K/Akt pathways. Therefore activation of these pathways through addition of polyplexes containing putative EGFR-targeting ligands was examined on U87MGwtEGFR glioblastoma and HuH7 human hepatocellular carcinoma cells (Fig. 2). A low level of the phosphorylated signal transducers Erk and Akt was present also in untreated cells, indicating that these pathways, resulting in activation and proliferation, are active in most tumor cells when cultivated in the presence of 10% serum. Polyplexes containing EGF or the targeting peptides MYI and CMY led to phosphorylation of the extracellular signal-regulated kinase (Erk, MAPK1), similar to free EGF. Apparently, both EGF and peptides from the second loop of EGF were able to activate EGFR when covalently coupled to LPEI and incorporated into a polyplex, as described for the free peptide (Komoriya et al., 1984). Clearly these peptide ligands seem to preserve their mitogenic potential even after incorporation into LPEI polyplexes.
After incubation with polyplexes containing GE11 peptides, p-Akt and p-Erk levels were not increased in U87MGwtEGFR or HuH7 cells compared with untreated control cells. This is in line with observations made by Li and colleagues, where GE11 did not promote the mitosis of hepatoma cells (Li et al., 2005). For this purpose, we further evaluated the GE11 peptide on other tumor types, that is, hepatoma and prostate carcinoma. HuH7 and HepG2 are two hepatoma cell lines differing considerably in their aggressiveness (HuH7 is highly tumorigenic in nude and SCID mice, whereas HepG2 cells must be implanted subcutaneously at a 10-fold higher cell dose to induce tumors in SCID mice; Wolschek et al., 2002). In addition, HuH7 cells express high EGFR levels comparable to the level on U87MGwtEGFR cells, whereas among HepG2 cells only about 20% are EGFR positive (Supplementary Fig. S1). Interestingly, in both cell lines EGFR-targeted polyplexes were superior in transfection compared with untargeted polyplexes, and in both lines GE11 polyplexes induced higher transgene expression when compared with EGF polyplexes. Murine epidermal growth factor (mEGF) exhibits 70% sequential identity with human EGF with a similar affinity for EGFR (Kd=1–2 nM; Lemmon et al., 1997). Although GE11 peptide has a 10-fold lower affinity for EGFR (Kd=20 nM; Li et al., 2005), GE11 polyplexes were able to induce similar or even higher transgene expression levels when compared with EGF polyplexes (Figs. 1 and 3). This can be explained by a cooperative binding effect, as several GE11 molecules are clustered onto one polyplex. On the other hand, it could be caused by improved intracellular performance of GE11 polyplexes. Indeed, when using proteins as ligands on PEI-based polyplexes, we often observed a negative effect on transfection efficiency, which can be explained by the negative interference of a bulky protein with the endosomal release process (Ogris et al., 2001, 2003; Walker et al., 2005). The GE11 peptide is rather small (1.2 kDa) when compared with EGF (6 kDa) and the small size could be advantageous in this respect. The specificity of GE11-mediated transfection via the EGFR was demonstrated by coincubating GE11 polyplexes with a 1000-fold molar excess of free EGF (Fig. 3C). It is important to note that only simultaneous coincubation of polyplexes and EGF decreased transgene expression to a level comparable with untargeted polyplexes, whereas preincubation with EGF followed by polyplexes, in contrast, increased both transgene expression and polyplex uptake (Supplementary Fig. S2). Another explanation for the enhanced transgene expression level of EGF-treated cells is the mitogenicity of EGF, as the transfection efficiency of polyplexes is higher in mitotically active cells (Brunner et al., 2000, 2002). The enhanced polyplex uptake points to an additional function of EGF, namely, the general increase in surface internalization and the induction of macropinocytosis due to EGFR activation (Bryant et al., 2007). When measuring total polyplex association after 30 min of transfection, EGF polyplexes were clearly superior compared with GE11 polyplexes (Fig. 4B, right). Apparently, internalization of GE11 polyplexes