Abstract
Quantum dots are bright, photostable fluorophores used extensively to investigate biological processes. Here, we report that bromocresol green at low micromolar concentrations rapidly, efficiently and reversibly quenches the fluorescence of commercial quantum dots having a wide range of functionalities. The broad utility of bromocresol green quenching of quantum dots in cell biology is demonstrated in quantitative assays of trafficking of the β2-adrenergic receptor and the cystic fibrosis transmembrane conductance regulator.
Keywords: Quantum dots, protein trafficking, CFTR, β2AR, fluorescence quenching, single molecule imaging
Introduction
Quantum dots (QDs) are used extensively for single molecule studies in live cells because of their high photostability and brightness (1, 2). Their small size (diameter ~20 nm) compared with intracellular vesicles such as endosomes and synaptic vesicles (~40–500 nm) would allow their application, in principle, to quantify cellular trafficking events such as endocytosis and exocytosis. However, analysis of cellular trafficking requires rapid and unambiguous resolution of internalized vs. surface-localized QDs. The few studies to date that have applied QDs to study trafficking have been based on irreversible, partial quenching of streptavidin- (SA-) conjugated QDs with fluorophore-labelled biotin analogs (3), mild pH-sensitivity of non-targeted QDs (~15 % decrease in fluorescence intensity from pH 7 to 5.5 (4)), or the restricted application of QD-labelled nerve growth factor to distal axons of neurons grown in compartmentalized Campenot chambers (5). In general, these methods are not widely applicable.
We reasoned that QD fluorescence quenching by an efficient, membrane-impermeant small-molecule quencher would readily resolve external vs. internal QDs. Although irreversible quenching of QDs has been described (3, 6, 7), efficient reversible QD quenching, as would be needed to follow cycles of protein internalization and exocytosis, has not been reported. In this study we report the identification and characterization of a reversible QD quencher that effectively reduces fluorescence emissions of commercial QDs with different functional groups. Quenching of QD fluorescence was used to measure rates of protein trafficking and to visualize single exocytic events.
Results and Discussion
Resonance energy transfer is a useful mechanism for rapid and reversible fluorescence quenching. Resonance energy transfer is most efficient when the acceptor, or quencher, has strong absorbance at the donor emission wavelength. Based on this requirement, an initial screen was done to identify polar, membrane-impermeant compounds with high absorbance that rapidly, efficiently and reversibly quench QD fluorescence, without having background fluorescence in the highly protein-rich biological milieu. We discovered that the inexpensive and common dye bromocresol green (BCG) showed remarkably and unexpectedly efficient QD fluorescence quenching over a wide wavelength range, and had all the requisite properties for use in live cells.
BCG has a broad absorbance spectrum with maximum at 617 nm (Figure 1A). Fluorescence quenching measurements showed that 3 µM BCG reduced fluorescence by ~50 % of SA-conjugated and F(ab’)2-conjugated QDs emitting at 605 nm, non-targeted and SA-conjugated QDs emitting at 565 nm, and SA-conjugated QDs emitting at 585 nm and 655 nm. BCG also efficiently quenched the fluorescence of SA-conjugated QDs emitting at 525 nm and 705 nm (~25 % at 3 µM BCG). In all cases, complete quenching was found at higher BCG concentrations.
