Abstract
Long-chain fatty acids (LCFA) serve as structural components for membrane biogenesis and as primary energy sources during mitochondrial-oxidation reactions. Hepatic LCFA uptake is complex, with characteristics suggestive of a dual-kinetic model manifested by rapid (carrier-assisted/facilitated) and delayed (passive diffusional) phases. Our previous work using mice deficient of the Iqgap2 gene established a highly novel link between IQGAP2, a putative GTPase-activating protein, and hepatocarcinogenesis. Now we report that Iqgap2 deficiency also results in selective loss of the facilitated phase of hepatocyte LCFA uptake with preservation of the diffusional component. This molecular defect was seen in Iqgap2-/- hepatocytes of all ages studied (1-, 4-, 8-months). The loss of facilitated LCFA uptake protected against development of hepatic triglyceride accumulation in Iqgap2-deficient mice fed high-fat diet, consistent with a fundamental role in physiological fat partitioning. These phenotypic changes could not be explained by genetic loss of fatty acid processing proteins known to regulate lipid uptake or metabolic processing pathways. Iqgap2-deficient livers also displayed enhanced insulin sensitivity.
Conclusion
These observations identify a novel property of the putative GTPase-activating protein IQGAP2 in LCFA uptake in vitro and in vivo, and implicate IQGAP2 in an intracellular signaling pathway necessary for functional fatty acid uptake, lipid processing, and, possibly, glucose homeostasis.
Keywords: GTPase-activating proteins, liver, hepatic steatosis, fatty acid uptake, mouse knockout model
1. INTRODUCTION
Three highly homologous members of IQGAP family have been identified in humans and mice – IQGAP1, IQGAP2, and, most recently, IQGAP3 [1]. All three are large (180 – 190 kDa) multidomain cytoplasmic scaffolding proteins that juxtapose Rho GTPases Rac1 and Cdc42, Ca2+/calmodulin signals and cytoskeletal reorganization events [2-4]. The GTPase binding domain in IQGAPs lacks an arginine finger residue essential for the GTP-hydrolysis, which may explain why none of IQGAPs demonstrated in vitro GAP activity toward GTPases [5]. To date, IQGAP1 is the most extensively studied member of the family. While IQGAP1 is expressed broadly, IQGAP2 expression is primarily limited to the liver and platelets [6]. To further characterize IQGAP2 physiological functions, we generated a conventional knockout mouse model and showed that Iqgap2 deficiency leads to a high incidence of hepatocellular carcinoma (HCC), establishing the first IQGAP2-specific phenotype [7].
Long-chain fatty acids (LCFAs) are the major components of phospholipids and provide the primary energy source in the form of triglyceride deposits. LCFA uptake by the liver follows one of two intracellular pathways: (i) a β-oxidative reaction that occurs within the mitochondrial matrix for ATP generation, or (ii) esterification that produces triacylglycerols for subsequent hepatic storage or incorporation into VLDL (very-low density lipoprotein) particles. Defects in either or both of these pathways can lead to hepatic steatosis, or can progress to steatohepatitis, a condition associated with development of end-stage liver disease [8]. Furthermore, hepatic triglyceride accumulation occurring in the setting of obesity typically is associated with the development of insulin resistance in humans [9].
Molecular control of lipid processing and metabolism occurs at the level of cellular uptake and transport, in parallel with transcriptional regulation of effector genes mediated by the family of peroxisome proliferator-activated receptors (PPAR). PPARs (α, β and γ) are ligand-dependent nuclear hormone receptors whose target genes variably participate in the catabolism, storage, and processing of fatty acids [reviewed in [10]]. Cellular uptake of unesterified LCFAs occurs through both passive (diffusional) and facilitated transport mechanisms [11]. At physiological serum to albumin ratios, the majority of LCFAs are taken up by the saturatable facilitated transport pathway, functional in a broad array of tissues including adipose tissue [12], cardiac myocytes [13], and hepatocytes [14]. Studies in both yeast and mammalian cells suggest that facilitated LCFA uptake is likely regulated by a multiprotein complex composed of fatty acid transport proteins (FATP) [15-17], fatty acid binding proteins (FABP) [18], long-chain acyl-CoA synthases [19], and the multifunctional adhesive glycoprotein CD36/FAT (fatty acid translocase) [20]. While CD36/FAT may concentrate fatty acids at the plasma membrane, FATP family members are primarily responsible for LCFA transport across the cellular membrane. Upon uptake, rapid esterification by acyl-CoA synthases presumably prevents cellular efflux, while binding to FABPs could facilitate cellular unloading [11].
FATP and FABP encompass numerous family members with distinct tissue-specific expression patterns. Based on the presence of a conserved 311-amino acid signature sequence [21], six fatp genes (fatp1-6) have been described to date. FATP2 and FATP5 are the predominant transporter proteins in the liver, demonstrating little to no expression in skeletal muscle or adipose tissue [11, 22].
FABPs is an extended family of genes (fabp1-10) displaying distinct binding affinities for long-chain fatty acids, retinols, eicosanoids, bile acids and heme [23]. FABPs enhance free fatty acid (FFA) uptake by increasing their concentration gradient in the cell, exchanging fatty acids with membrane structures via collisional transfer [24]. Cellular FABP concentration correlates with rates of fatty acid metabolism, generally highest in hepatocytes, cardiomyocytes and adipocytes. L-FABP or FABP1 (fabp1) is the predominant subtype of FABPs in liver, although recent evidence has suggested that a greater variety of the FAPBs may be found in hepatocytes [25]. FABP1 binds two fatty acid molecules and two molecules of their CoA-esters with comparable affinities, leading to an increase in fatty acid uptake, and rapid incorporation into triacylglycerols and phospholipids [26]. Despite the critical roles of FATPs and FABPs in the uptake and fate of intracellular FFAs, the molecular signals coordinating this process remain unknown.
We now provide evidence that murine IQGAP2 selectively regulates the facilitated phase of hepatic fatty acid uptake both in vitro and in vivo. Our data suggest a new physiologic function for IQGAP2, and implicate distinct signaling pathways in the normal regulation of hepatocyte FFA uptake and/or transport.
2. MATERIALS AND METHODS
2.1. Iqgap2-deficient mice
Iqgap2-/- mice were generated in this laboratory and described previously [7]. Mice were maintained on a 129J1 background and handled according to the National Institutes of Health and institutional guidelines for the humane care and use of experimental animals, with approval for all studies from the Stony Brook University Animal Care and Use Committee.
2.2. Quantification of mRNA expression levels
Total cellular RNA was isolated from livers of Iqgap2-/- and wild-type mice by immediate solubilization in Trizol reagent (Invitrogen, Carlsbad, CA) and serial isopropanol precipitations. RNA integrity was established using an Agilent 2100 Bioanalyzer, and qRT-PCR was performed using fluorescence-based real-time PCR technology [27]. Oligonucleotide primer pairs were generated using Primer3 software (www-genome.wi.mit.edu), designed to amplify ~200-bp PCR products at the same annealing temperature (71°C). Primer sequences are listed in Table 1. Purified total RNA (5 μg) from murine livers was used for first strand cDNA synthesis (reverse transcription reaction or RT) using oligo(dT) and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). For qRT-PCR analysis, the RT reaction was diluted at 1:15, equally divided among primer pairs and used in a PCR reaction with the following cycle: 95°C for 15 seconds, then 39 cycles of 94°C for 30 seconds, 55°C for 30 seconds and 72°C for 1 minute. The mRNA levels were quantified by monitoring real-time fluorimetric intensity of SYBR green I (Qiagen, Valencia, CA) at a reading temperature of 70°C. Relative mRNA abundance was determined from triplicate assays performed in parallel for each primer pair, calculated using the comparative threshold cycle number (Δ-Ct method) [28], and normalized to cellular β-actin mRNA, all as previously described [29].