in HuH7 cells is slower because of the absence of an additional internalization trigger. Still, after a 4-hr transfection period cellular association of EGF- and GE11-polyplex appeared similar (Fig. 5C for labeled conjugate; data not shown for labeled plasmid DNA), and transgene expression of GE11 polyplexes was as efficient as or even superior to that of EGF polyplexes (Figs. 1 and 3). To monitor the fate of the EFGR after polyplex incubation, we simultaneously analyzed the surface EGFR level and polyplex association both by laser scanning microscopy and flow cytometry (Figs. 4 and 5). After only 30 min of incubation with EGF polyplexes, no (LSM analysis) or low (FACS analysis) levels of surface-bound EGFR were found, suggesting the inability of cells to recover internalized EGFR. The absence of anti-EGFR signal may be partly due to competition between EGF and anti-EGFR antibody for binding to EGFR (K. von Gersdorff and M. Ogris, unpublished observations). Still, when incubating HuH7 cells for up to 4 hr with polyplexes containing fluorescently labeled LPEI-PEG-EGF conjugate, most of the Cy5 signal was found inside the cells, resembling a patchy, endosome-like pattern, while no signal was observed on the cell surface (Fig. 5), hence enabling the anti-EGFR antibody to bind EGFR. In sharp contrast, EGFR levels were totally unaffected by GE11 polyplexes. We conclude that EGF-triggered macropinocytosis further stimulated internalization of EGF polyplexes, either when bound to the polyplex or when pretreating the cells with free EGF (see Supplementary Fig. S2). This can also be seen in Fig. 5, where high amounts of fluorescently labeled LPEI-PEG-EGF conjugate were found in HuH7 cells after only 30 min of incubation, whereas LPEI-PEG-GE11 conjugate was internalized to a lower degree. The inability of EGFR to recycle to the cell surface after treatment with EGF polyplexes also bears a potential disadvantage: in the case of multiple treatment cycles, the disappearance of EGFR will inhibit EGFR-mediated uptake of polyplexes after the first round of transfection.
Having now a well-defined, efficient, and safe transfection system at hand, we applied GE11 polyplexes in a clinically relevant model of orthotopic prostate cancer (Fig. 6). Initial studies with fluorescently labeled polyplexes (plasmid DNA covalently labeled with Cy5) indicated that after intratumoral injection distribution of LPEI-PEG and LPEI-PEG-GE11 polyplexes within the tumor was better when compared with LPEI polyplexes (data not shown). This can be explained by the shielding properties of PEG reducing aggregation with proteinaceous blood components immediately after injection, whereas highly positively charged LPEI polyplexes tend to aggregate immediately and stay close to the injection site. For transgene expression studies, we have chosen a novel, in-house-developed plasmid based on a backbone completely devoid of potentially immune stimulatory sequences and an expression cassette containing an hCMV enhancer element combined with the strong, nonsilenced elongation factor 1α promoter (Navarro et al., 2010). Both bioluminescence imaging and the analysis of tumor lysates clearly showed the advantage of GE11-targeted polyplexes over nontargeted polyplexes, resulting in significantly elevated transgene expression levels.
Summary and outlook
In this paper we have presented the novel, well-defined nucleic acid carrier LPEI-PEG-GE11, where EGFR mediated gene delivery is achieved without activation of EGFR. In contrast to LPEI-PEG-EGF, there is no induction of macropinocytosis otherwise leading to loss of surface EGFR. In combination with a plasmid optimized for in vivo gene delivery, high transgene levels are observed after intratumoral injection into a clinically relevant, orthotopic prostate cancer model. Meanwhile, the LPEI-PEG-GE11 carrier has also been used for systemic delivery of a therapeutic gene in a HuH7 hepatoma model, enabling efficient treatment of hepatoma with combined gene and radiation therapy (Klutz et al., 2011). This, in combination with well-defined and upscalable conjugate synthesis, will allow us to move toward preclinical and clinical evaluation of this carrier.
