Abstract
Proteases are enzymes that cleave peptide bonds in protein substrates. This process can be important for regulated turnover of a target protein but it can also produce protein fragments that then perform other functions. Because the last few decades of protease research have confirmed that proteolysis is an essential regulatory process in both normal physiology and in multiple disease-associated conditions, there has been an increasing interest in developing methods to image protease activity. Proteases are also considered to be one of the few druggable classes of proteins and therefore a large number of small molecule based inhibitors of proteases have been reported. These compounds serve as a starting point for the design of probes that can be used to target active proteases for imaging applications. Currently, several classes of fluorescent probes have been developed to visualize protease activity in live cells and even whole organisms. The two primary classes of protease probes make use of either peptide/protein substrates or covalent inhibitors that produce a fluorescent signal when bound to an active protease target. This review outlines some of the most recent advances in the design of imaging probes for proteases. In particular, it highlights the strengths and weaknesses of both substrate- and activity- based probes and their applications for imaging cysteine proteases that are important biomarkers for multiple human diseases.
Introduction
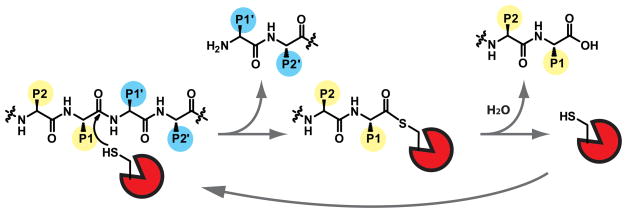
The protease family contains approximately 560 members, comprising nearly 2% of the human genome. The primary function of this diverse family of enzymes is to cleave specific peptide bonds of substrates. While this activity is important for normal cellular processes, it is also a critical regulatory mechanism for many pathologies including cancer, arthritis, atherosclerosis, and neurodegenerative disorders such as Alzheimer’s and Huntington’s Disease, among others. Proteases are classified into five sub-families, according to their mechanism of catalysis. Cysteine, serine, and threonine proteases use a nucleophilic amino acid side chain to catalyze the hydrolysis of the peptide substrate (Figure 1). Metallo and aspartic proteases, on the other hand, use active site residues to deprotonate a water molecule for substrate attack.
Figure 1.
Because unchecked proteolysis would be highly detrimental to the cell, proteases are subject to tight regulatory mechanisms. They are synthesized as inactive zymogens that can be activated by a number of mechanisms. Once activated, proteases are often negatively regulated by endogenous protein-based inhibitors. Therefore, to obtain a clear understanding of both the normal and pathological function of proteases, direct assessment of the regulation of their enzymatic activities is required. Traditional tools, such as antibodies or proteomic methods survey total protein levels and therefore do not provide information on the dynamic regulation of protease activity. For this reason, new biochemical tools to study protease activity have been in high demand. This review will primarily discuss two major classes of probes, substrate- and activity-based probes, and how these reagents have been applied to study the biological function of cysteine proteases biochemically and using optical imaging methods. We aim to provide a critical interpretation of the pros and cons of each type of probe and to provide insight regarding the future of this technology.
Substrate-based probes
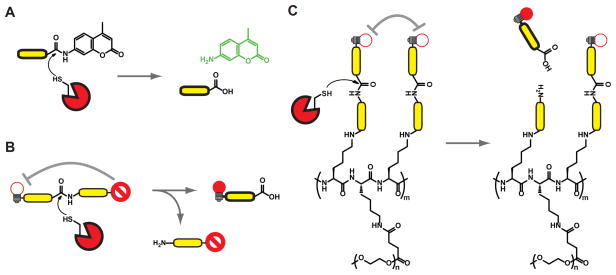
Although proteases were originally thought to completely degrade proteins in order to maintain homeostasis of protein levels in the cell, it is now clear that they perform limited proteolysis of substrates at defined cleavage sites. This allows proteases to regulate structure, function, and localization of substrates. Although the ability to cleave a specific site on a protein substrate can be controlled by a number of factors including tertiary structure and localization of target and protease, in many cases, substrate cleavage is controlled by the primary amino acid sequences surrounding the scissile amide bond. Therefore, it is possible to generate fluorescent substrate probes based on optimal peptide sequences, whose spectral properties change when cleaved by an active protease. The simplest and perhaps most widely used fluorogenic substrate probes consist of a peptide sequence attached at the c-terminus to a fluorophore, such as an aminomethyl coumarin (AMC) [1, 2](Figure 2A). In the presence of the active protease, the AMC is cleaved from the peptide, leading to a detectable shift in its fluorescent spectrum. Alternatively, it is also possible to make peptide substrates containing a fluorophore and quencher at opposite ends of the substrate (Figure 2B). These substrates can then be cleaved to liberate fluorescent fragments.