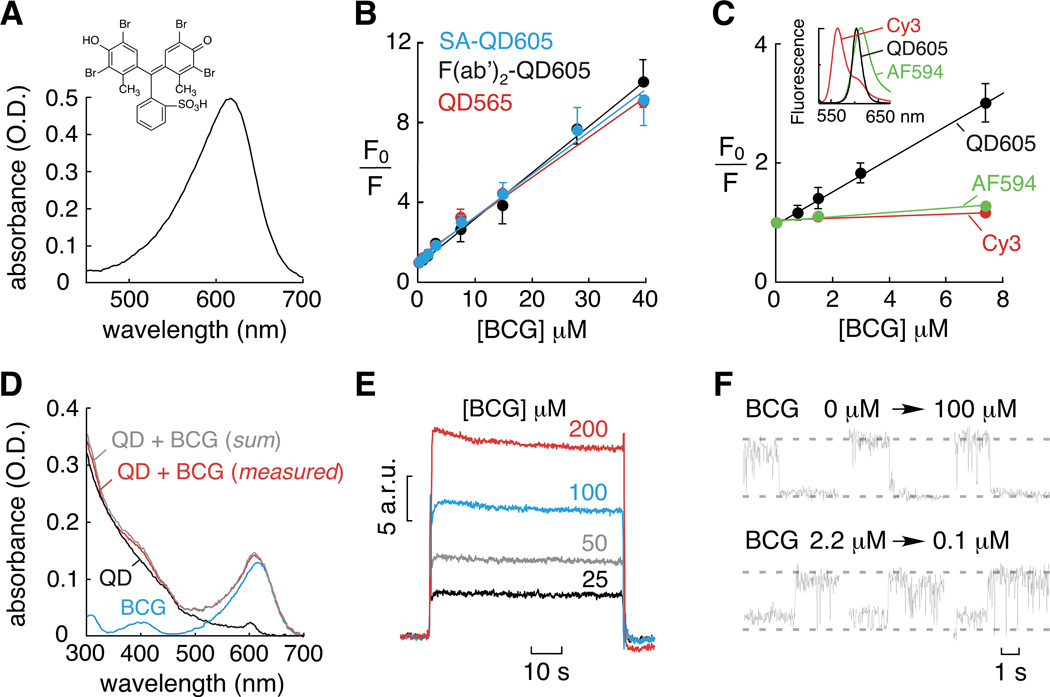
Figure 1. QD quenching by BCG.
A) BCG absorbance spectrum and chemical structure (inset). B) Stern-Volmer plot for BCG quenching of streptavidin- (SA-) and F(ab’)2-conjugated 605 nm QDs and unconjugated 565 nm QDs. C) Stern-Volmer plot for BCG quenching of 605 nm QDs (black), Cy3 (red) and Alexa Fluor 594 (AF594, green). (inset) Fluorescence emission spectra for Cy3, 605 nm QDs and AF594. D) QD absorbance is not altered by BCG. E) Surface plasmon resonance sensorgrams demonstrating direct binding of BCG to QDs. F) Fluorescence imaging of BCG quenching (top) and dequenching (bottom) of QD fluorescence.
Analysis of BCG quenching of QD fluorescence indicated near linear Stern-Volmer plots (Figure 1B) with quenching constants (Ksv) of ~ 2 × 105 M−1. For collisional quenching, where Ksv is the product of the bimolecular rate constant kq and fluorescence lifetime τ0, we computed kq ~1013 M−1s−1. This value is orders of magnitude greater than predicted for a diffusion-limited reaction, indicating that classical collisional quenching cannot account for the exceptionally efficient QD quenching by BCG. BCG quenching of Cy3 and Alexa Fluor 594 (organic fluorophores with emission wavelengths close to QDs studied (Figure 1C, inset)) was remarkably less efficient than that of QDs, supporting a different BCG quenching mechanism (Figure 1C). Collisional quenching was also ruled out by the insensitivity of Ksv to solution viscosity (data not shown). In addition, the absorbance spectra of QDs and BCG were additive, indicating that ground state effects were not responsible for QD quenching (Figure 1D).
We postulated that a binding interaction between QDs and BCG (static quenching) was responsible for the high quenching efficiency. Measurement of BCG binding to surface-immobilized QDs by surface plasmon resonance confirmed direct binding (Figure 1E). Very rapid BCG–QD association and dissociation were found by surface plasmon resonance, suggesting fast and reversible quenching. Single-molecule QD fluorescence imaging (verified by QD blinking) confirmed rapid and reversible quenching, in which the fluorescence of surface-immobilized QDs changed within one image frame (30 ms) following superfusion with BCG-containing and BCG-free solutions (Figure 1F). Therefore, the efficient quenching of QD fluorescence by BCG involves a static quenching mechanism.
The rapid, reversible quenching of QD fluorescence by BCG and its low membrane permeability (as experimentally validated below) were exploited to measure endocytosis and exocytosis in live cells. The general strategy to investigate trafficking of QD-labelled proteins such as receptors and transporters is shown schematically in Figure 2A. QD-labelled proteins are visualized at the cell surface and in vesicles in the absence of quencher, but only in vesicles in the presence of quencher. We studied the cellular trafficking of β2AR and CFTR engineered with external hemagglutinin- (HA-) epitope-tags (HA-β2AR and CFTR-3HA) to allow selective, antibody-mediated QD-labelling. These proteins have distinct trafficking patterns: β2AR undergoes agonist-dependent internalization, whereas CFTR undergoes constitutive endocytosis and recycling.