Supplementary Material
Acknowledgments
This work was supported by the EC Network GIANT, the Nanosystems Initiative Munich (NIM), DFG grant no. OG 63/4-1, and the DFG collaborative research center (SFB824).
Author Disclosure Statement
No competing financial interests exist.
References
- Blessing T. Kursa M. Holzhauser R., et al. Different strategies for formation of pegylated EGF-conjugated PEI/DNA complexes for targeted gene delivery. Bioconjug. Chem. 2001;12:529–537. doi: 10.1021/bc0001488. [DOI] [PubMed] [Google Scholar]
- Boeckle S. Von Gersdorff K. Van Der Piepen S., et al. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J. Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- Brunner S. Sauer T. Carotta S., et al. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- Brunner S. Furtbauer E. Sauer T., et al. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol. Ther. 2002;5:80–86. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- Bryant D.M. Kerr M.C. Hammond L.A., et al. EGF induces macropinocytosis and SNX1-modulated recycling of E-cadherin. J. Cell Sci. 2007;120:1818–1828. doi: 10.1242/jcs.000653. [DOI] [PubMed] [Google Scholar]
- de Bruin K. Ruthardt N. Von Gersdorff K., et al. Cellular dynamics of EGF receptor-targeted synthetic viruses. Mol. Ther. 2007;15:1297–1305. doi: 10.1038/sj.mt.6300176. [DOI] [PubMed] [Google Scholar]
- de Bruin K.G. Fella C. Ogris M., et al. Dynamics of photoinduced endosomal release of polyplexes. J. Control. Release. 2008;130:175–182. doi: 10.1016/j.jconrel.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Diagaradjane P. Orenstein-Cardona J.M. Colon-Casasnovas N.E., et al. Imaging epidermal growth factor receptor expression in vivo: Pharmacokinetic and biodistribution characterization of a bioconjugated quantum dot nanoprobe. Clin. Cancer Res. 2008;14:731–741. doi: 10.1158/1078-0432.CCR-07-1958. [DOI] [PubMed] [Google Scholar]
- Erbacher P. Bettinger T. Belguise-Valladier P., et al. Transfection and physical properties of various saccharide, poly(ethylene glycol), and antibody-derivatized polyethylenimines (PEI) J. Gene Med. 1999;1:210–222. doi: 10.1002/(SICI)1521-2254(199905/06)1:3<210::AID-JGM30>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Khalil M.Y. Grandis J.R. Shin D.M. Targeting epidermal growth factor receptor: novel therapeutics in the management of cancer. Expert Rev. Anticancer Ther. 2003;3:367–380. doi: 10.1586/14737140.3.3.367. [DOI] [PubMed] [Google Scholar]
- Kim H. Muller W.J. The role of the epidermal growth factor receptor family in mammary tumorigenesis and metastasis. Exp. Cell Res. 1999;253:78–87. doi: 10.1006/excr.1999.4706. [DOI] [PubMed] [Google Scholar]
- Kim I.Y. Kang Y.S. Lee D.S., et al. Antitumor activity of EGFR targeted pH-sensitive immunoliposomes encapsulating gemcitabine in A549 xenograft nude mice. J. Control. Release. 2009;140:55–60. doi: 10.1016/j.jconrel.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Klutz K. Schaffert D. Willhauck M.J., et al. Epidermal growth factor receptor-targeted 131I-therapy of liver cancer following systemic delivery of the sodium iodide symporter gene. Mol. Ther. 2011;19:676–685. doi: 10.1038/mt.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoriya A. Hortsch M. Meyers C., et al. Biologically active synthetic fragments of epidermal growth factor: localization of a major receptor-binding region. Proc. Natl. Acad. Sci. U.S.A. 1984;81:1351–1355. doi: 10.1073/pnas.81.5.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaij R. van Weerden W.M. De Ridder C.M., et al. Validation of transrectal ultrasonographic volumetry for orthotopic prostate tumours in mice. Lab. Anim. 2002;36:165–172. doi: 10.1258/0023677021912451. [DOI] [PubMed] [Google Scholar]