Figure 2.
Another important class of substrate-based probes for proteases uses two or more fluorophores, that are self-quenched when in close proximity [3–6]. Multiple fluorophores can be linked to graft polymers containing peptide substrate sequences (Figure 2C). When these linkers are cleaved by the protease, free fluorescent monomers are released. This class of probes has been used to study the activity of multiple classes of proteases in a number of disease models, and will be discussed in greater detail below.
While natural substrate sequences often serve as a starting point for probe development, these sites may also be engineered to enhance potency and sensitivity towards the protease target of choice. A common way to identify optimal substrate sequences is to generate positional scanning libraries, in which pools of fluorogenic peptide substrates are generated with one position held constant as a fixed amino acid, while the other positions are mixtures of all possible natural amino acids [7–10]. Each pool contains a different fixed amino acid, and the pools that yield the highest fluorescence in the presence of the enzyme indicate the optimal residues for that site. Subsequently, if multiple positions are scanned, the results can be combined to produce substrate recognition sequences for various proteases. However, the use of positional scanning methods has its drawbacks since it fails to provide information on the cooperativity of multiple substrate positions for binding in the active site of the protease target. Therefore, some have chosen to focus on libraries of individual protease substrates [11, 12]. Another more recent strategy for generating selective substrates uses a “reverse design” approach in which protease inhibitors that have been optimized by medicinal chemistry serve as a starting scaffold for design of substrate-based probes [13, 14].
Activity-based probes
As an alternative to substrate-based probes, it is also possible to monitor protease activity using activity-based probes (ABPs). ABPs are compounds that have been engineered to covalently modify enzyme targets in an activity dependent manner. ABPs typically contain a reactive functional group (often referred to as a warhead) linked to a targeting sequence and a tag for visualization or affinity purification (Figure 3A). ABPs have been developed for a number of enzyme families, including proteases, kinases, phosphatases, glycosidases, etc [15].
Figure 3.
Activity-based probes differ from substrate-based probes primarily in their mechanism of action. Rather than acting as a substrate that is processed by the protease, they act as direct covalent inhibitors of the protease. Like substrate-based probes, ABPs also contain primary recognition motifs that drive selectivity towards the protease of interest and away from others. ABPs can be tagged using a variety of modalities, including fluorescence, biotinylation, and radiolabels [16]. Because ABPs covalently modify the target enzyme, it is possible to directly identify and quantify the amount of labeled protein using standard biochemical methods. Fluorescent or radioactive tags can be visualized by scanning of the resulting SDS-PAGE gel. Biotin probes can be detected by western blotting and also allow direct affinity purification of target proteases [17].
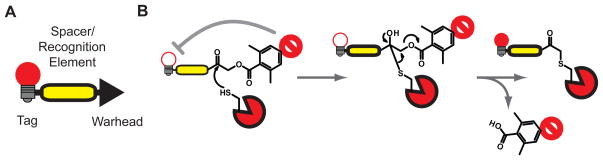
Some types of electrophiles also allow the use of fluorophore/quencher pairs. The most prominent example is the acyloxymethyl ketone (AOMK) group [18, 19], which releases a carboxylate leaving group upon modification of a protease target. If a quencher is linked to this leaving group, the result is a ‘smart probe’ that only produces a fluorescent signal when covalently bound to the protease target (Figure 3B). Like the substrate-based probes, quenched ABPs are ideal for live cell imaging applications as they allow monitoring of protease activation in real time. Furthermore, unlike the substrate probes, ABPs remain bound to the target protease allowing dynamic studies of enzyme activation and localization.
Applications of substrate- and activity-based probes
Cathepsins
Papain-like cysteine proteases, or cysteine cathepsins, were once thought to degrade proteins nonspecifically in the lysosome. However, their roles in normal cellular processes and disease pathologies have become increasingly apparent. Cysteine cathepsins are implicated in cancer progression, due to their roles in angiogenesis, apoptosis, and tumor cell invasion [20]. They are also key regulators of inflammation in diseases such as atherosclerosis, rheumatoid arthritis, and asthma. A number of probes have been developed for studying the functional roles of cysteine cathepsins. In addition to being valuable biochemical reagents, many of these probes have also become useful as contrast agents for imaging disease processes.