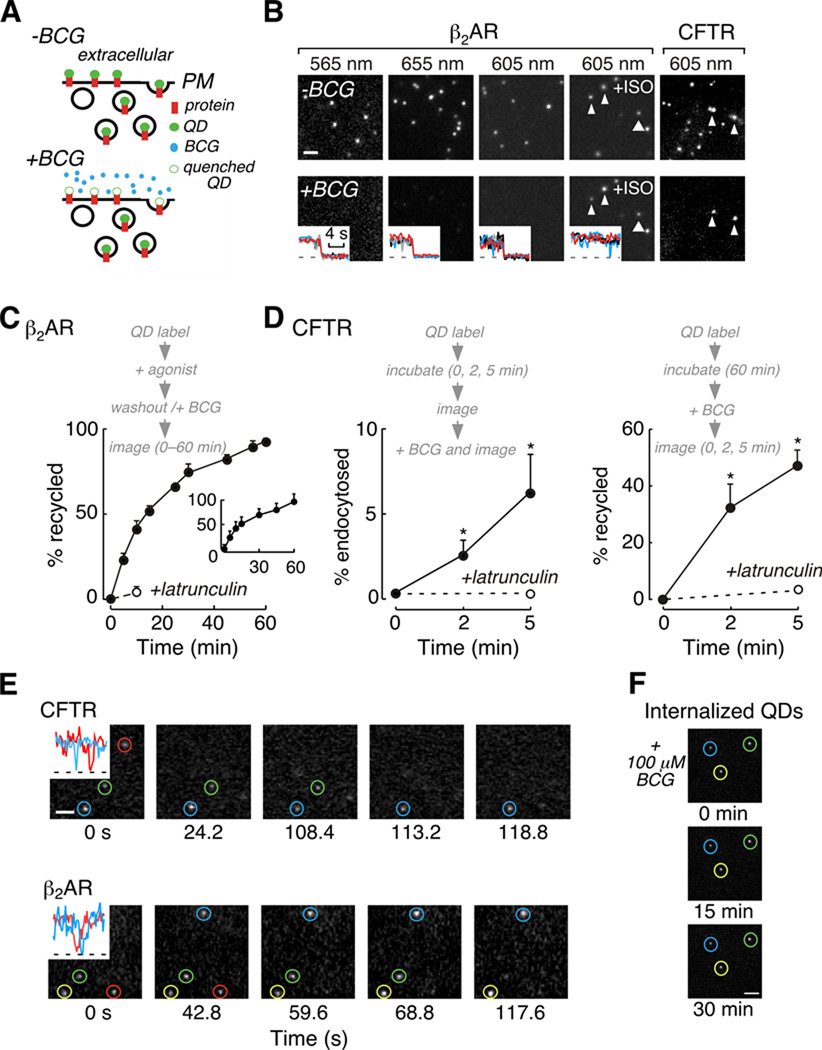
Figure 2. Application of QD quenching to study protein trafficking.
A) Schematic representation of the experimental strategy. B) Examples of BCG quenching of QD fluorescence at different emission wavelengths (scale bar, 2 µm). (insets) Area integrated fluorescence intensities for QD (black, red, grey and blue) and background areas (dashed line). C) β2AR recycling after stimulation and agonist wash-out measured by QD quenching and by (inset) TIRF imaging of Alexa Fluor 488-labelled receptors. D) CFTR endocytosis (left) and recycling (right) rates quantified by QD quenching. E) Real time visualization of CFTR and β2AR recycling assessed by QD quenching (scale bar, 1 µm). (insets) QD blinking indicates individual QDs were imaged. F) BCG cell permeability. MDCK-CFTR-3HA cells labelled with 605 nm QDs that were internalized were imaged at indicated times following addition of 100 µM BCG (scale bar, 2 µm).