Table 1.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Fatp-1 | 5’-AGACGGACGTGGCTGTGTAT-3’ | 5’-TGGATCTTGAAGGTGCCTGT-3’ |
| Fatp-2 | 5’-ATGCCGTGTCCGTCTTTTAC-3’ | 5’-GCGATGATGATTGATGGTTG-3’ |
| Fatp-3 | 5’-CTACCTCGCACTCCCACTGT-3’ | 5’-ATCTGCAGAGGTCCAAATCG-3’ |
| Fatp-4 | 5’-TCTGTCCTTCCTGCCTGTCT-3’ | 5’-GCTAAGGGCTTATCCCAAGG-3’ |
| Fatp-5 | 5’-CCAAGGTTCGAAAGAACCAA-3’ | 5’-AGATACGTTTTCGCCCTTCC-3’ |
| Fabp-1 | 5’-GCAGAGCCAGGAGAACTTTG-3’ | 5’-CAGTCATGGTCTCCAGTTCG-3’ |
| Fabp-2 | 5’-TTGCTGTCCGAGAGGTTTCT-3’ | 5’-TGGAGACCAGTGCTGATAGG-3’ |
| Fabp-4 | 5’-ACGACAGGAAGGTGAAGAGC-3’ | 5’-AAATTTCCATCCAGGCCTCT-3’ |
| Fabp-5 | 5’-ACTGAGACGGTCTGCACCTT-3’ | 5’-CTGGCAGCTAACTCCTGTCC-3’ |
| FAT/CD36 | 5’-TCCCTCACTGGAGGAAACTG-3’ | 5’-TATCTGGCCTTGCTGTAGCC-3’ |
| Cave-1 | 5’-GGCCAGCGTGTCTATTCAGT-3’ | 5’-CTTGGCAGTCTCGGTTTAGC-3’ |
| Acox1 | 5’-GAGGGGAACATCATCACAGG-3’ | 5’-GTTCAGTGGGGACTTCTTGG-3’ |
| Cpt1a | 5’-GCAGCAGGTGGAACTGTTT-3’ | 5’-GGCTTGTCTCAAGTGCTTCC-3’ |
| FAS | 5’-ACTGGGTGACCTTCATGGAC-3’ | 5’-ACCACCAGAGACCGTTATGC-3’ |
| PPARα | 5’-ACGGAGCTCACAGAATTTGC-3’ | 5’-AACGGCTTCCTCAGGTTCTT-3’ |
| PEPCK | 5’-ATGCTGATCCTGGGCATAAC-3’ | 5’-CCGTTTTCTGGGTTGATAGC-3’ |
| SREBP-1c | 5’-TACTTCTTGTGGCCCGTACC-3’ | 5’-GCTGTGGCCTCATGTAGGAA-3’ |
| SREBP-2 | 5’-GCTGCTGGAGCATAGCCTAC-3’ | 5’-GGTCACCCACCTTCTCCAG-3’ |
| β-actin | 5’-TACCACAGGCATTGTGATGG-3’ | 5’-TTTGATGTCACGCACGATTT-3’ |
2.3. Diet modification and metabolic measurements
Diet studies were completed using 16-week-old male mice that were housed in a full barrier facility with a 12-hour light/dark cycle, given full access to food and water. Mice were weaned onto fixed diets over a 1-week period (control chow diet contained 10% calories from fat, high fat diet contained 45% calories from fat; Research Diets, New Brunswick, NJ). Chow was provided ad libitum (and intake carefully monitored) for 6 weeks prior to termination of studies, at which point mice were weighed and sacrificed for blood and organ collection and processing. Livers were immediately harvested after cervical dislocation, snap-frozen in liquid nitrogen with a portion embedded in OCT medium (Sakura Finetek, Torrence, CA), and stored at -80°C until used. Frozen sections in OCT were cut on a Leica CM1900 cryostat (Leica Microsystems, Weltzar, Germany). For triglyceride detection, sections were stained with Oil Red O and counterstained with hematoxylin.
For insulin studies, 6-month-old male wild-type and Iqgap2-/- mice (n=3 each) fed regular chow were fasted for 4 hours prior to liver collection. Another set of fasted age-matched mice of both genotypes (n=3 each) were injected intraperitoneally with either 1U/kg or 5U/kg insulin (Novolin, Novo Nordisk), and livers were collected 10 minutes after injection. Total RNA was isolated using Trizol reagent for qRT-PCR. For glucose tolerance test (GTT), mice were fasted overnight followed by intraperitoneal injection of glucose (2 g/kg). Blood samples were collected from a tail vein immediately before and 10, 20, 30, 45, 60, 90, 120 and 180 minutes after glucose administration for glucose levels measurement.
For biochemical analyses, blood was collected by retroorbital bleeding and serum was immediately prepared. For hepatic triglyceride levels, liver lysates were prepared by lysis in 0.1N KOH. Triglyceride levels were measured using a kit from Wako Diagnostics (Richmond, VA), insulin with an ELISA kit from Mercodia (Uppsala, Sweden) and glucose using an Accu-Check® Instant Plus blood glucose monitor (Roche). Mouse total body fat was measured using nuclear magnetic resonance (NMR) at the NMR Facility, Department of Chemistry, University of Cincinnati, OH, and expressed as a percent of fat weight of a total body weight.
2.4. Immunoblotting
Protein lysates were isolated as previously described using lysis buffer (50 mM Tris, pH 7.4, 1% Triton X-100, 0.2% sodium deoxycholate, 0.2% sodium dodecylsulfate (SDS), 1 mM sodium EDTA) supplemented with 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml leupeptin and 10 μg/ml aprotinin [3]. Crude lysates were centrifuged at 14,000 g for 10 minutes at 4°C, and protein supernatants were quantified using bicinchoninic acid assay (BCA, Pierce, Rockford, IL) prior to SDS-PAGE using 4–15% gradient gels (Bio-Rad Laboratories, Hercules, CA). Immunoblot analysis was completed as previously described [30] using the following antibodies: anti-FAS, total Akt, total GSK3β, phospho-Akt (Ser473), phospho-GSK3β (Ser9) monoclonal, Cell Signaling Technology, Danvers, MA), anti-caveolin-1 (rabbit polyclonal, Cell Signaling Technology), anti-FABP1 (rabbit polyclonal, abcam Inc., Cambridge, MA), and anti-GAPDH (mouse monoclonal, Millipore, Billerica, MA), all at 1:1000 dilution.
2.5. Free fatty acid uptake studies
Primary hepatocytes were isolated from mice of various ages using modified in situ collagenase digestion of livers, as previously described [31]. Viable cells were identified and counted using trypan blue exclusion, with >80-85% viability observed using both wild-type and Iqgap2-/- mice, and cultured in DMEM (Invitrogen), supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 7 ng/ml glucagon, 7.5 μg/ml hydrocortisone, 0.5U/ml insulin and 20 ng/ml epidermal growth factor (EGF). MTT cell proliferation assay (Sigma) and ApoStrand ELISA Apoptosis Detection Kit (Enzo Life Sciences) were used to evaluate viability of isolated hepatocytes. Fatty acid uptake was quantified using the long-chain fatty acid analog BODIPY-FA (4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-S-indacene-3-dodecanoic acid; Molecular Devices, Sunnyvale, CA) in solution with the quenching agent Q-Red.1 to reduce non-cellular fluorescence [32]. This formulation allows for real-time monitoring of cellular fatty acid uptake and parallels LCFA uptake by intracellular fatty acid levels [33]. Serum-starved, freshly isolated hepatocytes were plated into 96-well microtiter plates (1 X 105/well), and loaded in a fixed concentration of albumin supplemented with 2 μM BODIPY-FA for 30 min at 37°C (QBT FA Uptake Assay Kit, Molecular Devices). FFA uptake was analyzed in real time using a Molecular Devices FLEX station. For some experiments, cells were incubated with various concentrations of FA to monitor the specificity and kinetics of LCFA uptake and diffusion. Kinetic data were analyzed using SoftMaxPro software [30], and data reported as the mean ± SEM from triplicate wells.
Alternatively, [3H]palmitate uptake was measured in primary mouse hepatocytes as follows. Palmitate-bovine serum albumin (BSA) complex was prepared by combining 5μM nonradioactive palmitate (Sigma-Aldrich) in Krebs-Ringer buffer with fatty acid free, low endotoxin BSA (Sigma-Aldrich) at the molar ratio 1:3. [9,10-3H(N)]palmitate (1.0mCi/mL) (PerkinElmer) was then added to the palmitate-BSA complex to the final concentration of 0.5μCi/mL and incubated on an orbital rotator at room temperature for 2 hours. Freshly isolated primary hepatocytes from 3-4 month old male wild-type and Iqgap2-/- mice were seeded in 12-well plates at 2 × 105 cells/well and allowed to attach for 18 hours. For uptake studies, hepatocytes were serum-starved for 2 hours, washed with PBS, and then incubated in triplicates in the [3H]Palmitate-BSA solution for the specified time points at room temperature. Uptake was stopped by addition of 200 μM phloretin (Sigma) in 0.1% BSA. Cells were washed three times with ice-cold PBS and lysed in 0.1N NaOH/0.03% SDS buffer. [3H] radioactivity of each lysate was counted using a Tri-Carb 3100TR Liquid Scintillation Analyzer (PerkinElmer) and QuantaSmart software. The uptake was normalized to lysate protein concentration determined by BCA assay. Primary hepatocyte isolations and subsequent FFA uptake assays for each genotype were performed side-by-side to replicate exact conditions and eliminate possible sources of error or discrepancies.