To this end, Blum et al. developed a series of AOMK-based ABPs that target cathepsin B, L, and S [18]. Both quenched and non-quenched versions are cell permeable and can be used to biochemically profile active cathepsins in live cells and lysates by fluorescent SDS-PAGE. Furthermore, near infrared versions of the AOMK probes can be used for whole-body non-invasive imaging. GB123 and GB137 (non-quenched and quenched, respectively) both accumulated specifically in xenografted tumors of nude mice upon systemic intravenous delivery [19]. The quenched probe showed specific signal in tumors in as little as 30 minutes, whereas the non-quenched version required longer times to generate contrast, due to the need to clear the unbound probe. Importantly, fluorescent signal in the tumors could be substantially reduced by pretreatment with a potent cathepsin inhibitor indicating the specific nature of the accumulation. Since its initial publication, GB123 has been applied in a number of additional studies. Most recently it was used to show the localization of Cathepsin B activity to caveolae of endothelial cells during tube formation in vitro, to investigate the role of cathepsins in VEGF-induced angiogenesis, and to image the effects of expression of neutrophilic granule protein (NGP) in tumors [21–23].
In addition to ABPs, there has been significant interest in the use of quenched substrate probes for non-invasive imaging applications. Polymer-based probes have been developed by VisEn Medical (now Perkin Elmer) and are commercially available. In particular, ProSense-680, a cathepsin targeted probe, has been used to assess the contribution of Cathepsin B activity to many inflammatory processes, including asthma [24], focal inflammation, angiogenesis, and growth of intestinal polyps [3], immune cell function following rejection of transplanted mouse hearts [25], and following myocardial infarction [5]. Substrate based probes have also been used to image cathepsin activities and assess their contributions in other heart-related conditions including early aortic valve disease [26] and atherosclerosis [27].
A variation of the Prosense cathepsin B probe containing a substrate sequence with selectivity towards Cathepsin K, a matrix-degrading elastase, was used to study atherosclerosis [28]. Using intravital fluorescent microscopy, Cathepsin K activity was identified in strong focal regions of atherotic lesions. Fluorescence was strongly enhanced in the macrophage population and colocalized with immunoreactive Cathepsin K. In this instance, the population of Cathepsin K was broader than the probe fluorescence, suggesting that only a fraction of the total protein was proteolytically active. The same substrate-based probe was also used to image Cathepsin K activity in osteoclasts in models of accelerated bone loss [4].
A new class of substrate probes was recently developed using a “reverse design” method in which potent inhibitors are converted back into cleavable reporter substrates [13, 14]. Because these inhibitors were optimized using extensive medicinal chemistry efforts, they are highly selective and have optimized pharmacokinetic properties making them likely to be better probes than standard peptide substrates. One substrate probe developed by this approach, AW-091, was used in vivo to image cathepsin S activity in a mouse model of paw inflammation. Comparison with ProSense680 in the same models indicated that AW-091 gave maximal signal-to-noise ratio after 3 hours, while ProSense680 required 24 hours, reflecting enhanced pharmacodynamic properties.
Caspases
Caspases are cysteine proteases that mediate a programmed form of cell death called apoptosis [29]. Apoptosis is critical for normal development and tissue homeostasis, as well as for a number of diseases including cancer. In addition, a sub-family of caspases plays a major role in regulating a pro-inflammatory form of cell death called pyroptosis [30]. Caspases have a unique reactivity compared to other cysteine proteases, in that they only cleave substrates containing aspartic acid residues in the P1 position. Therefore, all peptide-based probes for caspases have made use of this selectivity requirement.
As with cathepsins, one of the major challenges in developing probes for caspases is selectivity for unique proteases within the family (i.e. caspase-3 over caspase-7); however, an even bigger problem is the tendency for caspase probes to target other enzymes, such as cathepsins or legumain [31, 32]. Legumain, is a lysosomal cysteine protease that has roles in antigen processing [33], matrix degradation, and tumorigenesis[34]. Legumain has a preference for substrates containing asparagine in the P1 position, however, it is also capable of binding to activity-based AOMK probes containing a P1 aspartic acid (Asp) [35]. This is most likely due to the fact that the Asp side chain is protonated in the acidic environment of the lysosome, allowing it to fit into the S1 binding pocket of the active site.