- Lemmon M.A. Bu Z. Ladbury J.E., et al. Two EGF molecules contribute additively to stabilization of the EGFR dimer. EMBO J. 1997;16:281–294. doi: 10.1093/emboj/16.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Zhao R. Wu X., et al. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005;19:1978–1985. doi: 10.1096/fj.05-4058com. [DOI] [PubMed] [Google Scholar]
- Marques R.B. van Weerden W.M. Erkens-Schulze S., et al. The human PC346 xenograft and cell line panel: a model system for prostate cancer progression. Eur. Urol. 2006;49:245–257. doi: 10.1016/j.eururo.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Moon W.S. Park H.S. Yu K.H., et al. Expression of betacellulin and epidermal growth factor receptor in hepatocellular carcinoma: Implications for angiogenesis. Hum. Pathol. 2006;37:1324–1332. doi: 10.1016/j.humpath.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Navarro G. Maiwald G. Haase R., et al. Low generation PAMAM dendrimer and CpG free plasmids allow targeted and extended transgene expression in tumors after systemic delivery. J. Control. Release. 2010;146:99–105. doi: 10.1016/j.jconrel.2010.04.030. [DOI] [PubMed] [Google Scholar]
- Ogris M. Steinlein P. Carotta S., et al. DNA/polyethylenimine transfection particles: Influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3:E21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogris M. Walker G. Blessing T., et al. Tumor-targeted gene therapy: Strategies for the preparation of ligand–polyethylene glycol–polyethylenimine/DNA complexes. J. Control. Release. 2003;91:173–181. doi: 10.1016/s0168-3659(03)00230-x. [DOI] [PubMed] [Google Scholar]
- Peng K.W. Pham L. Ye H., et al. Organ distribution of gene expression after intravenous infusion of targeted and untargeted lentiviral vectors. Gene Ther. 2001;8:1456–1463. doi: 10.1038/sj.gt.3301552. [DOI] [PubMed] [Google Scholar]
- Pereboeva L. Komarova S. Roth J., et al. Targeting EGFR with metabolically biotinylated fiber-mosaic adenovirus. Gene Ther. 2007;14:627–637. doi: 10.1038/sj.gt.3302916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert D. Kiss M. Rödl W., et al. Poly(I:C)-mediated tumor growth suppression in EGF-receptor overexpressing tumors using EGF-polyethylene glycol-linear polyethylenimine as carrier. Pharm. Res. 2011;28:731–741. doi: 10.1007/s11095-010-0225-4. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- Shir A. Ogris M. Wagner E. Levitzki A. EGF receptor-targeted synthetic double-stranded RNA eliminates glioblastoma, breast cancer, and adenocarcinoma tumors in mice. PLoS Med. 2006;3:e6. doi: 10.1371/journal.pmed.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungaro F. De Rosa G. Miro A. Quaglia F. Spectrophotometric determination of polyethylenimine in the presence of an oligonucleotide for the characterization of controlled release formulations. J. Pharm. Biomed. Anal. 2003;31:143–149. doi: 10.1016/s0731-7085(02)00571-x. [DOI] [PubMed] [Google Scholar]
- van Weerden W.M. De Ridder C.M. Verdaasdonk C.L., et al. Development of seven new human prostate tumor xenograft models and their histopathological characterization. Am. J. Pathol. 1996;149:1055–1062. [PMC free article] [PubMed] [Google Scholar]
- Walker G.F. Fella C. Pelisek J., et al. Toward synthetic viruses: Endosomal pH-triggered deshielding of targeted polyplexes greatly enhances gene transfer in vitro and in vivo. Mol. Ther. 2005;11:418–425. doi: 10.1016/j.ymthe.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Wells A. EGF receptor. Int. J. Biochem. Cell Biol. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- Wolschek M.F. Thallinger C. Kursa M., et al. Specific systemic nonviral gene delivery to human hepatocellular carcinoma xenografts in SCID mice. Hepatology. 2002;36:1106–1114. doi: 10.1053/jhep.2002.36372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.