To study an effect of Ca2+ on LCFA uptake, human hepatocellular carcinoma HepG2 cell line (ATCC) was propagated in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Prior to uptake experiments, the media was replaced with serum-free DMEM and cells pre-incubated with 125 μM, 500 μM or 1 mM of cell-permeant 1,2-bis(2-aminophenoxy)ethane-N,N,N',N'-tetraacetic acid (acetoxymethyl) ester (BAPTA-AM, Molecular Probes, Eugene, OR), a Ca2+ chelator, for 30 min at 25°C, and uptake experiments conducted as described above using BODIPY-FA.
2.6. Statistical analysis
Data are presented as mean ± SD. Multiple comparisons between groups were performed using ANOVA. For two groups’ comparisons, probabilities of chance differences were calculated using Student's t-test assuming unequal variance with two-tailed analysis. For all comparisons, values of p < 0.05 were considered statistically significant. For qRT-PCR data analysis, the relative transcript expression was normalized to actin expression and calculated using the comparative threshold cycle number (Δ-Ct method) as described previously [28]. Mean expression levels and standard deviation were log10-transformed and plotted on a logarithmic scale. For densitometric analysis of immunoblots, the relative optical density of a protein band was normalized to the integrated optical density of the equal loading control protein (GAPDH).
3. RESULTS
3.1. Iqgap2-deficient mice are protected from diet-induced hepatic steatosis
Generation of Iqgap2-deficient mice has been previously described [7]. We have demonstrated that while healthy in the young age, the majority of Iqgap2-/- mice develop HCC by the age 18 – 24 months [7]. Our initial observations showed an elevated body weight of adult (12 weeks and older) Iqgap2-/- mice fed regular chow and an increased fat-to-body ratio, while plasma analysis revealed lower levels of non-fasted glucose in Iqgap2-/- mice and levels of insulin comparable to the wild-type mice (Table 2).
Table 2.
Metabolic characteristics of non-fasted male 6-month-old wild-type and Iqgap2-/- mice (n=10 per group).
| Metabolic parameters | Wild-type | Iqgap2-/- |
|---|---|---|
| Body weight, g | 26.54±4.71 | 29.85±4.33 |
| Food intake, g/day | 3.92±0.31 | 3.88±0.33 |
| Body fat, % | 11.74±0.72 | 15.28±0.61* |
| Plasma TG, mg/dl | 100.67±14.05 | 54.0±7.20* |
| Glucose, mg/dl | 120.9±5.08 | 87.2±4.46* |
| Insulin, μg/l | 0.502±0.041 | 0.54±0.062 |
Asterisk denotes p < 0.05.
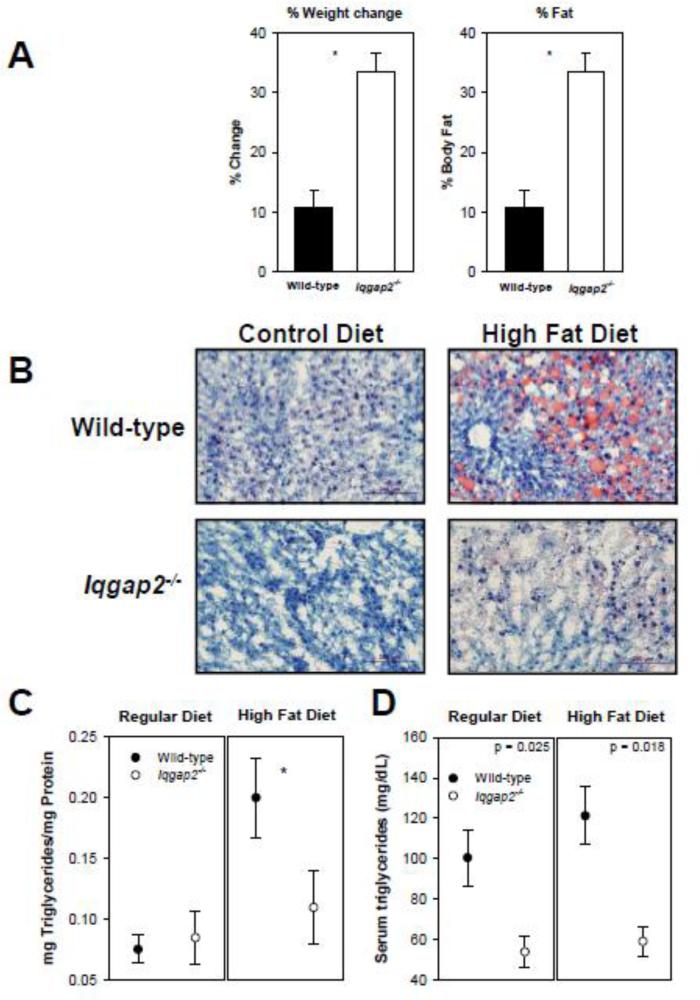
Since mitochondrial β-oxidation is the predominant pathway for disposition of FFAs under physiological conditions, we completed diet-modification studies in Iqgap2-/- and wild-type mice. When fed a high-fat diet (45% calories from fat), Iqgap2-/- mice demonstrated significantly greater weight gain that was associated with redistribution to the adipose tissue compared to wild-type (Figure 1A). Furthermore, there was clear evidence for decreased hepatic triglyceride accumulation as established using Oil Red O staining of liver sections and triglyceride measurement in liver extracts (Figure 1B, C). Iqgap2-/- mice also had lower plasma triglyceride levels when fed both control and high-fat diet compared to the wild-type mice (Figure 1D), suggesting either increased peripheral utilization or a defect in FFA uptake. When integrated with the Oil Red O staining, however, these data were more consistent with a requisite role for IQGAP2 in hepatic fatty acid uptake and/or intracellular transport.
Figure 1.
Defective fatty acid partitioning in diet-modified Iqgap2-/- mice. (A) Weight change and alterations in body composition for high-fat diet; asterisk indicates statistically significant differences (p < 0.05). % Fat was calculated as a percent of total body fat weight measured by NMR from total body weight. (B) Fresh-frozen liver sections stained with Oil Red O and counterstained with hematoxylin demonstrate protection against hepatic steatosis in Iqgap2-/- mice. A size bar denotes 200 micron. Hepatic (C) and serum (D) triglyceride content is expressed as the mean ± SEM (n=6 per genotype), hepatic triglyceride levels are expressed in mg per mg of total protein. Asterisk indicates statistically significant differences (p<0.05).
3.2. Selective loss of facilitated free fatty acid uptake in Igqap2-/- hepatocytes
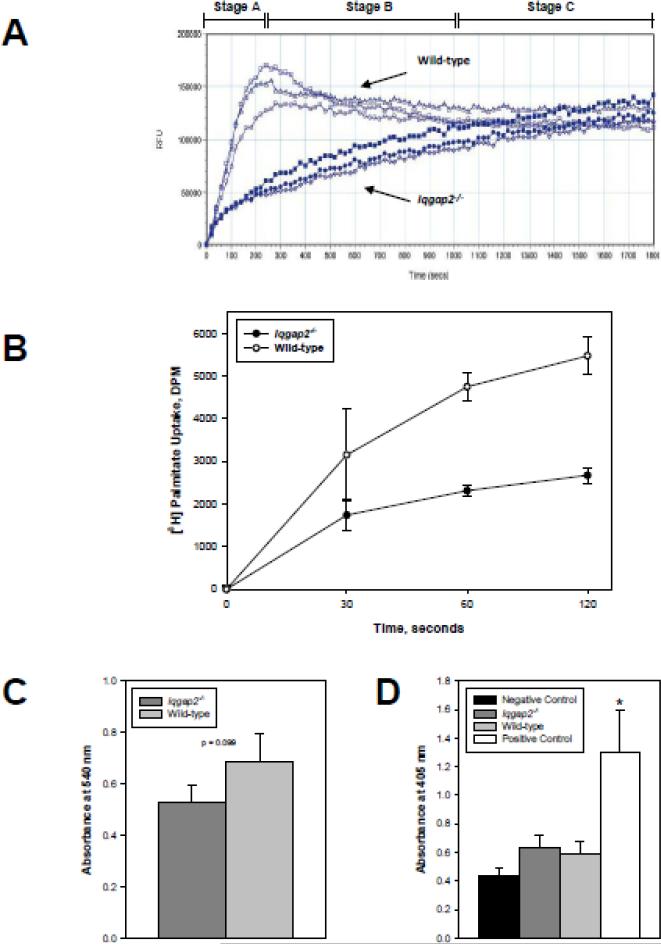
Cellular uptake of unesterified LCFA occurs through both passive (diffusional) and facilitated transport mechanisms [11]. To further dissect the mechanism for the altered diet-modified phenotype, hepatocytes were isolated from Iqgap2-/- and wild-type mice of various ages, and LCFA uptake studied utilizing a novel Quencher-Based Technology (QBT) fluorescence assay [32]. This method employs a fluorescently-labeled LCFA (BODIPY-FA) and a cell-impermeable quenching agent that allows direct measurement of FFA uptake in real time using a Flexstation plate reader. While in solution outside the cell, the quenching agent suppresses BODIPY-FA fluorescence. When BODIPY-FA enters the cell, it becomes unquenched and its fluorescence can be measured on a plate reader. As shown in Figure 2A, FFA uptake dynamics differ dramatically between wild-type and Iqgap2-/- hepatocytes. It can be empirically divided into three stages. Stage A, which corresponds to the first 250 seconds from the time of LCFA addition to cells, displays an extremely high rate of uptake by wild-type hepatocytes, while Iqgap2-/- hepatocytes uptake LCFA significantly slower, reaching only 1/3 of the total FFA taken-up by wild-type hepatocytes. During Stage B (250 – 1000 seconds), wild-type LCFA uptake arrests, while Iqgap2-/- hepatocytes continue uptaking LCFA at a steady rate. In Stage C (1000 – 1800 seconds), representing the saturated phase of uptake where LCFA inside the cell reaches the maximum level, results are indistinguishable between the two genotypes. This uptake trend has been shown in mice of all age groups and both sexes. These data were confirmed by measuring [3H]palmitate uptake in isolated hepatocytes (Figure 2B). Thus, Iqgap2-/- hepatocytes demonstrate a 2-fold reduction in palmitate uptake rate over initial period of 2 minutes compared to the age-matched wild-type control. Both wild-type and Iqgap2-/- hepatocytes showed comparable viability and a lack of apoptotic cell death (Figure 2C, D). Collectively, Iqgap2-/- hepatocytes demonstrate selective loss of the facilitated phase of LCFA uptake, retaining the intact passive phase of LCFA uptake.