In an attempt to develop selective probes for caspases, Berger et al. used a positional scanning library approach [36]. Optimal sequences were identified and converted to biotinylated probes, which were evaluated in kinetic studies using cell free extracts and intact cells. In subsequent studies, the most selective caspase ABPs were tagged with NIRF fluorophores for use in non-invasive imaging applications[32]. Interestingly, the most potent in vitro probe exhibited substantial cross-reactivity with capthepsin B and legumain. To avoid cathepsin reactivity, a proline was substituted at the P2 position. This new probe, AB50 (Cy5-EPD-AOMK), showed significantly improved in vivo selectivity properties and was applied in vivo in two mouse models of apoptosis. These studies show that, in addition to being valuable for non-invasive imaging applications, AB50 can also be used to assess apoptosis by microscopy, flow cytometry, and ex vivo fluorescence imaging. Importantly, since the probes are covalent labels, caspase modification can be confirmed biochemically using SDS-PAGE. These studies also confirmed that AB50 suffers from cross-reactivity with legumain. Efforts were made to reduce legumain reactivity, but increased selectivity came at the cost of reduced caspase potency.
Addition of the transporter peptide Tat to AB50 (tAB50) significantly enhanced the fluorescent signal in apoptotic cells. Unfortunately, the Tat labeled probes increased labeling of both legumain and Cathepsin B, due to uptake of the Tat-labeled probes by endocytosis. Hence, transporter peptides such as Tat may not be ideal for delivery of probes to cytosolic protease targets, especially when primary off-targets are lysosomal enzymes.
One of the most widely used classes of activity-based probes for caspases contains a fluoromethyl ketone (FMK) electrophile. Carboxyfluorescein and sulphorhodamine-labeled versions are commercially available and marketed under such names as FLICA (fluorochrome-labeled inhibitor of caspases) and CaspaTag. FLICA has been used to assess the kinetics of cell death in response to several stimuli by flow cytometry [37] and microscopy [38]. FAM-YVAD-FMK is also a common reagent to visualize caspase-1 activity during inflammasome-dependent cell death [39]. New versions called FLIVO are currently being marketed for use in in vivo studies [40, 41].
CaspaTags for caspase 3/7 (SR-DEVD-FMK) and caspase 9 (FAM-LEHD-FMK) were used in a head-to-head comparison with cleaved caspase antibodies for immunofluorescence microscopy of gentamicin-treated chick cochlea [42]. The overall trend and timing of labeling with both antibodies and the CaspaTag probes were similar; however, at later time points the CaspaTag showed more caspase-3 positive cells than the antibodies. The authors concluded that antibodies showed the activated caspases present at a given time point, whereas the CaspaTag could track cells that had already completed cell death in addition to those currently dying, giving a more complete assessment of apoptotic cells. Alternatively, the enhanced signal from CaspaTag may be due to cross-reactivity with other proteases. Specifically, FMK- and CMK- based inhibitors have been shown to block the activity of cathepsins and legumain [31]. The increase in fluorescence at late stages of apoptosis in chick cochlea may reflect the involvement of lysosomal proteases in cell death [43]. This cross-reactivity could also explain the results of studies that show that FLICA signal in flow cytometry could not be decreased by pre-treating cells with concentrations of untagged Z-DEVD-FMK or Z-VAD-FMK sufficient to block caspase activity [44, 45]. Interestingly, there have been no reported biochemical data regarding the selectivity of the FLICA reagents when used in cells. Inhibitor versions marketed as selective for specific caspases (i.e. z-DEVD-FMK for caspase-3/7, z-LEHD-FMK for caspase-9, z-LETD-FMK, etc.) were demonstrated to have broad reactivity in competition assays [9]. Given these findings, one should interpret all data obtained using activity-based probes with extreme caution. Data analysis should always be paired with careful biochemical analyses, such that observations of enzyme activity and function can be assigned to the correct enzyme.
Likewise, the same practice should be applied to the use of substrate-based probes, which also suffer from a lack of specificity that is even more difficult to track. The most widely used substrate probes are the fluorogenic substrates, which typically contain a tetrapeptide caspase cleavage site and aminomethylcoumarin (AMC) or aminofluorocoumarin (AFC). These reagents are commercially available and, like FMK probes, are usually marketed as specific for one caspase. The specificity regions have been optimized based on reported data from positional scanning studies [1]; however it is important to keep in mind that just because an enzyme prefers one substrate over others does not mean it cannot cleave the others, especially at higher substrate concentrations.