HA-β2AR trafficking was studied in a fibroblast cell line that expresses physiologically relevant receptor levels. In unstimulated, HA-β2AR expressing cells, the fluorescence of receptor-bound QDs of different emission wavelengths was strongly quenched by 10–20 µM BCG (Figure 2B, first to third panels). Following addition of the agonist isoproterenol (10 µM, 15 min), there was significant endocytosis of QD-labelled HA-β2AR such that internalized QDs were not accessible to BCG and hence remained visible (Figure 2B, fourth panel). CFTR-3HA was studied in an epithelial cell that expresses low, physiologically appropriate levels of CFTR (8). After a brief incubation (5 min, 37 °C), external vs. constitutively endocytosed CFTR-3HA labelled with 605 nm QDs was resolved following addition of 10 µM BCG (Figure 2B, fifth panel). Control experiments showed that BCG as used here did not affect β2AR or CFTR functions (see Materials and Methods and data not shown).
Kinetics of HA-β2AR and CFTR-3HA trafficking were quantified by QD fluorescence quenching. Experiments were done using wide-field illumination at low magnification (20×, NA 0.75) for detection of QD fluorescence from multiple cells with a large depth of focus. To quantify agonist-induced HA-β2AR internalization, receptors were labelled with QDs, stimulated (isoproterenol, 10 µM, 10 min), and fluorescence intensity was measured before and after addition of 20 µM BCG. HA-β2AR internalization was 33 ± 6 % in isoproterenol-treated cells vs. 0.3 % in control cells (data not shown). Recycling of internalized QD-labelled HA-β2AR after stimulation was also quantified (Figure 2C). Cells were QD-labelled, stimulated, washed (to remove agonist), and exposed to 20 µM BCG to quench fluorescence of QDs associated with HA-β2AR remaining at or recycled to the cell surface. Recycling was near complete within 1 hour, and inhibited by the actin-disrupting compound latrunculin. There was close agreement between agonist-mediated HA-β2AR internalization (data not shown) and recycling kinetics determined by QD quenching and by an alternative method involving total internal reflection fluorescence microscopy (Figure 2C, inset and see Materials and Methods).
To quantify CFTR-3HA endocytosis cells were QD-labelled, incubated for defined times (0, 2 and 5 min) and imaged before and after BCG addition (Figure 2D, left). To quantify CFTR-3HA recycling, cells were QD-labelled, incubated to generate a cohort of internalized QD-labelled CFTR-3HA, BCG added, and QD fluorescence measured over time (Figure 2D, right). The extent of CFTR internalization and recycling assessed by QD-quenching was in agreement with trafficking rates previously determined for CFTR by alternative methods (9). As shown in Figure 2E recycling events could also be observed in real-time using low magnification imaging (20×, NA 0.75), in which loss of QD fluorescence results from their surface exposure due to recycling. In these experiments, individual QDs were imaged, as confirmed by fluorescence blinking (Figure 2E, insets).
Finally, control experiments were performed to verify that BCG was cell impermeable as used here. MDCK-CFTR-3HA cells were labelled with QDs emitting at 605 nm, incubated for 30 min to generate a pool of internalized QDs and treated with jasplakinolide to inhibit recycling. Subsequent addition of 100 µM BCG resulted in quenching of external QDs, however, fluorescence from internalized QDs was not altered over 30 min (Figure 2F).
In summary, BCG is a rapid, efficient and reversible fluorescence quencher of QDs over a broad wavelength range. As demonstrated for a receptor (β2AR) and a channel (CFTR), QD fluorescence quenching allows quantitative measurement of membrane protein internalization and recycling at low, physiologically relevant receptor/channel density. The trafficking rates determined for β2AR and CFTR were independently validated or comparable to literature data. Confirmation that QD-labelling does not alter trafficking should be formally shown for applications to other membrane proteins. Individual recycling events are observable in real-time, allowing the resolution of vesicle trafficking events with higher spatial and temporal resolution than those afforded by confocal or total internal reflectance fluorescence microscopy. Given the reversible nature of QD quenching by BCG, labelled proteins could be followed through multiple cycles of trafficking events by sequential application and removal of BCG. In addition, platereader-based high-throughput screening to identify modulators of membrane trafficking is possible (data not shown).
Materials and Methods
Reagents
SA-conjugated and non-targeted QDs, Alexa Fluor 594-conjugated biocytin, and Alexa Fluor 594-conjugated antibody were purchased from Invitrogen. QDs of 605 nm emission wavelength conjugated to F(ab’)2 fragments were purchased from Chemicon International. Fab fragments conjugated to Cy3 were purchased from Jackson ImmunoResearch. Bromocresol green was purchased from Sigma.