Figure 2.
Selective loss of facilitated fatty acid uptake in Iqgap2-/- hepatocytes. (A) LCFA uptake by isolated hepatocytes from Iqgap2-/- and wild-type mice was measured in real time using BODIPY-FA/Q-Red.1 [32]. Data plots reflect those of a complete set of paired experiments representative of six identical experiments. All data points are mean relative fluorescence units (RFU) per 105 cells/well from triplicate wells (SEM not shown to minimize plot complexity). Three empirical phases of uptake (A, B, C) are shown.
(B) [3H]palmitate uptake by Iqgap2-/- and wild-type hepatocytes. Uptake is expressed as mean decay per minute (DPM) per 2 × 105 cells ± SEM from triplicate wells. p < 0.05 for 60 and 120 seconds time points. Plot is representative of four independent experiments.
(C) Viability of isolated hepatocytes was assessed by MTT assay. Data are expressed as mean relative absorbance at 540 nm per 2 x 105 cells ± SEM from triplicate wells. Difference in absorbance between Iqgap2-/- and wild-type is not statistically significant (p = 0.099).
(D) Apoptosis in isolated hepatocytes was quantified by ApoStrand ELISA and expressed as mean relative absorbance at 405 nm per 104 cells ± SEM from triplicate wells. Asterisk indicates p < 0.05 compared with all other groups.
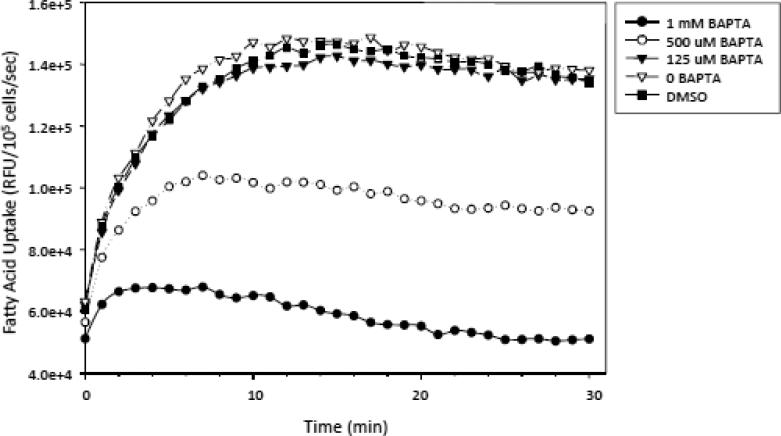
Previous observations have identified complex relations among Ca2+, calmodulin, and IQGAP IQ domains. Ca2+ can directly associate with IQGAP1 [34], and Ca2+/calmodulin binding modulates IQGAP1's functional interactions with actin and cdc42 [35-36], which prompted us to dissect the role of intracytoplasmic calcium [Ca2+]i in the regulation of FFA uptake. We showed that pretreatment of human hepatocellular carcinoma HepG2 cells with 500 μM of BAPTA-AM, a Ca2+ chelator, mimicked all the characteristics of FFA uptake by Iqgap2-/- hepatocytes, while 1 mM BAPTA-AM completely inhibited FFA uptake by HepG2 cells, suggesting that the facilitated phase of cellular FFA uptake might be Ca2+-dependent (Figure 3). In every experiment, we used trypan blue staining to ensure that these relatively high doses of BAPTA-AM did not affect cell viability. The percent of viable cells was consistently above 90% after each BAPTA-AM pretreatment.
Figure 3.
Effect of BAPTA-AM on LCFA uptake by HepG2 cells. HepG2 cells were pre-incubated with 125 M, 500 M and 1 mM BAPTA-AM for 30 minutes at 25°C prior to real time LCFA uptake quantification using BODIPY-FA/Q-Red.1. All plots represent the mean ± SEM from triplicate wells from one representative set of complete experiments (repeated once). DMSO treatment was used as a positive control. The results are expressed as relative fluorescent units (RFU) per 105 cells per second.
3.3. Lipid catabolism pathway analysis in Iqgap2-/- mice
The binding of fatty acids to PPARs is known to transcriptionally regulate gene expression. PPARα, primarily expressed in brown adipose tissue and hepatocytes, regulates the expression of a heterogeneous repertoire of target genes that control phases of lipid catabolism, including FFA uptake, intracellular binding, and processing [10]. Membrane-bound sterol regulatory element-binding proteins (SREBPs) are another family of transcription factors involved in lipid homeostasis [37].
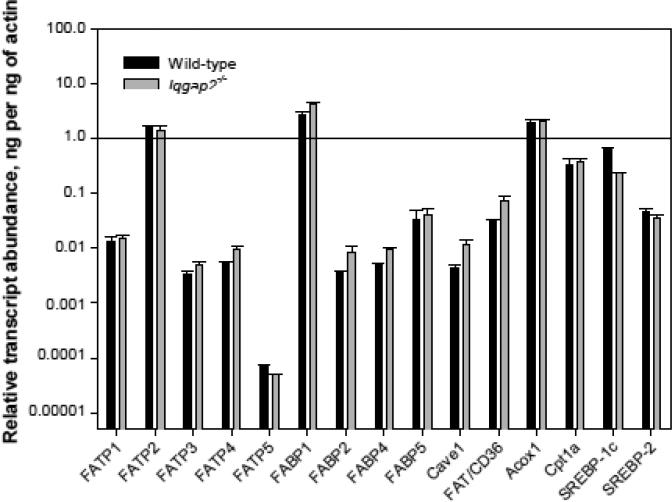
To exclude the possibility that the observed selective defect in FFA uptake was due to a more global abnormality of genes regulating the FA processing pathway, we compared RNA transcript expression profiles of the genes encoding fatty acid transport proteins FATP 1-5, fatty acid binding proteins FABP 1, 2, 4, 5, caveolin-1, fatty acid translocase FAT/CD36, acyl-Coenzyme A oxidase 1, carnitine palmitoyltransferase 1a (Cpt1a), and SREBP-1c and SREBP-2. As shown in Figure 4, we observed a modest (~ 2-fold or less) increase in hepatic transcript levels of fabp1, fabp2, caveolin-1 and FAT/CD36 (p < 0.05). Only fatp2 and fatp5 genes were mildly downregulated in Iqgap2-/- liver, although these transcript changes were statistically significant. The differences in fabp1 and caveolin-1 expression between the genotypes were not apparent at the protein level (Figure 5B). Interestingly, Iqgap2-/- livers also displayed a 3-fold decrease in SREBP-1c transcript levels (p < 0.05), while changes in SREBP-2 levels were not statistically significant (Figure 4). SREBP-1c upregulates transcription of genes involved in fatty acid synthesis, whereas SREBP-2 is mostly responsible for cholesterol synthesis regulation [37].
Figure 4.