In 2005, Bullok and Piwnica-Worms first described the synthesis of a novel substrate based probe for imaging caspase activity during apoptosis [46]. This probe, TcapQ647, contains the caspase substrate DEVD, flanked by a fluorophore/quencher pair, Alexa Fluor 647/QSY 21. Similar to the tAB50 activity-based probe [32], TcapQ647 contains the tat peptide sequence. Initial studies with this probe verified that it was 92 – 99% quenched, and that it could be unquenched in the presence of active caspases-7 and -9. Apoptotic cells could be detected by flow cytometry and fluorescence microscopy. Later, the kinetic properties of this probe towards other caspases were reported, and a non-cleavable control probe was shown to be inactive in apoptotic cells [47]. These probes were also used in vivo in a model of parasite-induced apoptosis in human colon xenografts. The degree of fluorescence in tumors was shown to correlate with the rate of apoptosis, as assessed by TUNEL assay.
A second generation probe called KcapQ was later introduced, in which the tat peptide was replaced by the Lys-Arg-rich sequence KKKRKV [48]. Efficacy of KcapQ was assessed in a mouse model of retinal ganglion cell apoptosis induced by N-methyl-D-aspartate (NMDA), a clinically relevant model of glaucoma. NMDA-treated eyecups showed an increase in fluorescence when compared with PBS-treated controls and localization corresponded with TUNEL-positive cells. TcapQ was also tested in this model [49]. Pre-treatment with Z-DEVD-FMK reduced the number of probe-positive cells by approximately 60%.
Pros and cons of substrate- and activity-based probes
One of the potential drawbacks of the quenched activity-based probes is that, due to their covalent nature, there is a one-to-one reaction between probe and enzyme, preventing signal amplification. Because the substrate-based probes do not render the protease inactive, one molecule of enzyme is theoretically able to cleave many probe molecules, potentially leading to enhanced fluorescent signal. In order to assess the potential contribution of signal amplification, Blum et al. performed a comparative assessment of the ProSense substrate-based probes and the AOMK ABPs by non-invasive imaging of tumors [50]. The data indicate that ProSense probes are less bright and slower to produce signal than the corresponding ABPs. These results suggest that signal amplification may not be a major factor in overall signal strength and probe sensitivity. Another explanation is that the large polymer probes have slower penetration into tissues followed by more rapid diffusion of the cleaved fluorophores after proteolysis. As a result, VisEn Medical has introduced a new class of “fast” probes that are based on smaller substrate scaffolds.
One of the other major drawbacks of substrate-based probes is the assessment of their selectivity. Peptide substrates can be cleaved by any number of proteases including members of multiple families (i.e. metallo, aspartyl, cysteine, etc.). Furthermore, since the probe does not stay bound to the target, it is difficult or impossible to identify the protease responsible for generation of signal. Activity-based probes, on the other hand, make use of reactive functional groups that have a defined reactivity within a general protease class. For example electrophiles that target cysteine proteases are different from electrophiles that would be used for serine proteases. Thus, by controlling the reactive group, it is possible to generate a significant degree of class selectivity before any efforts are made to identify selective binding sequences.
On the other hand, a potential disadvantage of ABPs is the fact that they inhibit the target protease, thus potentially altering the biology of the system. To address this issue, Blum et al. removed tumors that had been non-invasively imaged with an ABP and labeled extracts ex vivo using a radiolabeled general cathepsin probe 125I-JPM-OEt. Cathepsin B activity in tumors from probe treated mice was not significantly reduced compared to vehicle-treated control tumors, suggesting that ABPs only inhibit a small fraction of active enzyme in vivo, therefore they are not likely to alter the biology being studied.
Conclusions
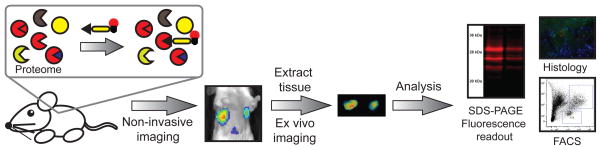
Cysteine proteases have been targeted by a significant number of probes, resulting in a wealth of tools that can be used to monitor their activity. Here we have described key advances in the development of fluorescent probes for imaging cysteine proteases. We have highlighted some of the key differences between substrate- and activity-based probes, providing insight into the strengths and weaknesses of each. Both classes of probes have utility across a wide range of applications, allowing analysis of protease activity at organismal, tissue, single-cell and biochemical levels (Figure 4).
