Abstract
Selenite is an inorganic form of selenium that has a cytotoxic effect against several human cancer cell lines: one or more selenite metabolites are considered to be responsible for its toxicity. X-ray absorption spectroscopy was used to monitor Se speciation in A549 human lung cancer cells incubated with selenite over 72 h. As anticipated, selenodiglutathione and elemental Se both comprised a large proportion of Se in the cells between 4 and 72 h after treatment, which is in accordance with the reductive metabolism of selenite in the presence of glutathione and glutathione reductase/NADPH system. Selenocystine was also present in the cells, but was only detected as a significant component between 24 h and 48 h concomitant with a decrease in the proportion of selenocysteine and the viability of the cells. The change in speciation from the selenol, selenocysteine, to the diselenide, selenocystine, is indicative of a change in the redox status of the cells to a more oxidizing environment, likely brought about by metabolites of selenite. X-ray fluorescence microscopy of single cells treated with selenite for 24 h revealed a punctate distribution of Se in the cytoplasm. The accumulation of Se was associated with a greater than two-fold increase in Cu, which was colocalized with Se. Selenium K-edge extended X-ray absorption fine structure (EXAFS) spectroscopy revealed Se-Se and Se-S bonding, but not Se-Cu bonding, despite the spatial association of Se and Cu. Microprobe X-ray absorption near-edge structure spectroscopy (μ-XANES) showed that the highly localized Se species was mostly elemental Se.
INTRODUCTION
Selenium is an essential element that is incorporated into selenoproteins as selenocysteine (SeCys). Supranutritional doses of Se have been linked to the prevention of cancers in animal models and in humans largely through an associated toxicity.1 Different forms of Se follow different metabolic pathways, and it is generally accepted that Se metabolites are responsible for the biological activities of dietary Se supplements.2
Selenite, commonly used as an inorganic dietary Se supplement, exhibits toxicity towards a variety of cancer cell lines through multiple mechanisms.3–6 Selenite is readily reduced by glutathione to selenodiglutathione (GSSeSG),7 a substrate for glutathione reductase, that further reduces GSSeSG to selenopersulfide (GSSeH) and elemental Se.8 Hsieh et al. demonstrated that isolated glutathione reductase ultimately reduces GSSeSG to hydrogen selenide (HSe−) in the presence of NADPH.9 Interestingly, the reaction of selenite with glutathione (GSH) produces the superoxide radical anion (•O2−), presumably by the reaction of HSe- and oxygen to give elemental Se.10 HSe− is the purported common metabolite of dietary Se compounds2 and is thought to be the form of Se utilized in selenoprotein synthesis.11
The toxicity and biological activity of several metabolites of selenite have been investigated. For example, GSSeSG is slightly more toxic to cells than selenite and is a substrate of both glutaredoxin and thioredoxin and their respective oxidoreductase systems. The glutaredoxin system appears to contribute to the toxicity of selenite and GSSeSG whereas the thioredoxin system protects against selenium-induced toxicity.6,12 HSe− is toxic to the yeast Saccharomyces cerevisiae, possibly by perturbing intracellular redox due to the consumption of intracellular GSH.13 Elemental Se is generally considered non-toxic due to its insolubility.13 The generation of •O2− from the reaction of oxygen with HSe− has also been implicated in the toxicity of selenite. Shen et al. demonstrated that selenite produced •O2− in human hepatoma cells, leading to oxidative stress and apoptosis.14 Other studies have shown that the production of •O2− in cancer cells causes mitochondrial damage leading to apoptotic4 or autophagic3 cell death.
The reductive metabolism of selenite in the presence of glutathione and the glutathione reductase/NADPH system has been established by in vitro experimentation. The few studies that have been conducted on the speciation of Se compounds in mammalian cells or tissues have commonly employed liquid chromatography-inductively coupled mass spectrometry (LC-ICP-MS) techniques. Thus, Braga et al. identified GSSeSG and other selenotrisulfides in liver homogenates obtained from rats administered selenite.15 Gabel-Jensen et al. found large amounts of unidentified protein-bound Se and small amounts of selenite and other unidentified small molecule metabolites in hepatocytes treated with selenite.16 Studies of selenite metabolism in the organs of rats given oral doses of isotopically labeled Se compounds have focused on the production of selenoproteins, methylated Se compounds and selenosugars.17,18 While LC-ICP-MS is capable of the separation and detection of trace, minor and major Se metabolites in biological samples, the sample preparation required can lead to adventitial oxidation of Se compounds and loss of volatile metabolites.19
By contrast, synchrotron X-ray techniques can be used to determine the speciation of major and minor, but not trace, Se species in biological matrices with minimal sample preparation. X-ray absorption and fluorescence methods are ideal probes for investigating the speciation and distribution of elements heavier than Si. X-ray absorption spectroscopy (XAS) and X-ray fluorescence (XRF) microscopy has previously been used to study the speciation and distribution of Se in human lung cancer cells incubated with selenomethionine and methylselenocysteine,20 Cr in human lung cancer cells21 and As in human hepatoma cells22 and the aggregation of Pt and Gd in human lung cancer cells.23 In the work reported here, we conduct a time-dependent analysis of the speciation of Se in human lung cancer cells incubated with selenite and map the distribution of Se in intact single cells after incubation with selenite for up to 24 h.
EXPERIMENTAL
Materials
Sodium selenite (approx. 98%) was used as purchased from Sigma Aldrich. Solutions of selenite in phosphate-buffered saline (PBS) solution (prepared using MilliQ water) were prepared immediately before use.
Cell Culture
A549 human lung adenocarcinoma epithelial cells, originally purchased from the American Tissue Culture Collection, were a gift from Aviva Levina (University of Sydney). Cells were cultured as monolayers in complete Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with foetal bovine serum (2% v/v), L-glutamine (2 mM), antibiotic-antimycotic mixture (100 mg mL−1 penicillin and 100 U mL−1 streptomycin) and non-essential amino acids (100 U mL−1) at 310 K in a 5% CO2-humidified incubator and were subcultured every 3–7 days.