Spectroscopy
Absorbance measurements were made using a Nanodrop 2000c (Thermo Scientific). Fluorescence spectroscopy and fluorimetry was performed using a FluoroMax-3 spectrofluorimeter (Jobin Yvon Horiba). A quartz fluorometer cell with 1 mm emission path-length (Starna Cells) was used for fluorimetry.
Analysis of BCG quenching of QD fluorescence was performed using 40–200 pM suspensions of QDs, and 440 nm excitation light and emission wavelengths centred at QD emission peaks. Absorption of excitation light (at 440 nm) by BCG was negligible at the BCG concentrations used. Absorption of emitted fluorescence by BCG was wavelength dependent (maximal for QDs emitting at 605 nm) and was corrected for a 1 mm path-length according to the Beer-Lambert law and BCG optical density. The maximum reduction in QD emission intensity at the highest BCG concentration was ~30 %. There was no effect of pH in the range 6.5 to 7.4 on QD fluorescence for the QDs used in this study. BCG quenching of Cy3-conjugated Fab fragment (500 nm excitation and 580 nm emission) and Alexa Fluor 594-conjugated antibody (580 nm excitation and 620 nm emission) was done using 20 µg/ml protein. Overall corrections for BCG absorption of excitation and emission light were less than 15 %.
Surface plasmon resonance
Surface plasmon resonance experiments were performed using a Biacore T100. SA-conjugated QDs emitting at 585 nm were immobilized on the surface of a carboxymethylate-functionalized C1 sensor chip using standard carbodiimide coupling. BCG binding experiments were performed at pH 7.4 in Hepes buffered saline and sensorgrams were corrected by subtraction of responses observed for unloaded C1 sensor chips. Qualitatively similar data was obtained for F(ab’)2-conjugated 605 nm QDs immobilized on the surface of a C1 sensor chip (data not shown).
Cell culture and QD labelling
A CV-1 monkey fibroblast cell line stably expressing HA-tagged human β2AR was generated using the Flp-In system (Invitrogen). Western blot analysis indicated that HA-β2AR expression levels were approximately three times higher that β2AR levels in human airway epithelial cells (data not shown). An MDCK cell lines stably expressing CFTR with a triple HA epitope inserted into the 4th extracellular loop (MDCK-CFTR-3HA) was previously described and expresses CFTR at physiologically appropriate levels (8).
For assays of protein trafficking, CFTR-3HA and HA-β2AR were labelled by sequential incubation with anti-HA antibody (Covance), biotin conjugated Fab fragment (Jackson ImmunoResearch) and streptavidin-conjugated QDs, as described for prior single particle tracking studies (8, 10). To visualize single QDs, cells were labelled by 2–5 min incubations with 0.05–0.1 µg/ml anti-HA antibody and Fab fragment and 0.1 nM QDs. To assess CFTR-3HA and HA-β2AR trafficking in cell populations, concentrations of anti-HA antibody, Fab fragment and QDs were increased 10–fold. For microscopy, cells were mounted in a custom perfusion chamber and maintained at 37 °C during imaging by a microincubator (Harvard Instruments).
Microscopy
Fluorescence imaging experiments were performed using a Nikon TE2000S equipped with an EXFO X-cite light source and a Hamamatsu EM-CCD or a Nikon TE2000E equipped with an EXFO X-cite light source, Photometrics QuantEM EMCCD and Perfect Focus System. QDs were imaged using a D440/20 excitation filter, 470dcxr dichroic and standard QD emission filters (QD emission maxima ± 20 nm) (Chroma). Images were acquired using a Nikon 100×, NA 1.49 TIRF objective lens or a Nikon 20×, NA 0.75 S Fluor objective lens. Data acquisition times for all experiments were 30–200 ms. Image processing for trafficking assays was performed using ImageJ. Background-corrected, area-integrated fluorescence intensities were determined before and after BCG quenching. For single QD experiments, in which intermittency of fluorescence (blinking) confirmed that individual QDs were imaged, data was analysed using NIS Elements AR software.
In control experiments to image internalized QDs in BCG treated cells, CFTR-3HA was labelled with 605 nm QDs, cells were incubated at 37 °C for 30 min and then treated with jasplakinolide (2.5 µM, 15 min). Cells were imaged over 30 min following addition of 100 µM BCG.