Figure 4.
Many factors go into the making of the ideal protease probe. Not only must a probe detect activity, it must do so selectively in order to distinguish between family members with similar substrate specificity. Probes must be reactive, but not so reactive that they bind every cysteine they encounter. Probes must yield a detectable and sustained signal, so they must also be stable in vivo. One of the major hurdles in developing useful probes is the need for cell permeability. Probes must freely enter the cell, but they also need to be directed to the proper location such that they come into contact with the desired proteases. As discussed for the caspases, this can be especially difficult for cytoplasmic proteases. The future of protease probes will be focused on improving selectivity and improving cellular delivery and pharmacodynamic properties for in vivo studies.
HIGHLIGHTS.
Explanation of the need for tools to image protease activity
Examples of recently developed substrate-based imaging probes for cysteine proteases
Examples of recently developed activity-based imaging probes for cysteine proteases
Discussion of the pros and cons of substrate and activity based probes
Acknowledgments
The authors thank Edgar Deu for helpful discussions of the manuscript. This work was funded by National Institutes of Health grants R01 AI-078947 and R01 EB005011 (both to M.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thornberry NA, Chapman KT, Nicholson DW. Determination of caspase specificities using a peptide combinatorial library. Methods Enzymol. 2000;322:100–10. doi: 10.1016/s0076-6879(00)22011-9. [DOI] [PubMed] [Google Scholar]
- 2.Los M, et al. Fluorogenic substrates as detectors of caspase activity during natural killer cell-induced apoptosis. Methods Mol Biol. 2000;121:155–62. doi: 10.1385/1-59259-044-6:155. [DOI] [PubMed] [Google Scholar]
- 3**.Gounaris E, et al. Live imaging of cysteine-cathepsin activity reveals dynamics of focal inflammation, angiogenesis, and polyp growth. PLoS One. 2008;3(8):e2916. doi: 10.1371/journal.pone.0002916. ProSense680 was used to image cathepsin activity by intravital microscopy of intestinal lesions in mouse model of hereditary polyposis. Results indicate a link between protease activity, inflammation, and polyp growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Kozloff KM, et al. Non-invasive optical detection of cathepsin K-mediated fluorescence reveals osteoclast activity in vitro and in vivo. Bone. 2009;44(2):190–8. doi: 10.1016/j.bone.2008.10.036. A polymer-based substrate probe was used to image Cathepsin K activity in osteoclast cultures and in mouse models of accelerated bone loss. Cathepsin K up-regulation could be imaged prior to bone loss detected by micro-CT, indicating its potential as a biomarker for early disease detection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nahrendorf M, et al. Dual channel optical tomographic imaging of leukocyte recruitment and protease activity in the healing myocardial infarct. Circ Res. 2007;100(8):1218–25. doi: 10.1161/01.RES.0000265064.46075.31. [DOI] [PubMed] [Google Scholar]
- 6.Pham W, et al. Developing a peptide-based near-infrared molecular probe for protease sensing. Bioconjug Chem. 2004;15(6):1403–7. doi: 10.1021/bc049924s. [DOI] [PubMed] [Google Scholar]
- 7.Choe Y, et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281(18):12824–32. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- 8.Maly DJ, Huang L, Ellman JA. Combinatorial strategies for targeting protein families: application to the proteases. Chembiochem. 2002;3(1):16–37. doi: 10.1002/1439-7633(20020104)3:1<16::AID-CBIC16>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 9.Berger AB, Sexton KB, Bogyo M. Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res. 2006;16(12):961–3. doi: 10.1038/sj.cr.7310112. [DOI] [PubMed] [Google Scholar]
- 10.Cuerrier D, et al. Development of calpain-specific inactivators by screening of positional scanning epoxide libraries. J Biol Chem. 2007;282(13):9600–11. doi: 10.1074/jbc.M610372200. [DOI] [PubMed] [Google Scholar]
- 11.Patterson AW, Wood WJ, Ellman JA. Substrate activity screening (SAS): a general procedure for the preparation and screening of a fragment-based non-peptidic protease substrate library for inhibitor discovery. Nat Protoc. 2007;2(2):424–33. doi: 10.1038/nprot.2007.28. [DOI] [PubMed] [Google Scholar]