Cytotoxicity assay
Cell viability was assessed using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay.24 Briefly, cells were seeded at a density of 1 × 105 cells per well in a 96-well plate for 24 hours at 310 K in a 5% CO2-humidified incubator. Solutions of selenite prepared by serial dilution in PBS (10 μL) were added to complete DMEM (100 μL). After treatment, the cells were incubated with MTT solution (0.25 mg mL−1 in serum free DMEM) for 3 h. The MTT solution was then replaced with dimethyl sulfoxide (100 μL) and the formation of formazan was measured at 560 nm using a microplate spectrophotometer (BMG Lab Tech Fluostar Galaxy). Cell viability was reported as percentage absorbance relative to the control as a mean of three independent experiments (with eight replicates per experiment). IC50 values were determined by curve-fitting plots of cell viability against the log of selenite concentration (data not shown).
Sample Preparation
Bulk cell pellets from treated cultures were prepared for X-ray absorption spectroscopy. Cells were grown over 5 days to ~ 90% confluence in 75 cm2 culture flasks in complete DMEM and were treated with selenite (300 μL addition in PBS, making 1 μM or 5 μM final concentration) or PBS (300 μL) for 24 h. Cells were also treated for 1, 4, 48 and 72 h with 5 μM selenite. A cell pellet was isolated after scraping and centrifugation (600 × g). The supernatant was removed and the cells were rinsed by resuspension in PBS (3 × 5 mL) and centrifugation before the pellet was collected and stored at 203 K, then vacuum-dried for 3 h.
Cells used in XRF imaging were grown on 1.5 × 1.5 mm2 × 500 nm silicon nitride windows (Silson, UK) in 6-well plates as described previously.20,25 Briefly, the plates were seeded at 1.8 × 105 cells/well in complete DMEM and were incubated at 310 K in a 5% CO2-humidified incubator for 24 h prior to treatment. Cells were treated with 5 μM selenite for 20 minutes or PBS (1 h) as a vehicle-alone control or 5 μM selenite or PBS for 24 h. At the end of the treatment time the medium was removed and the cells were washed in PBS and fixed by dipping in cold methanol until residual PBS was removed. Samples were stored at 277 K.
X-ray Absorption Spectroscopy and Data Analyses
Se K-edge X-ray absorption spectra of the bulk cell pellets were recorded at the Stanford Synchrotron Radiation Lightsource (SSRL), California, USA on beamline 9-3. The X-ray beam was monochromated by diffraction from a pair of Si(220) crystals. Harmonic rejection was achieved by setting the cut-off energy of a Rh-coated mirror to 15 keV. Cell pellets were compressed to approximately 3 mm in diameter, secured between Kapton tape and cooled to ~ 10 K in a flowing He cryostat. Spectra were recorded in fluorescence mode on a 30-element Ge detector array (Canberra) at 90° to the incident beam. The energy ranges used for X-ray absorption near edge structure (XANES) data collection were: pre-edge region 12425 to 12635 eV (10 eV steps); XANES region 12635 to 12685 eV (0.25 eV steps); and post-edge region 12685 to 12872 eV (0.05 Å−1 steps in k-space). Extended X-ray absorption fine structure (EXAFS) spectra were collected at the following energy ranges: pre-edge region 12435 to 12635 eV (10 eV steps); XANES region 12635 to 12685 eV (0.25 eV steps); and EXAFS region 12685 to 13443 eV (0.05 Å−1 steps in k-space to 14 Å−1). A hexagonal Se foil standard was used to calibrate the energy scale to the first peak of the first derivative of the Se edge (12658 eV).
Data analysis, including calibration, averaging and background subtraction of all spectra and principal component analysis (PCA), target and linear regression analyses of XANES spectra were performed using the EXAFSPAK software package (G. N. George, SSRL). Linear combination fits of XANES spectra were performed over the region 12600 eV to 12750 eV. Spectra of model Se compounds for target and linear regression analyses were provided by G. N. George (University of Saskatchewan) except for that of methylselenocysteine (MeSeCys), which was obtained during these experiments.
X-ray Fluorescence Imaging, μ-XANES Spectra and Data Analyses
XRF elemental distribution maps of single cells were recorded on beamline 2-ID-E at the Advanced Photon Source (APS), Argonne National Laboratory, Illinois, USA and on the X-ray fluorescence microprobe (XFM) beamline26 at the Australian Synchrotron (AS), Victoria, Australia. At the APS, the beam was tuned to an incident energy of 13.0 keV using a beam splitting Si(220) monochromator and was focused to a diameter of 1 μm using a ‘high-flux’ zone plate. A single element silicon drift energy dispersive detector (Vortex EX, SII Nanotechnology, Northridge, CA), at 90° to the incident beam, was used to collect the fluorescence signal for 1 s per spatial point from samples under a He atmosphere.
At the AS, a monochromatic 12.9 keV X-ray beam was focused (to a spot size of about 1 μm × 4 μm) using a zone plate and the fluorescence signal was collected using a single element silicon drift energy dispersive detector (Vortex EM, SII Nanotechnology, Northridge, CA) oriented at 107° to the incident beam, for 3 s per spatial point. Individual cells were located using images obtained from an optical microscope positioned in the beamline downstream from the sample.
The fluorescence spectrum at each spatial point was fit to Gaussians, modified by the addition of a step function and a tailing function to describe mostly incomplete charge collection and other detector artifacts.27 For XRF images collected at the APS, two regions of interest corresponding to the whole cell (identified using optical images and the elemental distribution maps of P, S, Cl, K and Zn) and nuclear regions (identified using optical images and regions of P and Zn colocalization) were selected. The integrated fluorescence spectra extracted from these regions were also fit with modified Gaussians to determine average elemental area densities (in units of μg cm−2). Quantification was performed by comparison to the corresponding measurements on the thin-film standards NBS-1832 and NBS-1833 from the National Bureau of Standards (Gaithersburg, MD, USA). The analysis was performed using MAPS software.28 At the AS, several regions of high Se concentration in the high-resolution XRF maps of Se distribution were selected for microprobe X-ray absorption near edge structure (μ-XANES) analysis. Data were collected over the following energy ranges: pre-edge region, 12500 to 12640 eV (10 eV steps); XANES region, 12640 to 12690 eV (0.5 eV steps); and post-edge region, 12690 to 12900 eV (10 eV steps) with a dwell time of 1 s per point. The sample stage was repositioned in the x, y and z planes while scanning through the energy range in order to keep the position of interest in the focal point of the X-ray beam as its focal length changed. A Se foil was used to calibrate the energy scale to the first peak of the first derivative of the elemental Se edge (12658 eV).