Hepatic expression of genes involved in FA processing in Iqgap2-/- and wild-type mice. Quantitative RT-PCR was completed using oligonucleotide primers specific for: fatty acid transport proteins (fatp1-5), fatty acid binding proteins (fabp 1-5), caveolin-1 (cave1), FAT/CD36, acyl-Coenzyme A oxidase 1 (Acox1), carnitine palmitoyltransferase 1a (Cpt1a) and sterol regulatory element-binding protein SREBP-1c and SREBP-2 genes (see Table 1). The relative transcript expression was normalized to actin expression, calculated using the comparative threshold cycle number (Δ-Ct method) and expressed as relative transcript abundance in ng mRNA per 1 ng of β-actin mRNA. Mean expression levels from triplicate wells and standard deviation were log10-transformed and plotted on a logarithmic scale.
Figure 5.
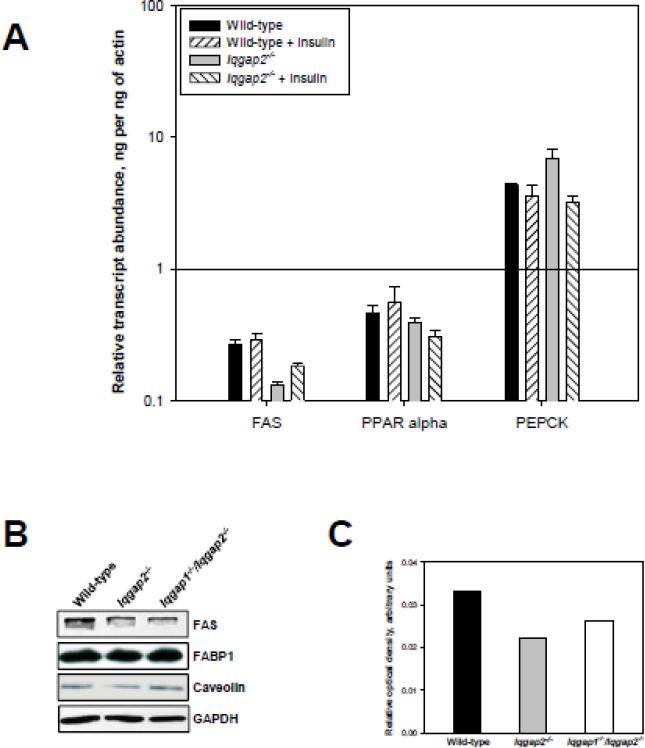
Expression levels of lipogenesis-related genes in IQGAP2-deficient livers. (A) Hepatic FAS, PPARα and PEPCK RNA transcript levels measured by qRT-PCR. RNA transcript levels of fatty acid synthase (FAS), peroxisome proliferator-activated receptor α (PPARα) and phosphoenolpyruvate carboxykinase (PEPCK) were measured in response to a single intraperitoneal insulin injection (5U/kg) by qRT-PCR. The relative transcript expression was calculated as described for Figure 4. (B) Immunoblot confirming protein expression of FAS, FABP1 and caveolin-1 in wild-type, Iqgap2-/- and Iqgap1-/-/Iqgap2-/- livers. The blots are representative of three independent experiments analyzing three different groups of mice. (C)Densitometric quantification of hepatic FAS protein expression normalized to GAPDH levels.
[14C]palmitate oxidation rates in isolated liver explants were indistinguishable between wild-type and Iqgap2-/- livers, and even more remarkably, a high fat diet did not affect oxidation rates in Iqgap2-/- liver (not shown), suggesting that protection from steatosis is not explained by an exaggerated hepatic liver metabolism. Based on our SREBPs data, we next hypothesized that a lack of triglyceride accumulation by Iqgap2-/- livers in mice fed a high fat diet may be due to impaired de novo lipogenesis in hepatocytes, in addition to impaired FFA uptake. To evaluate this possibility, we measured RNA transcript levels of fatty acid synthase (FAS), PPARα and phosphoenolpyruvate carboxykinase (PEPCK) in livers from wild-type and Iqgap2-/- mice by qRT-PCR (Figure 5A). We found that, compared to wild-type controls, Iqgap2-/- livers expressed 2-fold lower levels of FAS (p<0.05), while no statistically significant difference was observed in both PPARα and PEPCK mRNA levels in Iqgap2-/- liver compared to wild-type. Downregulation of FAS expression in Iqgap2-/- livers was also evident at the protein level (Figure 5B, C).
3.4. Iqgap2-/-livers display enhanced insulin sensitivity
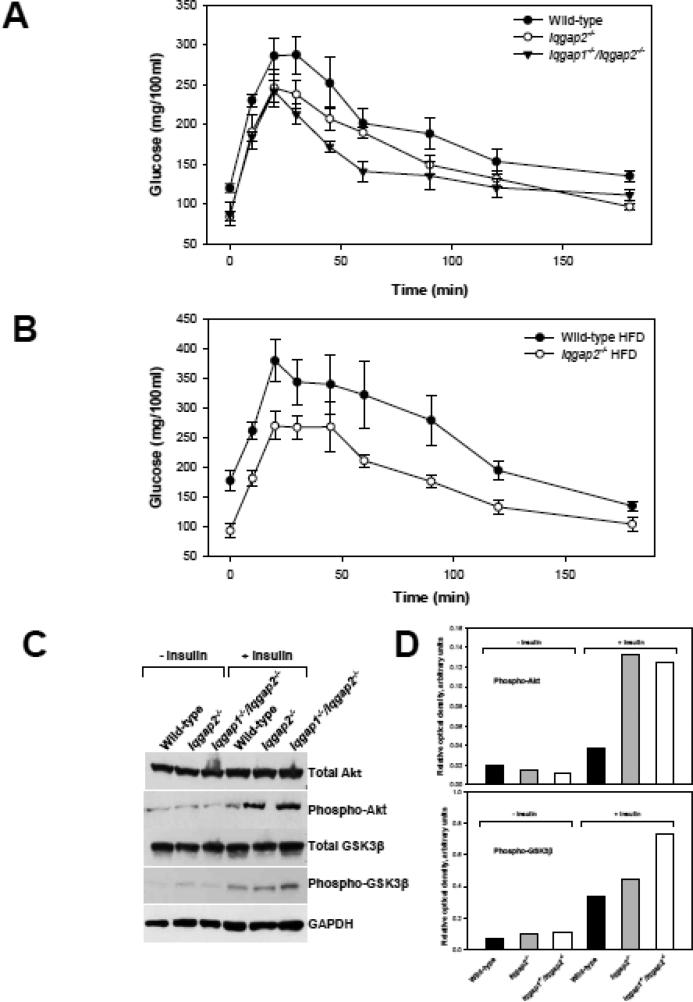
Apparent hypoglycemia of Iqgap2-/- mice indicates possible involvement of IQGAP2 in insulin signaling. To test for insulin sensitivity, a glucose tolerance test was conducted using 6-month-old male mice fed a regular diet and fasted overnight. It confirmed a marked increase in glucose tolerance of both Iqgap2-/- and Iqgap1-/-/ Iqgap2-/- (Figure 6A). Mice deficient in both Iqgap1and Iqgap2 genes were generated in this laboratory by interbreeding of Iqgap1-/- [38] and our Iqgap2-/- mice, and their phenotype was described previously [7]. Similarly to Iqgap2-/- mice, Iqgap1-/-/ Iqgap2-/- mice show hypoglycemia (an average non-fasted glucose is 98.88 7.97 mg/dl) and levels of insulin comparable to wild-type (0.603±0.071 μg/l, p = 0.099). Moreover, unlike in wild-type mice, a high fat diet did not have an effect on glucose clearance rate in Iqgap2-/- mice (Figure 6B). To further explore Iqgap2-/- metabolic phenotype, insulin stimulation experiments were performed with wild-type, Iqgap2-/- and Iqgap1-/-/Iqgap2-/- mice. Recent data [39] report that hepatic Akt2 kinase is a requisite component in the development of steatosis in insulin resistant mice. We demonstrate here that both Iqgap2-/- and Iqgap1-/-/Iqgap2-/- mice had elevated (~3-fold) hepatic levels of the phosphorylated (at Ser473) form of Akt kinase after insulin stimulation compared to wild-type (Figure 6C, D). The levels of the phosphorylated (at Ser9) form of GSK3β, a substrate of Akt, were elevated in these mice as well, and the difference was especially pronounced in Iqgap1-/-/Iqgap2-/- livers (2-fold), suggesting that in IQGAP2-deficiency, GSK3β kinase activity is inhibited.
Figure 6.