- 12.Wood WJ, et al. Substrate activity screening: a fragment-based method for the rapid identification of nonpeptidic protease inhibitors. J Am Chem Soc. 2005;127(44):15521–7. doi: 10.1021/ja0547230. [DOI] [PubMed] [Google Scholar]
- 13.Watzke A, et al. Selective activity-based probes for cysteine cathepsins. Angew Chem Int Ed Engl. 2008;47(2):406–9. doi: 10.1002/anie.200702811. [DOI] [PubMed] [Google Scholar]
- 14*.Caglic D, et al. Functional in vivo imaging of cysteine cathepsin activity in murine model of inflammation. Bioorg Med Chem. 2011;19(3):1055–61. doi: 10.1016/j.bmc.2010.10.028. The authors used ‘reverse design’ to convert a potent cathepsin inhibitor to a quenched substrate probe, which was used to non-invasively image cathepsin up-regulation in a mouse model of inflammation. [DOI] [PubMed] [Google Scholar]
- 15.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 16.Sadaghiani AM, Verhelst SH, Bogyo M. Tagging and detection strategies for activity-based proteomics. Curr Opin Chem Biol. 2007;11(1):20–8. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 17.Greenbaum D, et al. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol. 2000;7(8):569–81. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 18*.Blum G, et al. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol. 2005;1(4):203–9. doi: 10.1038/nchembio728. This work describes the first example of a quenched activity-based probe for the cysteine cathepsins. [DOI] [PubMed] [Google Scholar]
- 19**.Blum G, et al. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol. 2007;3(10):668–77. doi: 10.1038/nchembio.2007.26. Quenched activity based probes for cathepsins were applied to non-invasive imaging of mouse tumors. [DOI] [PubMed] [Google Scholar]
- 20.Gocheva V, Joyce JA. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6(1):60–4. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- 21.Cavallo-Medved D, et al. Live-cell imaging demonstrates extracellular matrix degradation in association with active cathepsin B in caveolae of endothelial cells during tube formation. Exp Cell Res. 2009;315(7):1234–46. doi: 10.1016/j.yexcr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boutte AM, et al. Identification of a myeloid-derived suppressor cell cystatin-like protein that inhibits metastasis. Faseb J. doi: 10.1096/fj.10-180604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang SH, et al. VEGF-A induces angiogenesis by perturbing the cathepsin-cysteine protease inhibitor balance in venules, causing basement membrane degradation and mother vessel formation. Cancer Res. 2009;69(10):4537–44. doi: 10.1158/0008-5472.CAN-08-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortez-Retamozo V, et al. Real-time assessment of inflammation and treatment response in a mouse model of allergic airway inflammation. J Clin Invest. 2008;118(12):4058–66. doi: 10.1172/JCI36335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christen T, et al. Molecular imaging of innate immune cell function in transplant rejection. Circulation. 2009;119(14):1925–32. doi: 10.1161/CIRCULATIONAHA.108.796888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aikawa E, et al. Multimodality molecular imaging identifies proteolytic and osteogenic activities in early aortic valve disease. Circulation. 2007;115(3):377–86. doi: 10.1161/CIRCULATIONAHA.106.654913. [DOI] [PubMed] [Google Scholar]
- 27.Nahrendorf M, et al. Hybrid in vivo FMT-CT imaging of protease activity in atherosclerosis with customized nanosensors. Arterioscler Thromb Vasc Biol. 2009;29 (10):1444–51. doi: 10.1161/ATVBAHA.109.193086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaffer FA, et al. Optical visualization of cathepsin K activity in atherosclerosis with a novel, protease-activatable fluorescence sensor. Circulation. 2007;115(17):2292–8. doi: 10.1161/CIRCULATIONAHA.106.660340. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14(1):32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 30.Schroder K, Tschopp J. The inflammasomes. Cell. 140(6):821–32. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 31*.Rozman-Pungercar J, et al. Inhibition of papain-like cysteine proteases and legumain by caspase-specific inhibitors: when reaction mechanism is more important than specificity. Cell Death Differ. 2003;10(8):881–8. doi: 10.1038/sj.cdd.4401247. This work demonstrates that fluoromethyl ketones designed to target caspases are also reactive against lysosomal cysteine proteases and encourages cautious data analysis. [DOI] [PubMed] [Google Scholar]