Data analysis, including the calibration, averaging and background subtraction of μ-XANES spectra were performed using EXAFSPAK software. Fitting of multiple model compounds using multiple linear regression analysis was not attempted due to the high levels of noise in the spectra. Elemental area densities are presented as the mean and the standard deviation in the mean. Student’s t-test assuming unequal variance was used to determine significant difference between control and selenite treatments. Differences were considered significant at P < 0.05.
RESULTS
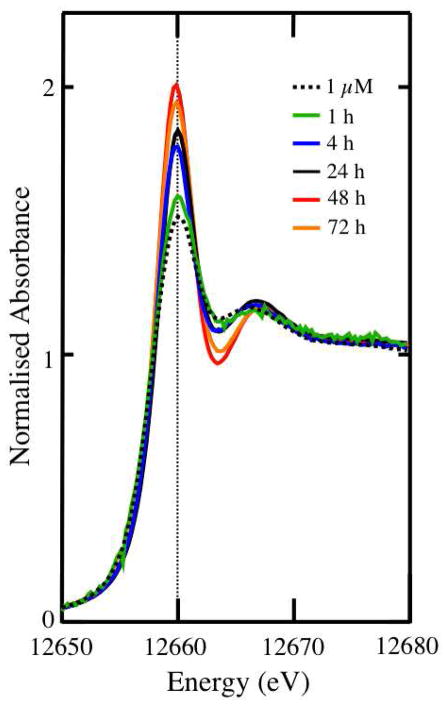
The speciation of Se in A549 cells treated with 5 μM selenite changed gradually between 1 and 24 h after treatment: more substantial changes in speciation were recorded between 24 and 48 h after treatment. Data shown in Figure 1 demonstrates how the Se K-edge XANES spectra of the treated cells changed with time; most significant is the changing intensity and position of the low-energy peak. The ratios of the high-energy to low-energy peak intensities decreased between 1 and 48 h, from 0.74 to 0.59 and increased slightly to 0.60 at 72 h. Similarly the position of the low-energy peak shifted from 12660.1 eV at 1 h to 12659.8 eV at 48 h, with a slight increase to 12659.9 eV at 72 h. The greatest shift in the positions of the peaks occurred between 24 h and 48 h when the low-energy peak shifted 0.2 eV to lower energy, while the high-energy peak shifted 0.5 eV to higher energy (see Figure S1 of the Supporting Information for a difference plot).
Figure 1.
Se K-edge X-ray absorption near-edge spectra of A549 cells treated with selenite. Cells were cultured to yield a monolayer as described in the Methods section. Next, the monolayers were treated with 1 μM selenite for 24 h or 5 μM selenite for 1, 4, 24, 48 or 72 h, then worked up and X-ray absorption near-edge spectral analysis performed as described in detail in the Methods section.
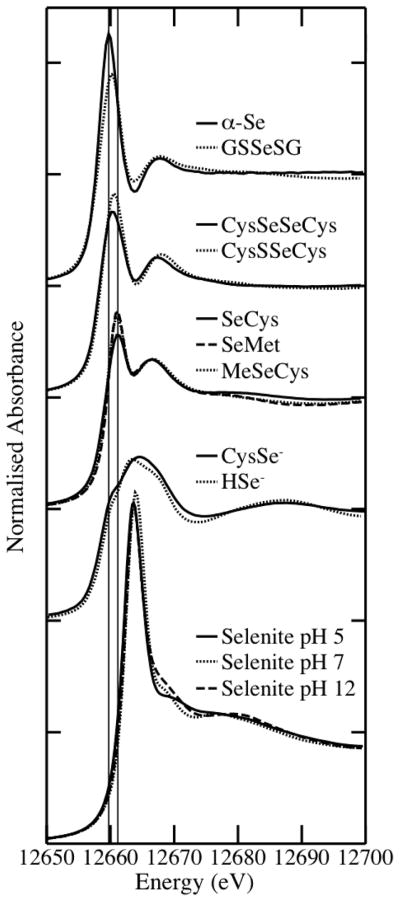
The fitting of Se model compounds to the experimental XANES spectra reflects the significant change in the spectra between 24 and 48 h. The results of the linear combination fitting of model Se compounds to the XANES spectra are summarized in Table 1. The models were selected from the model compound library (Figure 2) following PCA and target analyses. Combinations of SeCys, selenocystine (CysSeSeCys), red elemental Se (α-Se, elemental Se) and GSSeSG were fit to the spectra. The fits to the spectra generated from cells exposed to 1 μM selenite for 24 h and 5 μM selenite for 1 h were very poor (as denoted by the high residual value for the fit, see Table 1) and therefore inconclusive, but it is clear that at 1 h no selenite, which has a strong peak at 12663.6 eV, remained. GSSeSG and α-Se are fit to the remaining 5 μM selenite-treated cells spectra; the percentage of GSSeSG in each component remained constant at about 25%, while the percentage of elemental Se increased from 37% at 4 h to 55% at 48 h, before decreasing to 49% at 72 h. Notably, while about a third of the Se in cells treated for 4 and 24 h was detected in the SeCys form, no SeCys was found in the spectra collected at 48 h: the decrease in the proportion of SeCys between 24 and 48 h was concomitant with the appearance of CysSeSeCys at detectable levels. At 72 h, approximately equal levels of SeCys and CysSeSeCys were present in the cells.
Table 1.
Percent Se species in A549 cells treated with selenite, as estimated by a linear combination of model compound spectra.a
| Treatment (μM) | Time (h) | Cell Viability (%) | Percentage (%) of component fitted
|
Ntot | residual (× 10−3) | |||
|---|---|---|---|---|---|---|---|---|
| SeCys | CysSeSeCys | GSSeSG | α-Se | |||||
| 1 | 24 | 95 ± 3 | - | 98(3)b | - | - | 0.98 | 1.69b |
| 5 | 1 | n. d.c | - | 68(5)b | 33(5)b | - | 1.01 | 2.22b |
| 5 | 4 | 98 ± 5 | 37(1) | - | 26(4) | 37(3) | 1.00 | 0.42 |
| 5 | 24 | 85 ± 6 | 34(2) | - | 23(5) | 44(3) | 1.01 | 0.55 |
| 5 | 48 | 53 ± 8 | - | 19(4) | 26(6) | 55(3) | 1.00 | 0.70 |
| 5 | 72 | 51 ± 1 | 12(3) | 13(6) | 27(4) | 49(3) | 1.01 | 0.31 |
Values in parentheses are the estimated standard deviation derived from the diagonal elements of the covariance matrix and are a measure of precision. Ntot is the sum of the fractions.