Enhanced insulin sensitivity of IQGAP2-deficient liver. (A) Glucose tolerance test (GTT) using 6-month-old male wild-type, Iqgap2-/- and Iqgap1-/-/Iqgap2-/- mice fed regular diet and fasted overnight. Glucose (2 g/kg) was administered as a single intraperitoneal injection and blood samples were collected from a tail vein at the specified times. Data are presented as mean SEM, n = 5 in each group. The graph is representative of two independent experiments using different mice. (B) GTT using 6-month-old male wild-type and Iqgap2-/- mice fed high fat diet (HFD) for 8 weeks and fasted overnight prior test. Data presented as mean SEM, n = 5 in each group. The graph is representative of three independent experiments using different mice. (C) Immunoblot analysis of hepatic total levels and the phosphorylated forms of Akt (Ser473) and GSK3β (Ser9) in response to insulin stimulation in 6-month-old male wild-type, Iqgap2-/- and Iqgap1-/-/Iqgap2-/- mice. The blot is representative of four independent studies. (D) Densitometric quantification of hepatic levels of phospho-Akt and phospho- GSK3β in response to insulin stimulation normalized to GAPDH levels.
4. DISCUSSION
This work establishes a novel, physiologically- relevant function for IQGAP2 in selective regulation of the facilitated phase of hepatic fatty acid uptake, with implications for the molecular regulation of fat partitioning. Evidence in support of this conclusion is predicated both on studies of LCFA uptake in vitro and correlative in vivo studies in diet-modified mice. Cellular LCFA processing is complex and regulated at multiple levels, but is clearly dependent on the extracellular composition and concentration of FFAs bound to serum albumin, coupled with steps controlling uptake, membrane translocation, cytoplasmic diffusion, and metabolic targeting [40]. Studies in hepatocytes, adipocytes, and cardiac myocytes have demonstrated that transmembrane FFA diffusion occurs in conjunction with a saturable phase that exhibits kinetic properties consistent with facilitated transport [41-42]. At physiologic ranges of albumin and unbound FFAs [≤ 50 – 500 nM [12]], the T½ for saturable uptake (~ 1 second) is less than that for passive uptake (~14 – 100 seconds), resulting in >90% of hepatocellular FFA uptake occurring through the saturable pathway [12]. An expanding family of FATPs (fatp1-fatp6) [11] and FABPs (fabp1-fabp10) [23] regulate the facilitated pathway of FFA uptake, with evidence for tissue-restricted expression among various members and within species. Although FATP2 and FATP5 are generally regarded as important for hepatocyte-mediated FFA uptake, recent data would suggest that liver expresses a wider repertoire of FATPs (FATP1 and FATP4), in addition to FAT/CD36 [43]. The demonstration that isolated IQGAP2 deficiency selectively affects hepatic FFA uptake identifies IQGAP2 as an independent target irrespective of the expression pattern(s) of previously characterized receptors and cytoplasmic proteins known to be important for FFA uptake and/or transport. Finally, although IQGAP2 is clearly the predominant homologue found in hepatocytes, our data suggest that IQGAP1 is reciprocally induced in Iqgap2-/- livers [7]. While the IQGAP1 induction may be subtle, ongoing studies in Iqgap1-/-/Iqgap2-/- mice [7] should clarify whether the two homologues have redundant, cooperative, or exclusive functions in regulating FFA uptake.
To date, targeted deletion of fatp1 [9], fatp4 [14], FAT/CD36 [44], and fabp1 [18, 43] genes have been described [9, 14], allowing for initial comparison with the Iqgap2-/- phenotype. Neonatal death of fatp4-/- mice was accompanied by a lethal restrictive dermopathy with no defects in adipocyte differentiation or organ lipid composition, although it remains unclear if the neonatal death obscured the development of an associated defect of fatty acid metabolism. Interestingly, fatp1-/- mice were protected against fat-induced insulin resistance in skeletal muscle, although there was no evidence for impaired fat partitioning to adipose tissue when subjected to either regular or high-fat diet; detailed fatty acid uptake studies were not described [9]. Nonetheless, given the apparent redundancy in hepatic FATP expression [43], a selective defect in FFA uptake would not be anticipated. FAT/CD36-/- mice demonstrate increased levels of fasting cholesterol, nonesterified FFAs, and attenuated [3H]-oleate uptake in adipocytes [44], although the hepatic triglyceride content was increased [45]. This is distinct from the Iqgap2-/- phenotype.
In contrast, the fabp1-/- mice share similarities with Iqgap2-/- mice. Comparable to our murine model, fabp1-/- mice demonstrate no changes or alterations in organ and tissue morphology [18, 43], with similar evidence for limited hepatic triglyceride accumulation in a model of enhanced lipolysis [43]. Moreover, isolated hepatocytes from fabp1-/- mice displayed defective [3H]oleate uptake, however the difference was more pronounced by 45 seconds than at 15 seconds [43]. We would speculate that both FABP1 and IQGAP2 may function in a comparable cytoplasmic pathway of hepatic FFA uptake, plasma membrane desorption, or cytosolic trafficking.
How does IQGAP2 regulate FFA uptake? Despite clear evidence supporting an active-transport mechanism of cellular FFA uptake, there is a paucity of evidence (either for or against) that this process is coupled to a signal transduction mechanism [40]. As extrapolated from studies focusing on IQGAP1, it is likely that IQGAP2 functions to integrate intracellular signaling pathways with cell-surface events, although these have been largely studied within the context of cellular adhesion and motility. IQGAP1's interaction with CLIP-170 regulates cell polarization via microtubule reorganization, a process that occurs downstream of [GTP]-charged Rac1 and/or cdc42 [46]. Similarly, IQGAP1's modulatory function on E-cadherin homophilic adhesion may be dually regulated both by Rho-GTPases [47] and calmodulin/Ca2+-signaling [48]. While less is known about the function of IQGAP2, recent data would suggest that its downstream effects in human blood platelets may be [GTP]Rac1-restricted, to the exclusion of [GTP]cdc42 [3]. To date, there is no evidence that cellular FFA uptake is regulated by Rho-GTPases; furthermore, both IQGAPs lack the critical arginine finger residue within their RasGAP homology domain known to be necessary for GAP activity [5, 49-50]. Rather, IQGAP1 and IQGAP2 appear to uniquely interact with cdc42 and Rac1 to inhibit their intrinsic and RhoGAP-stimulated GTPase activities [49, 51-52], a process that may prolong effector functions if IQGAPs are subsequently shown to regulate FFA transport and processing to mitochondria.
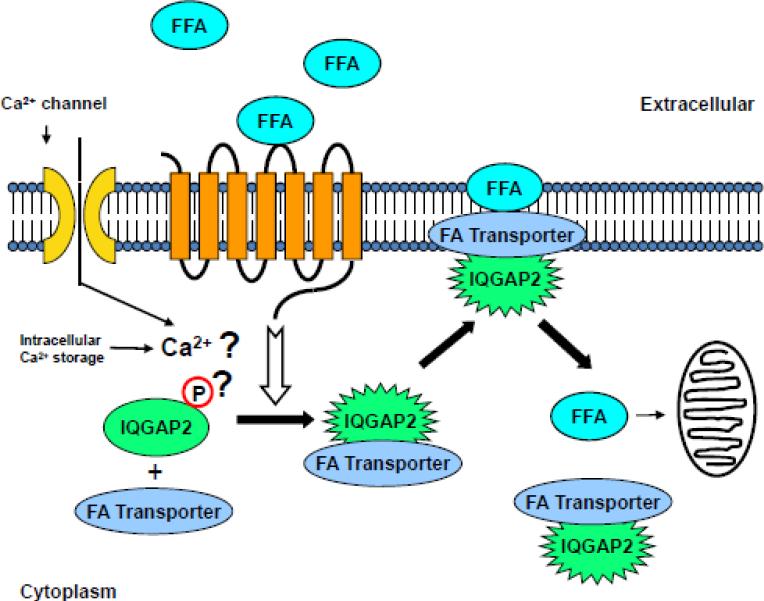
With relevance to FFA uptake, data using an enteroendocrine cell line (STC-1) have suggested that LCFA's (≥C12) can directly stimulate cytosolic calcium release ([Ca2+]i) from intracellular stores, with a more sustained [Ca2+]i response in the presence of extracellular calcium [53]. Also, the rapid onset of the [Ca2+]i release conforms to the timing of a signal required for facilitated FFA uptake [54]. Interestingly, the mechanism of [Ca2+]i release in STC-1 cells appeared distinct from G protein coupled-, tyrosine kinase-, or IP3 (inositol trisphosphate)-dependent pathways, suggesting a unique mechanism of LCFA-induced [Ca2+]i mobilization. We speculate that conformational change in IQGAP2 may be induced by such events as Ca2+ interaction with IQGAP2's calmodulin-binding domains (IQ) or IQGAP2 phosphorylation, providing the requisite molecular engine for effector functions, possibly inducing cellular FFA uptake and/or transport (Figure 7). The phosphorylation scenario may be more likely based on the recent compelling evidence for both IQGAP1 and IQGAP2. It has been shown that IQGAP1 can undergo signal-induced conformational changes and the process is regulated by phophorylation at Ser1443 [55-56]. Similarly, the cAMP-dependent protein kinase (PKA) is capable of phosphorylating IQGAP2 at Thr716, and this in turn enhances IQGAP2' binding ability to the active form of Rac1 GTPase [57]. We hypothesize that at the surface of hepatocyte, FFA bind a receptor (possibly a member of the GPR family) and trigger a signal for conformational change in IQGAP2, which, in turn, serves as a scaffolding protein binding a FA transporter (yet to be identified), thereby facilitating FA cellular entry. Based on data in fabp1-/- mouse [43], FABP1 protein seems one of plausible candidates for such transporter.