- 32*.Edgington LE, et al. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15(8):967–73. doi: 10.1038/nm.1938. This work describes the first application of fluorescent activity-based probes to non-invasive imaging of caspase activity in mouse models of apoptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maehr R, et al. Asparagine endopeptidase is not essential for class II MHC antigen presentation but is required for processing of cathepsin L in mice. J Immunol. 2005;174(11):7066–74. doi: 10.4049/jimmunol.174.11.7066. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, et al. Overexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapy. Cancer Res. 2003;63(11):2957–64. [PubMed] [Google Scholar]
- 35.Kato D, et al. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol. 2005;1(1):33–8. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 36.Berger AB, et al. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23(4):509–21. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Wolbers F, et al. Apoptotic cell death kinetics in vitro depend on the cell types and the inducers used. Apoptosis. 2004;9(3):385–92. doi: 10.1023/b:appt.0000025816.16399.7a. [DOI] [PubMed] [Google Scholar]
- 38.Bedner E, et al. Activation of caspases measured in situ by binding of fluorochrome-labeled inhibitors of caspases (FLICA): correlation with DNA fragmentation. Exp Cell Res. 2000;259(1):308–13. doi: 10.1006/excr.2000.4955. [DOI] [PubMed] [Google Scholar]
- 39.Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci U S A. 2008;105(11):4312–7. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cursio R, et al. Liver apoptosis following normothermic ischemia-reperfusion: in vivo evaluation of caspase activity by FLIVO assay in rats. Transplant Proc. 2008;40(6):2038–41. doi: 10.1016/j.transproceed.2008.05.039. [DOI] [PubMed] [Google Scholar]
- 41.Griffin RJ, et al. Use of a fluorescently labeled poly-caspase inhibitor for in vivo detection of apoptosis related to vascular-targeting agent arsenic trioxide for cancer therapy. Technol Cancer Res Treat. 2007;6(6):651–4. doi: 10.1177/153303460700600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser CL, et al. Comparison of activated caspase detection methods in the gentamicin-treated chick cochlea. Hear Res. 2008;240(1–2):1–11. doi: 10.1016/j.heares.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoka V, Turk V, Turk B. Lysosomal cysteine cathepsins: signaling pathways in apoptosis. Biol Chem. 2007;388(6):555–60. doi: 10.1515/BC.2007.064. [DOI] [PubMed] [Google Scholar]
- 44.Darzynkiewicz Z, Pozarowski P. All that glitters is not gold: all that FLICA binds is not caspase. A caution in data interpretation--and new opportunities. Cytometry A. 2007;71(8):536–7. doi: 10.1002/cyto.a.20425. [DOI] [PubMed] [Google Scholar]
- 45.Pozarowski P, et al. Interactions of fluorochrome-labeled caspase inhibitors with apoptotic cells: a caution in data interpretation. Cytometry A. 2003;55(1):50–60. doi: 10.1002/cyto.a.10074. [DOI] [PubMed] [Google Scholar]
- 46.Bullok K, Piwnica-Worms D. Synthesis and characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. J Med Chem. 2005;48(17):5404–7. doi: 10.1021/jm050008p. [DOI] [PubMed] [Google Scholar]
- 47**.Bullok KE, et al. Biochemical and in vivo characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. Biochemistry. 2007;46(13):4055–65. doi: 10.1021/bi061959n. This study applies a newly developed class of substrate probes for caspases to non-invasive imaging of apoptosis. [DOI] [PubMed] [Google Scholar]
- 48.Maxwell D, et al. An improved cell-penetrating, caspase-activatable, near-infrared fluorescent peptide for apoptosis imaging. Bioconjug Chem. 2009;20(4):702–9. doi: 10.1021/bc800516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnett EM, et al. Single-cell imaging of retinal ganglion cell apoptosis with a cell-penetrating, activatable peptide probe in an in vivo glaucoma model. Proc Natl Acad Sci U S A. 2009;106(23):9391–6. doi: 10.1073/pnas.0812884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Blum G, et al. Comparative assessment of substrates and activity based probes as tools for non-invasive optical imaging of cysteine protease activity. PLoS One. 2009;4(7):e6374. doi: 10.1371/journal.pone.0006374. A head-to-head comparison of activity- and substrate- based probes in a mouse tumor model, that highlights the pros and cons of each probe type. [DOI] [PMC free article] [PubMed] [Google Scholar]