High residuals (>1 × 10−3) indicate poor fits that do not give an accurate indication of Se speciation.
Cell viability was not determined at this time point.
Figure 2.
Se K-edge X-ray absorption near-edge spectra of Se model compounds. α-Se, red elemental selenium; GSSeSG, selenodiglutathione; CysSeSeCys, selenocystine; CysSSeCys, sulfoselenocystine; SeCys, selenocysteine; SeMet, selenomethionine; MeSeCys, methylselenocysteine; CysSe−, deprotonated selenocysteine; HSe−, hydrogen selenide. Reprinted with permission from ref 20. Copyright 2011 American Chemical Society.
A comparison between Se speciation at different selenite concentrations was also made. The spectra of the cells treated for 24 h with 1 μM and 5 μM selenite clearly differ in the relative intensities of the high- and low-energy peaks, which decrease from 0.78 in the 1 μM-treated cells to 0.65 in the 5 μM-treated cells. The difference in Se speciation is accompanied by a small difference in cell viability after 24 h, which was 95 ± 3% in 1 μM selenite-treated cells compared to 85 ± 6% in 5 μM selenite-treated cells. After 72 h there was little toxicity associated with 1 μM selenite (93 ± 8% viability), whereas 5 μM selenite showed significant cell death (51 ± 1% viability). XANES fitting to the 1 μM-treated cell spectrum was inconclusive due to the poor fit, which, considering the good signal to noise ratio, suggests that a major component of this fit was missing from the model compound library. Nonetheless, the good fits to the spectra collected from cells treated with 5 μM selenite for between 4 h and 72 h indicates that the model compound library is sufficient for determining the major selenium compounds present in these cells.
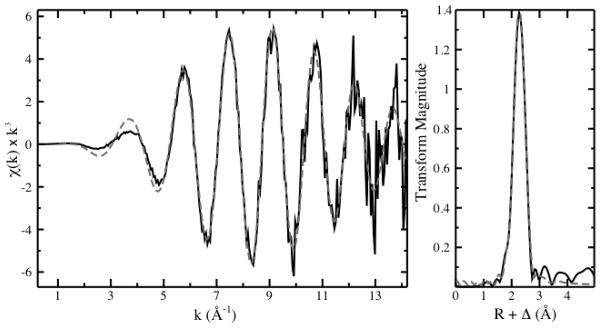
EXAFS spectra were collected for the cells treated with 5 μM selenite for 24 h (Figure 3) and the data was fit with a Se and a S scatterer (Table 2), with bond lengths of 2.370 Å and 2.195 Å, respectively. Fitting a Se or a S scatterer alone gave poor fits, while fitting both a S and a Cu scatterer produced physically unrealistic values (Table S1). The Se-Se bond length is similar to the bond length of 2.35 Å for elemental Se reported by Zhao et al.29 While the Se-Se bond length in CysSeSeCys has not previously been reported, a bond length of 2.33 Å was measured using EXAFS (Figure S2). The similarity between the Se-Se bond lengths in elemental Se and CysSeSeCys precludes the use of bond length in determining the species containing the Se-Se moiety present in the cells. A Se-S bond length of 2.19 Å was measured for GSSeSG (Figure S3), which is in agreement with the Se-S bond length of 2.195 Å measured in the cells. The coordination number for Se-Se (N = 0.7) determined from the EXAFS spectrum is lower than expected given the results of the multiple linear regression fitting of the XANES spectrum (44% α-Se and 23% GSSeSG). We ascribe this to the absence of C scatterers from the fit, which artificially increases the Se-S coordination number. Taking the increase in Se-S coordination number into account as well as the high degree of error in determining coordination numbers from EXAFS spectra (± 20%), the coordination numbers for the Se and S scatterers determined by EXAFS are in agreement with the percentage of α-Se and GSSeSG fit to the corresponding XANES spectrum.
Figure 3.
EXAFS spectrum (left) and corresponding Fourier Transform (right) of A549 cells treated with 5 μM selenite for 24 h (black solid line). The calculated fit to the spectrum is shown (grey dashed line). Fit parameters are listed in Table 2.
Table 2.
Parameters fit to EXAFS spectra of A549 cells treated with 5 μM selenite for 24 h.a
| Treatment (μM) | scatterer | coordination Number (N) | interatomic distance (R, Å) | Debye-Waller factor (σ2, Å2) | −ΔE0 (eV) | fit error |
|---|---|---|---|---|---|---|
| 5 | S | 1.3 | 2.195(5) | 0.0023(3) | 9(1) | 0.30 |
| Se | 0.7 | 2.370(4) | 0.0017(1) |
The k-range was 1 – 14.2 Å−1 and a scale factor (S02) of 0.9 was used for all fits. ΔE0 = E0 − 12658 (eV) where E0 is the threshold energy. Values in parentheses are the estimated standard deviation derived from the diagonal elements of the covariance matrix and are a measure of precision. The fit-error is defined as [Σk6(χexp−χcalc)2/Σk6χexp2]½.
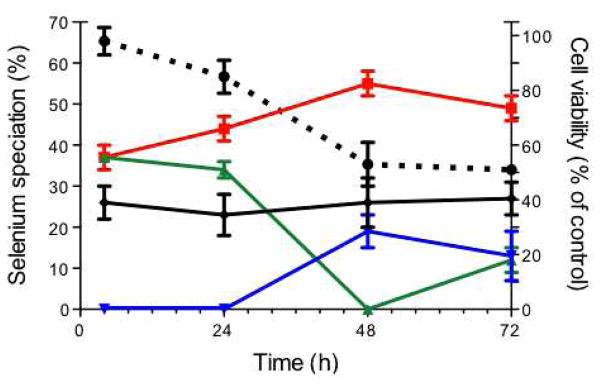
Measurements of the cytotoxicity of 5 μM selenite show that the viability of the cells decreased substantially from 85% to 53% between 24 and 48 h, coinciding with the significant visual changes observed in the X-ray absorption spectra collected, as well as the parallel increase in levels of elemental Se and the switch between SeCys and CysSeSeCys components (Figure 4). Smaller decreases in cytotoxicity between 4 and 24, and 48 and 72 h were accompanied by smaller changes in the speciation of Se between those times.