Figure 7.
Schema describing the proposed IQGAP2-mediated mechanism of facilitated FFA uptake by hepatocytes. Conformational change of the IQGAP2 protein in response to a signaling cue (possibly Ca2+ or phosphorylation) is shown.
IQGAP2 may be also involved in transcriptional regulation of lipid metabolism signaling pathways. We have demonstrated that Iqgap2-/- livers have diminished levels of both SREBP-1c and FAS, suggesting decreased levels of hepatic de novo lipogenesis. In addition to impaired FA uptake, this might be another factor contributing to the protection of these mice from hepatic steatosis. While presently it is unclear how IQGAP2 may take part in transcriptional regulation, evidence for IQGAP1 suggests a possibility of regulating protein synthesis and membrane traffic through binding to cdc42 and mTOR [58]. Recently, nuclear localization of IQGAP1 has been confirmed in various cell types where it is reported to regulate DNA replication and cell cycle progression [59].
Lastly, improved glucose tolerance and enhanced hepatic insulin sensitivity of Iqgap2-/- mice suggest IQGAP2' involvement in glucose homeostasis. While there is not enough evidence to speculate about the mechanism of such involvement, it will be of interest to evaluate hepatic glycogenesis, glucose disposal and also insulin sensitivity of other peripheral tissues in Iqgap2-/- mice. The present study demonstrates that IQGAP2-deficiency results in a complex metabolic phenotype, affecting hepatic free fatty acid uptake, glucose metabolism and also possibly de novo fatty acid synthesis and lipogenesis. While IQGAP2 is expressed predominantly in the liver and the current research presents hepatic aberrations caused by IQGAP2 disruption, it seems feasible that IQGAP2' role in other key metabolic tissues should be evaluated. While Iqgap2-/- mice displayed increased intra-abdominal adiposity when fed high fat diet, their plasma triglyceride levels and glucose remained low compared to the wild-type controls. We propose that low levels of plasma triglycerides in these mice can be explained by several factors, including enhanced insulin sensitivity, impaired hepatic de novo lipogenesis and, as a result, decreased hepatic triglyceride secretion, combined with an improved ability of the adipose tissue to store fat. The latter may be due to a possible increase in FFA uptake or decreased FA oxidation in Iqgap2-/- adipocytes, a notion which will require further studies. While it is well established that high intratissue triglycerides can induce states of insulin resistance, it is clearly not the case in Iqgap2-/- mice and other, yet unidentified, factors may be at play in compensating for the effect of the increased adiposity. It is also of interest to further evaluate the metabolic phenotype of Iqgap1-/-/ Iqgap2-/- mice to determine an understudied role of IQGAP1in lipid and glucose metabolism.
An array of emerging evidence implicates disruptions in lipid metabolism among causative factors in HCC development and progression. It is estimated that up to 30% of all HCC cases are associated with obesity and related metabolic diseases [60-61]. There is also experimental evidence in mouse models showing that a high fat diet alone induces suseptibility to HCC [62]. We have demonstrated for the first time a link between IQGAP2 and HCC development using Iqgap2-/- mice [7]. Here we show that these mice are also protected from hepatic steatosis. We believe this is the first evidence verifying that not only hepatic steatosis, but also perturbations in hepatic fatty acid uptake and lipogenesis may lead to HCC. Although we cannot exclude the possibility that IQGAP2 realizes its functions as a tumor suppressor and a regulator of lipid metabolism via distinct signaling pathways, there may be points of convergence. Thus, variable rates of fatty acid uptake have been shown for different HCC tumors [63], as have fluctuations in FAS expression, an established target for anti-cancer therapy [64]. Further research is needed to identify signaling pathways mediating hepatic lipid metabolism in the setting of IQGAP2 deficiency.
5. CONCLUSION
We report that Iqgap2-/- mice exhibit impaired facilitated phase of hepatic palmitate uptake and absence of diet-induced hepatic steatosis. Our findings point toward a novel role of IQGAP2 in fatty acid transport and, possibly, lipid metabolism. We also demonstrate enhanced hepatic insulin sensitivity associated with Iqgap2-deficiency. These findings may be relevant to both obesity and diabetes research.
Highlights.
Putative GTPase-activating protein IQGAP2 has a novel role in fatty acid metabolism
Iqgap2-deficient mice are protected against diet-induced fatty liver
Iqgap2-/- hepatocytes display a loss of the facilitated phase of fatty acid uptake
ACKNOWLEDGEMENTS
We thank Lesley Scudder (Stony Brook University) for expert technical assistance, Dr. Encarnación Capilla (University of Barcelona) for help with insulin studies, and Dr. Nada Abumrad (Washington University, St. Louis) for insightful discussions. Grant support: This work was supported by NIH grants DK62040 (VAS) and HL49141 (WFB) and an American Cancer Society Research Scholar Award (VAS). The funders had no role in study design, data collection and analysis, decision to publish, or the preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: CSC, VAS, WFB. Performed the experiments: CSC, JFL, VAS. Analyzed the data: CSC, VAS. Co-wrote the manuscript: VAS, WFB. All authors have approved the article in its final form.
REFERENCES
- 1.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–24. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–4. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt VA, Scudder L, Devoe CE, Bernards A, Cupit LD, Bahou WF. IQGAP2 functions as a GTP-dependent effector protein in thrombin-induced platelet cytoskeletal reorganization. Blood. 2003;101:3021–8. doi: 10.1182/blood-2002-09-2807. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Watanabe T, Noritake J, Fukata M, Yoshimura T, Itoh N, Harada T, Nakagawa M, Matsuura Y, Arimura N, Kaibuchi K. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J Cell Sci. 2007;120:567–77. doi: 10.1242/jcs.03356. [DOI] [PubMed] [Google Scholar]
- 5.Scheffzek K, Ahmadian M, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–62. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 6.Cupit LD, Schmidt VA, Miller F, Bahou WF. Distinct PAR/IQGAP expression patterns during murine development: implications for thrombin-associated cytoskeletal reorganization. Mamm Genome. 2004;15:618–29. doi: 10.1007/s00335-004-2370-8. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt VA, Chiariello CS, Capilla E, Miller F, Bahou WF. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol. 2008;28:1489–502. doi: 10.1128/MCB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–52. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JK, Gimeno RE, Higashimori T, Kim HJ, Choi H, Punreddy S, Mozell RL, Tan G, Stricker-Krongrad A, Hirsch DJ, Fillmore JJ, Liu ZX, Dong J, Cline G, Stahl A, Lodish HF, Shulman GI. Inactivation of fatty acid transport protein 1 prevents fat-induced insulin resistance in skeletal muscle. J Clin Invest. 2004;113:756–63. doi: 10.1172/JCI18917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–4. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 11.Stahl A. A current review of fatty acid transport proteins (SLC27). Pflugers Arch. 2004;447:722–7. doi: 10.1007/s00424-003-1106-z. [DOI] [PubMed] [Google Scholar]
- 12.Berk PD, Stump DD. Mechanisms of cellular uptake of long chain free fatty acids. Mol Cell Biochem. 1999;192:17–31. [PubMed] [Google Scholar]
- 13.Sorrentino BP, Brandt SJ, Bodine D, Gottesman M, Pastan I, Cline A, Nienhuis AW. Selection of drug-resistant bone marrow cells in vivo after retroviral transfer of human MDR1. Science. 1992;257:99–103. doi: 10.1126/science.1352414. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann T, van der Hoeven F, Grone HJ, Stewart AF, Langbein L, Kaiser I, Liebisch G, Gosch I, Buchkremer F, Drobnik W, Schmitz G, Stremmel W. Mice with targeted disruption of the fatty acid transport protein 4 (Fatp 4, Slc27a4) gene show features of lethal restrictive dermopathy. J Cell Biol. 2003;161:1105–15. doi: 10.1083/jcb.200207080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faergeman NJ, DiRusso CC, Elberger A, Knudsen J, Black PN. Disruption of the Saccharomyces cerevisiae homologue to the murine fatty acid transport protein impairs uptake and growth on long-chain fatty acids. J Biol Chem. 1997;272:8531–8. doi: 10.1074/jbc.272.13.8531. [DOI] [PubMed] [Google Scholar]