HA-β2AR trafficking determined by labelling surface receptors with Alexa Flour 488
An assay based on total internal reflection fluorescence (TIRF) microscopy was also used to characterize HA-β2AR trafficking in CV-1 cells. Surface HA-β2AR was labelled by incubating cells with Alexa Fluor 488-conjugated anti-HA antibody (Invitrogen) for 15 min at 4 °C prior to paraformaldehyde fixation. To assess agonist-mediated receptor internalization, cells were stimulated (isoproterenol, 10 µM, 10 min), labelled and fixed. To assess HA-β2AR recycling, cells were stimulated (isoproterenol, 10 µM, 10 min), washed to remove agonist, incubated at 37 °C for set time periods (to allow receptor recycling), labelled and fixed. Given the low expression level of HA-β2AR, Alexa Fluor 488 fluorescence levels were assessed by TIRF microscopy using a Nikon TE2000E equipped with a 488 nm argon-ion laser, a Photometrics QuantEM EMCCD and a Nikon 100×, NA 1.49 TIRF objective lens.
Analysis of CFTR function
Assays of CFTR function were performed using a Fischer rat thyroid cell line stably expressing CFTR and a halide sensitive YFP (11). Cells were stimulated with forskolin (20 µM, 15 min) in PBS and CFTR activity was assessed by application of iodide substituted PBS to generate a 100 mM I− gradient. Experiments were performed at 37 °C and no difference in CFTR activity was observed in the absence or presence of 1 mM BCG.
Acknowledgments
This work was supported by grants from the NIH (DK081355 to P.M.H and DK072517 and EB000415 to A.S.V.) and the Cystic Fibrosis Foundation (R613 to A.S.V.). The authors thank Peter Hwang, Ph.D. (U.C.S.F. Center for Advanced Technology) for assistance in performing and interpreting surface plasmon resonance experiments and Gergely Lukacs (McGill) for providing the MDCK-CFTR-3HA cell line.
Abbreviations
- BCG
bromocresol green
- β2AR
β2 adrenergic receptor
- CFTR
cystic fibrosis transmembrane conductance regulator
- QDs
quantum dots
- SA
streptavidin
References
- 1.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal JK, Simon SM. Imaging single events at the cell membrane. Nat. Chem. Biol. 2007;3:92–98. doi: 10.1038/nchembio855. [DOI] [PubMed] [Google Scholar]
- 3.Lidke DS, Lidke KA, Reiger B, Jovin TM, Arndt-Jovin D. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J. Cell Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Q, Li Y, Tsien RW. The dynamic control of kiss-and-run and vesicular reuse probed with single nanoparticles. Science. 2009;323:1448–1453. doi: 10.1126/science.1167373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui B, Wiu C, Chen L, Ramirez A, Bearer E, Li WP, Mobley WC, Chu S. One at a time, live trafficking of NGF axonal transport using quantum dots. Proc. Natl. Acad. Sci. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Proteolytic activity monitored by fluorescence energy transfer through quantum-dot-peptide conjugates. Nat. Mat. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 7.Funston AM, Jasieniak JJ, Mulvaney P. Complete quenching of CdSe nanocrystal photoluminescence by single dye molecules. Adv. Mater. 2008;20:4274–4280. [Google Scholar]
- 8.Haggie PM, Kim JK, Lukacs GL, Verkman AS. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol. Biol. Cell. 2006;17:4937–4945. doi: 10.1091/mbc.E06-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma M, Pampinella F, Nemes C, Benharouga M, So J, Du K, Bache KG, Papsin B, Zerangue N, Stenmark H, Lukacs GL. Misfolding diverts CFTR from recycling to degradation; quality control at early endosomes. J. Cell Biol. 2004;164:923–933. doi: 10.1083/jcb.200312018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valentine CD, Haggie PM. Confinement of β1- and β2-adrenergic receptors in the plasma membrane of cardiomyocyte-like H9c2 cells is mediated by selective interactions with PDZ domain and A-kinase anchoring proteins but not caveolae. Mol. Biol. Cell. 2011;22:2970–2982. doi: 10.1091/mbc.E11-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma T, Thiagarajah JR, Yang H, Sonawane ND, Folli C, Galietta LJ, Verkman AS. Thiazolidinone CFTR inhibitor identified by high-throughput screening blocks cholera toxin-induced intestinal fluid secretion. J. Clin. Invest. 2002;110:1651–1658. doi: 10.1172/JCI16112. [DOI] [PMC free article] [PubMed] [Google Scholar]