Figure 4.
Cell viability and speciation of Se in cells treated with 5 μM selenite for 4 h to 72 h. Percentage cell viability (dotted black line) is shown as the mean ± standard deviation of three independent experiments, each with 8 replicates. Selenium speciation is shown as a percentage of the component fitted to the Se K-edge XANES spectrum ± estimated standard deviation as recorded in Table 1. The components are elemental Se (red), GSSeSG (solid black), SeCys (green) and CysSeSeCys (blue).
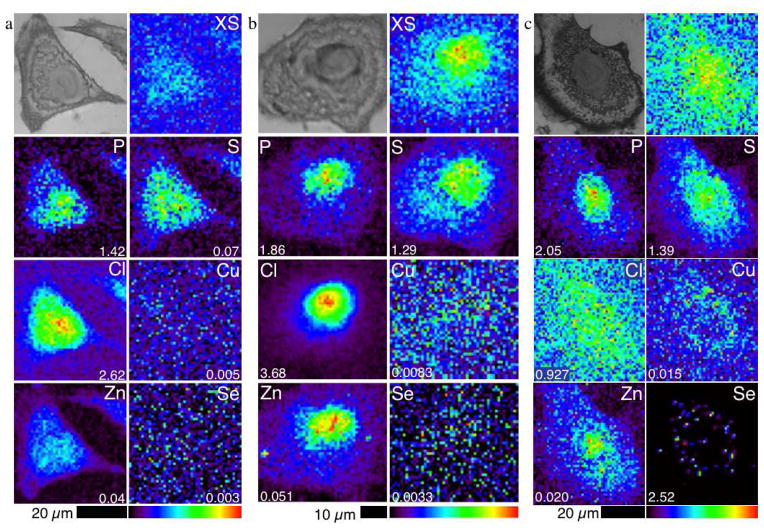
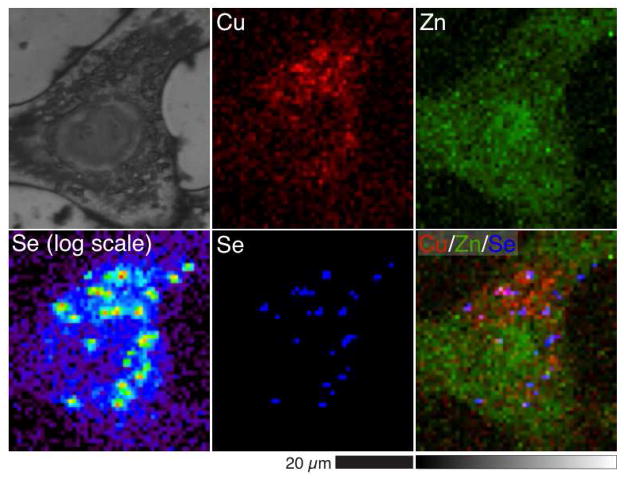
In addition to the collection of X-ray absorption spectra to determine the speciation of Se in cells, the distribution of Se was studied by XRF imaging. A treatment time of 20 minutes was chosen to observe the distribution of Se in the cells soon after treatment and the treatment time of 24 h was chosen to observe the distribution of Se in the cells at high Se concentrations while a high proportion of cells remained viable as compared to the longer time point where a 50% decrease in viability was noted. Cells treated with 5 μM selenite for 20 min had a mean Se content only slightly higher than the Se content of the vehicle-alone control A549 cells (Supplementary Information, Table S2). This Se content was too low to distinguish intracellular Se from background levels in the XRF image (Figures 5a and 5b). In contrast, the mean Se content of cells treated with 5 μM selenite for 24 h was 250-fold that of the control cells (Figure S4, Table S6). Figure 5c shows the Se distribution in the 5 μM selenite-treated cells, where Se is highly concentrated in areas less than 5 μm2 in the cytoplasm. This punctate distribution coincides with regions of high Cu content (Figure 6). The Cu content of the cells was 2.5-fold higher than in the control cells, while Zn content was reduced, although not significantly (Table S6).
Figure 5.
Optical micrographs (top left) and scattered X-ray (XS) and XRF elemental distribution maps of P, S, Cl, Cu, Zn, and Se of an A549 cell treated with (a) PBS as a vehicle control for 1 h, (b) 5 μM selenite for 20 min and (c) 5 μM selenite for 24 h. The maximal elemental area density (in micrograms per square centimeter) is given in the bottom corner of each map.
Figure 6.
Optical micrograph (top left) and scattered X-ray (XS) and XRF elemental distribution maps of Cu, Zn, and Se (log scale and linear scale) of an A549 cell treated with 5 μM selenite for 24 h. Cu (red), Zn (green), and Se (blue) maps are overlaid to show the colocalization of the elements (Cu/Zn/Se).
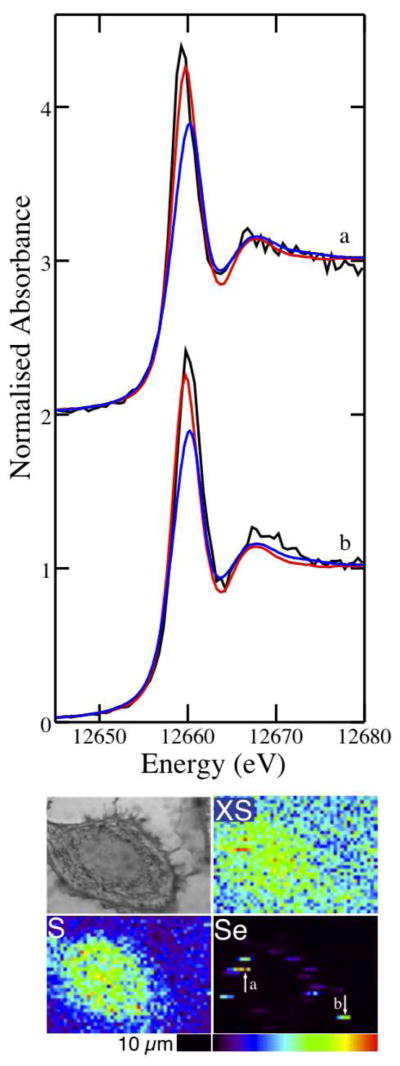
Microprobe XANES were collected at Se hotspots in cells treated with 5 μM selenite for 24 h and spectra are shown in Figure 7 and Figures S5 and S6. Despite some noise in the spectra, which precluded multiple linear regression analysis, direct comparison of the μ-XANES spectra to the spectra of model compounds suggests that the majority of Se in the hotspots is in the form of elemental Se. The mean peak position of the five μ-XANES spectra collected from Se hotspots was 12659.9 eV, compared to the peak positions of elemental Se at 12659.8 eV and GSSeSG at 12660.2 eV (Figure S5). However, the standard deviation in the mean of the peak positions was 0.7 eV due to the high noise levels of the spectra. The ratios of the high-energy to low-energy peak intensities were consistently low, between 0.49 and 0.56, with some of the variability in intensity and peak position arising from noise. In comparison, the ratio of the peak intensities in elemental Se and GSSeSG are 0.50 and 0.62, respectively, while the ratio of the peak intensities in the spectrum of bulk cells treated with selenite for 24 h was 0.65. While some of the large difference in the ratio of the peak intensities of the spectra from bulk cells and Se hotspots in cells treated with selenite for 24 h may be due to their collection at different beamlines, the 15–30% increase in the ratio of the peak intensities suggests that the speciation of Se in the hotspots differs substantially from the speciation in the remainder of the cell. The peak intensity ratios of the μ-XANES spectra are lower than in spectra of the bulk cell pellets, which contain 44% elemental Se, and are the same as the peak intensity ratio of elemental Se. On the basis of peak position and peak intensity ratios it is likely that elemental Se constitutes the majority of the Se species in the hotspots observed in the XRF images.
Figure 7.
Se K-edge μ-XANES spectra of Se hotspots in an A549 cell treated with 5 μm selenite. The experimental spectra (a and b, black) are overlaid with the spectra of elemental Se (red) and GSSeSG (blue). The optical micrograph (top left) and scattered X-ray (XS) and elemental distribution maps of S and Se of the cell are shown with arrows indicating the locations from which spectra (a) and (b) were collected.
DISCUSSION
Selenite was rapidly metabolized by cultured A549 cells such that within 1 h of treatment no selenite was detected suggesting a rapid metabolism of the supplement. Within 4 h of treatment with 5 μM selenite, large proportions of GSSeSG and elemental Se were present in the cells with GSSeSG consistently accounting for ~ 25% of the species present in Se-supplemented cells between 4 to 72 h. There was an increase in the proportion of elemental Se between 4 and 48 h, with a slight decrease between 48 and 72 h. The presence of GSSeSG is consistent with the reductive metabolism of selenite by GSH and glutathione reductase and has previously been1515 identified in liver tissue from rats injected with Se(IV) solution.15 Elemental Se has been identified as a major metabolite of trout hepatocytes treated with 100 μM selenite30 and is likely the insoluble Se species that accounted for 28% of Se in selenite-treated rat hepatocytes studied by Gabel-Jensen et al.16 Elemental Se may be produced either by the oxidation of HSe− by oxygen13 or by the decomposition of GSSeH.8 HSe− was present in the library of model Se compounds used to fit the Se XANES spectra collected from the cell pellets, but was not identified as a component of any of the spectra. Therefore, if HSe− is present in the cell it is present below the level of detection. This result is not surprising considering its labile nature: HSe− is readily oxidized to elemental Se by disulfides and other biological oxidants.31
In addition to the inorganic species, the organic Se species, SeCys and CysSeSeCys were present in the cells. Free SeCys, with a pKa of 5.24,32 would be expected to exist in its deprotonated form at physiological pH, but CysSe− was not identified as a component of the spectra. The microenvironment within proteins can differ from the intracellular environment and so the identification of SeCys in the cells may reflect the presence of selenoproteins rather than free SeCys.
The differences between the XANES spectra of 1 μM and 5 μM selenite-treated cells at 24 h indicate the speciation of Se in the cells treated at the two concentrations are substantially different. Unfortunately, the compounds in the current model compound library do not produce a good fit to the XANES spectrum of 1 μM selenite-treated cells, but the smaller peak intensity of the 1 μM-treated cells compared to the 5 μM-treated cells indicates that lower proportions of elemental Se and GSSeSG are present. The much lower toxicity of 1 μM selenite compared to 5 μM selenite suggests that the lower concentration of selenite is not sufficient to generate enough of the toxic Se metabolite(s) to significantly impact on cell viability.
Generation of •O2− is considered a primary mechanism by which selenite exerts its cytotoxicity. While •O2− was not directly measured in these experiments, the production of elemental Se after 4 h of treatment with selenite is consistent with •O2− being generated at that time point. Elevated levels of ROS formation have been observed in human hepatoma cells within 10 minutes of treatment with 10 μM selenite.14 The change in speciation of Se in the cells from a large SeCys component at 24 h to a substantial CysSeSeCys component at 48 h after treatment is indicative of a changing intracellular redox environment. Assuming that •O2− generation is initiated in the cells as early as 4 h after treatment with 5 μM selenite, there appears to be a substantial delay in the widespread oxidation of SeCys to CysSeSeCys. Some of this apparent delay may be accounted for by levels of CysSeSeCys being too low for detection at early time points. The formation of diselenide bonds in selenoproteins would lead to a loss of function that may explain the documented loss of cell viability. It was only as high levels of diselenide species were formed between 24 and 48 h after treatment that the viability of the cells suffered a substantial reduction.
The punctate distribution of elemental Se in the cytoplasm, observed in all cells 24 h after treatment with 5 μM selenite, may be a cellular defense mechanism. There is very little information available on the toxicity or biological activity of elemental Se and it is assumed to be non-toxic due to its insolubility. The toxicity of elemental Se to yeast cells has been shown to be negligible,13 while elemental Se at nano size is known to be biologically active, but it also has a low toxicity.33 As there is no evidence that elemental Se is toxic, the compartmentalization of elemental Se may be in response to the generation of •O2− at the site of elemental Se formation from the oxidation of HSe−.
The association of Cu and Se observed in cells treated with 5 μM selenite for 24 h may be related to attempts to protect the cell from damage caused by •O2− generation. Shen et al. demonstrated that co-treatment of normal human keratinocytes with selenite and CuSO4 for 24 h protected the cells against selenite-induced toxicity,34 although this protective effect may be as a result of extracellular interactions of Cu and Se.35 Cu2+ has been shown to inhibit the generation of •O2− from the reaction of selenite and GSH in vitro, presumably by the complexation of Cu2+ with selenide anions, preventing selenide oxidation to elemental Se.36 Despite the spatial association of Cu and Se in the cells, there is no evidence of Se-Cu bonding in the Se K-edge EXAFS, although it should be noted that the Se EXAFS can neither prove nor disprove the presence of a minor component of Se-Cu bonding due to the 13-fold higher levels of Se in the cells compared to Cu. Another explanation for the colocalization of Cu and Se may be the upregulation of Cu,Zn-superoxide dismutase (Cu,Zn-SOD) in response to increased oxidative stress. Human glioma cells overexpressing Cu,Zn-SOD and manganese-superoxide dismutase (Mn-SOD) significantly reduced selenite-induced cell death,3 however in human prostate cells cell death was inhibited by Mn-SOD overexpression, but not Cu,Zn-SOD overexpression.4 While it is plausible that Cu,Zn-SOD may be upregulated to counteract enhanced production of •O2− by the metabolism of selenite there is no direct evidence for this from these experiments and further experiments are warranted to elucidate this potential pathway.
An interaction of Se with proteins may also offer an explanation for the colocalization of Se and Cu in the absence of Se-Cu bonds. Metallothioneins, which usually bind Zn, are also known to bind other transition metals, including Cu, and have a well-established role in heavy metal detoxification.37 The metallothionein redox cycle is coupled to the GSH/GSSG redox cycle via catalytic selenols, which maintain thionein in the reduced form that binds Zn.38 Selenium compounds with a range of oxidation states have also been shown to cause the oxidation of metallothionein, the release of Zn and the formation of Se-S bonds under physiological conditions.39 The oxidation of metallothioneins by Se compounds may be enhanced under oxidizing conditions.38 The ability of Cu to displace Zn from metallothioneins under high Cu load40 and the important role Se plays in the redox cycle of metallothioneins suggests that this protein may also be involved in the transport of Se and Cu. While μ-XANES analysis showed that Se in the hotspots was in the form of elemental Se, and therefore not bound to metallothionein, its colocalization with Cu in small regions of the cell suggests a detoxification mechanism, which may involve transport of Se and Cu bound to metallothioneins.
In summary, the XAS time-course study of Se speciation has shown that selenite is metabolized to GSSeSG and elemental Se within 4 h of treatment. The formation of diselenide species between 24 and 48 h after exposure of the cells to selenite is evidence of intracellular oxidizing conditions that is in agreement with the generation of •O2− as a cytotoxic mechanism of selenite. For the first time, the distribution of Se in single cells treated with selenite has been mapped using XRF microscopy, revealing the accumulation of elemental Se in highly localized regions of the cytoplasm and an association between Se and Cu. Further investigation is required to determine the exact nature of the Se localization and Se and Cu association, although it is likely a response to the toxicity of Se. Insights into the speciation and distribution of Se in human cancer cells gained from this work show that XAS and XRF microscopy is a powerful combination in the study of the metabolism of Se compounds in biological systems.
Supplementary Material
Acknowledgments
A549 cells were a gift from Aviva Levina (The University of Sydney). Graham N. George (University of Saskatchewan) provided Se K-edge X-ray absorption spectra of model Se compounds. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Part of this research was undertaken at the X-ray Fluorescence Microprobe beamline at the Australian Synchrotron, Victoria, Australia. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209). We acknowledge travel funding provided by the International Synchrotron Access Program (ISAP) managed by the Australian Synchrotron and funded by the Australian Government, and research funding from the Australian Research Council (DP0985807).
ABBREVIATIONS
- SeCys
selenocysteine
- GSSeSG
selenodiglutathione
- GSSeH
selenopersulfide
- HSe−
hydrogen selenide
- •O2−
superoxide radical anion
- GSH
glutathione
- LC-ICP-MS
liquid chromatography-inductively coupled mass spectrometry
- XAS
X-ray absorption spectroscopy
- XRF
X-ray fluorescence spectroscopy
- PBS
phosphate-buffered saline
- DMEM
Dulbecco’s Modified Eagle’s Medium
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- XANES
X-ray absorption near edge structure
- EXAFS
extended X-ray absorption fine structure
- PCA
principal component analysis
- SSRL
Stanford Synchrotron Radiation Lightsource
- MeSeCys
Se-methylselenocysteine
- APS
Advanced Photon Source
- AS
Australian Synchrotron
- μ-XANES
microprobe X-ray absorption near edge structure
- CysSeSeCys
selenocystine
- α-Se
red elemental selenium
- Cu,Zn-SOD
Cu,Zn-superoxide dismutase
- Mn-SOD
Mn-superoxide dismutase
Footnotes
SUPPORTING INFORMATION AVAILABLE
Complete ref 25, alternative EXAFS fit parameters (Table S1), XANES spectra of cells treated with 5 μM selenite for 24 h and 48 h (Figure S1), EXAFS of model compounds (Figures S2 and S3), representative XRF image of a control cell at 24 h (Figure S4), μ-XANES spectra of Se hotspots in cells (Figure S5 and S6), and elemental area densities of cells imaged using XRF microscopy (Tables S2–S9). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ganther HE. Carcinogenesis. 1999;20:1657–1666. doi: 10.1093/carcin/20.9.1657. [DOI] [PubMed] [Google Scholar]
- 2.Combs G, Gray W. Pharmacol Ther. 1998;79:179–192. doi: 10.1016/s0163-7258(98)00014-x. [DOI] [PubMed] [Google Scholar]
- 3.Kim E, Sohn S, Kwon H, Kim S, Kim M. Cancer Res. 2007 [Google Scholar]
- 4.Xiang N, Zhao R, Zhong W. Cancer Chemother Pharmacol. 2009;63:351–362. doi: 10.1007/s00280-008-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olm E, Fernandes AP, Hebert C, Rundlöf AK, Larsen EH, Danielsson O, Björnstedt M. Proc Natl Acad Sci U S A. 2009;106:11400–11405. doi: 10.1073/pnas.0902204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallenberg M, Olm E, Hebert C, Bjornstedt M, Fernandes AP. Biochem J. 2010;429:85–93. doi: 10.1042/BJ20100368. [DOI] [PubMed] [Google Scholar]
- 7.Painter EP. Chem Rev. 1941;28:179–213. [Google Scholar]
- 8.Ganther HE. Biochemistry. 1971;10:4089–4098. doi: 10.1021/bi00798a013. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh HS, Ganther HE. Biochemistry. 1975;14:1632–1636. doi: 10.1021/bi00679a014. [DOI] [PubMed] [Google Scholar]
- 10.Yan L, Spallholz JE. Biochem Pharmacol. 1993;45:429–437. [PubMed] [Google Scholar]
- 11.Gromer S, Eubel JK, Lee BL, Jacob J. Cell Mol Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Björnstedt M, Kumar S, Holmgren A. J Biol Chem. 1992;267:8030–8034. [PubMed] [Google Scholar]
- 13.Tarze A, Dauplais M, Grigoras I, Lazard M, Ha-Duong NT, Barbier F, Blanquet S, Plateau P. J Biol Chem. 2007;282:8759–8767. doi: 10.1074/jbc.M610078200. [DOI] [PubMed] [Google Scholar]
- 14.Shen H, Yang C, Ong C. Int J Cancer. 1999;81:820–828. doi: 10.1002/(sici)1097-0215(19990531)81:5<820::aid-ijc25>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Braga P, Montes-Bayón M, Alvarez J, López JM, Sanz-Medel A. J Anal At Spectrom. 2004;19:1128. [Google Scholar]
- 16.Gabel-Jensen C, Gammelgaard B. J Anal At Spectrom. 2010;25:414–418. [Google Scholar]
- 17.Suzuki KT, Ohta Y, Suzuki N. Toxicol Appl Pharmacol. 2006;217:51–62. doi: 10.1016/j.taap.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki KT, Doi C, Suzuki N. Toxicol Appl Pharmacol. 2006;217:185–195. doi: 10.1016/j.taap.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 19.B’Hymer C, Caruso JA. J Chromatogr A. 2006;1114:1–20. doi: 10.1016/j.chroma.2006.02.063. [DOI] [PubMed] [Google Scholar]
- 20.Weekley CMM, Aitken JB, Vogt S, Finney LA, Paterson DJ, de Jonge MD, Howard DL, Musgrave IF, Harris HH. Biochemistry. 2011;50:1641–1650. doi: 10.1021/bi101678a. [DOI] [PubMed] [Google Scholar]
- 21.Harris HH, Levina A, Dillon CT, Mulyani I, Lai B, Cai Z, Lay PA. J Biol Inorg Chem. 2005;10:105–118. doi: 10.1007/s00775-004-0617-1. [DOI] [PubMed] [Google Scholar]
- 22.Munro KL, Mariana A, Klavins AI, Foster AJ, Lai B, Vogt S, Cai Z, Harris HH, Dillon CT. Chem Res Toxicol. 2008;21:1760–1769. doi: 10.1021/tx800128d. [DOI] [PubMed] [Google Scholar]
- 23.Crossley EL, Aitken JB, Vogt S, Harris HH, Rendina LM. Angew Chem, Int Edit. 2010;49:1231–1233. doi: 10.1002/anie.200902309. [DOI] [PubMed] [Google Scholar]
- 24.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 25.Carter EA, et al. Mol BioSyst. 2010;6:1316–1322. doi: 10.1039/c001499k. [DOI] [PubMed] [Google Scholar]
- 26.Paterson D, Boldeman J, Cohen D, Ryan C. AIP Conf Proc. 2007;879:864–867. [Google Scholar]
- 27.Espen P. In: Handbook of X-ray Spectrometry: Second Edition, Revised and Expanded. Grieken R, Markowicz A, editors. Marcel Dekker, Inc; New York: 2002. [Google Scholar]
- 28.Vogt S. J Phys IV. 2003;104:635–638. [Google Scholar]
- 29.Zhao Y, Lu K. Phys Rev B. 1999;59:11117–11120. [Google Scholar]
- 30.Misra S, Peak D, Niyogi S. Metallomics. 2010;2:710–717. doi: 10.1039/c0mt00008f. [DOI] [PubMed] [Google Scholar]
- 31.Nuttall KL, Allen FS. Inorg Chim Acta. 1984;92:33–36. [Google Scholar]
- 32.Huber RE, Criddle RS. Arch Biochem Biophys. 1967;122:164–173. doi: 10.1016/0003-9861(67)90136-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Gao X, Zhang L. BioFactors. 2001;15:27–38. doi: 10.1002/biof.5520150103. [DOI] [PubMed] [Google Scholar]
- 34.Shen CL, Song W, Pence BC. Cancer Epidemiol Biomark Prev. 2001;10:385–390. [PubMed] [Google Scholar]
- 35.Davis R, Spallholz J, Pence B. Nutr Cancer. 1998;32:181–189. doi: 10.1080/01635589809514738. [DOI] [PubMed] [Google Scholar]
- 36.Davis R, Spallholz JE. Biochem Pharmacol. 1996;51:1015–1020. doi: 10.1016/0006-2952(95)02435-2. [DOI] [PubMed] [Google Scholar]
- 37.Kang YJ. Exp Biol Med. 2006;231:1459–1467. doi: 10.1177/153537020623100903. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Maret W. Eur J Biochem. 2001;268:3346–3353. doi: 10.1046/j.1432-1327.2001.02250.x. [DOI] [PubMed] [Google Scholar]
- 39.Jacob C, Maret W, Vallee BL. Proc Natl Acad Sci U S A. 1999;96:1910–1914. doi: 10.1073/pnas.96.5.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winge D. Methods Enzymol. 1991;205:458–469. doi: 10.1016/0076-6879(91)05131-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.