- 16.Zou Z, Tong F, Faergeman NJ, Borsting C, Black PN, DiRusso CC. Vectorial acylation in Saccharomyces cerevisiae. Fat1p and fatty acyl-CoA synthetase are interacting components of a fatty acid import complex. J Biol Chem. 2003;278:16414–22. doi: 10.1074/jbc.M210557200. [DOI] [PubMed] [Google Scholar]
- 17.Schaffer JE, Lodish HF. Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell. 1994;79:427–36. doi: 10.1016/0092-8674(94)90252-6. [DOI] [PubMed] [Google Scholar]
- 18.Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M, Schroeder F, Binas B. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:21429–38. doi: 10.1074/jbc.M300287200. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, Hashimoto T, Yamamoto T. Structure and regulation of rat long-chain acyl-CoA synthetase. J Biol Chem. 1990;265:8681–5. [PubMed] [Google Scholar]
- 20.Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J Biol Chem. 1993;268:17665–8. [PubMed] [Google Scholar]
- 21.Hirsch D, Stahl A, Lodish HF. A family of fatty acid transporters conserved from mycobacterium to man. Proc Natl Acad Sci U S A. 1998;95:8625–9. doi: 10.1073/pnas.95.15.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gimeno RE, Ortegon AM, Patel S, Punreddy S, Ge P, Sun Y, Lodish HF, Stahl A. Characterization of a heart-specific fatty acid transport protein. J Biol Chem. 2003;278:16039–44. doi: 10.1074/jbc.M211412200. [DOI] [PubMed] [Google Scholar]
- 23.Haunerland NH, Spener F. Fatty acid-binding proteins--insights from genetic manipulations. Prog Lipid Res. 2004;43:328–49. doi: 10.1016/j.plipres.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Storch J, Veerkamp JH, Hsu KT. Similar mechanisms of fatty acid transfer from human anal rodent fatty acid-binding proteins to membranes: liver, intestine, heart muscle, and adipose tissue FABPs. Mol Cell Biochem. 2002;239:25–33. [PubMed] [Google Scholar]
- 25.Yu LR, Zeng R, Shao XX, Wang N, Xu YH, Xia QC. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis. 2000;21:3058–68. doi: 10.1002/1522-2683(20000801)21:14<3058::AID-ELPS3058>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Prows DR, Murphy EJ, Schroeder F. Intestinal and liver fatty acid binding proteins differentially affect fatty acid uptake and esterification in L-cells. Lipids. 1995;30:907–10. doi: 10.1007/BF02537481. [DOI] [PubMed] [Google Scholar]
- 27.Gnatenko DV, Dunn JJ, McCorkle SR, Weissmann D, Perrotta PL, Bahou WF. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101:2285–93. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- 28.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 29.Gnatenko DV, Cupit LD, Huang EC, Dhundale A, Perrotta PL, Bahou WF. Platelets express steroidogenic 17beta-hydroxysteroid dehydrogenases. Distinct profiles predict the essential thrombocythemic phenotype. Thromb Haemost. 2005;94:412–21. doi: 10.1160/TH05-01-0037. [DOI] [PubMed] [Google Scholar]
- 30.Bahou WF, Scudder L, Rubenstein D, Jesty J. A shear-restricted pathway of platelet procoagulant activity is regulated by IQGAP1. J Biol Chem. 2004;279:22571–7. doi: 10.1074/jbc.M402561200. [DOI] [PubMed] [Google Scholar]
- 31.Seglen PO. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 32.Liao J, Sportsman R, Harris J, Stahl A. Real-time quantification of fatty acid uptake using a novel fluorescence assay. J Lipid Res. 2005;46:597–602. doi: 10.1194/jlr.D400023-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Kampf JP, Kleinfeld AM. Fatty acid transport in adipocytes monitored by imaging intracellular free fatty acid levels. J Biol Chem. 2004;279:35775–80. doi: 10.1074/jbc.M403630200. [DOI] [PubMed] [Google Scholar]
- 34.Ho YD, Joyal JL, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem. 1999;274:464–70. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 35.Bashour A, Fullerton A, Hart MJ, Bloom GS. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links actin. J Cell Biol. 1997;137:1555–66. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyal JL, Annan RS, Ho YD, Huddleston ME, Carr SA, Hart MJ, Sacks DB. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J Biol Chem. 1997;272:15419–25. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- 37.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Wang Q, Chakladar A, Bronson R, Bernards A. Gastric hyperplasia in mice lacking the putative Cdc42 effector IQGAP1. Mol.Cell Biol. 2000;20:697–701. doi: 10.1128/mcb.20.2.697-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leavens KF, Easton RM, Shulman GI, Previs SF, Birnbaum MJ. Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell Metab. 2009;10:405–18. doi: 10.1016/j.cmet.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res. 1999;40:1371–83. [PubMed] [Google Scholar]
- 41.Weisiger R, Gollan J, Ockner R. Receptor for albumin on the liver cell surface may mediate uptake of fatty acids and other albumin-bound substances. Science. 1981;211:1048–51. doi: 10.1126/science.6258226. [DOI] [PubMed] [Google Scholar]
- 42.Abumrad NA, Perkins RC, Park JH, Park CR. Mechanism of long chain fatty acid permeation in the isolated adipocyte. J Biol Chem. 1981;256:9183–91. [PubMed] [Google Scholar]
- 43.Newberry EP, Xie Y, Kennedy S, Han X, Buhman KK, Luo J, Gross RW, Davidson NO. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:51664–72. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- 44.Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SF, Silverstein RL. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–62. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 45.Hajri T, Han XX, Bonen A, Abumrad NA. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J Clin Invest. 2002;109:1381–9. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–85. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 47.Fukata M, Kuroda S, Nakagawa M, Kawajiri A, Itoh N, Shoji I, Matsuura Y, Yonehara S, Fujisawa H, Kikuchi A, Kaibuchi K. Cdc42 and rac1 regulate the interaction of IQGAP1 with beta-catenin. J Biol.Chem. 1999;274:26044–50. doi: 10.1074/jbc.274.37.26044. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Kim SH, Higgins JM, Brenner MB, Sacks DB. IQGAP1 and calmodulin modulate E-cadherin function. J Biol Chem. 1999;274:37885–92. doi: 10.1074/jbc.274.53.37885. [DOI] [PubMed] [Google Scholar]
- 49.Brill S, Li S, Lyman C, Church D, Wasmuth J, Weissbach L, Bernards A, Snijders A. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol.Cell Biol. 1996;16:4869–78. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector. EMBO J. 1996;15:2997–300. [PMC free article] [PubMed] [Google Scholar]
- 51.Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol.Chem. 1996;271:23363–7. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- 52.McCallum S, Wu W, Cerione R. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol.Chem. 1996;271:21732–7. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- 53.Hira T, Elliott AC, Thompson DG, Case RM, McLaughlin JT. Multiple fatty acid sensing mechanisms operate in enteroendocrine cells: novel evidence for direct mobilization of stored calcium by cytosolic fatty acid. J Biol Chem. 2004;279:26082–9. doi: 10.1074/jbc.M400098200. [DOI] [PubMed] [Google Scholar]
- 54.Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–25. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 55.Grohmanova K, Schlaepfer D, Hess D, Gutierrez P, Beck M, Kroschewski R. Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of rho-GTPase regulator. J Biol Chem. 2004;279:48495–504. doi: 10.1074/jbc.M408113200. [DOI] [PubMed] [Google Scholar]
- 56.Rittmeyer EN, Daniel S, Hsu SC, Osman MA. A dual role for IQGAP1 in regulating exocytosis. J Cell Sci. 2008;121:391–403. doi: 10.1242/jcs.016881. [DOI] [PubMed] [Google Scholar]
- 57.Logue JS, Whiting JL, Tunquist B, Langeberg LK, Scott JD. Anchored PKA recruitment of active RAC. J Biol Chem. 2011 doi: 10.1074/jbc.M111.232660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang JB, Sonn R, Tekletsadik YK, Samorodnitsky D, Osman MA. IQGAP1 regulates cell proliferation through a novel CDC42-mTOR pathway. J Cell Sci. 2009;122:2024–33. doi: 10.1242/jcs.044644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson M, Sharma M, Brocardo MG, Henderson BR. IQGAP1 translocates to the nucleus in early S-phase and contributes to cell cycle progression after DNA replication arrest. Int J Biochem Cell Biol. 2011;43:65–73. doi: 10.1016/j.biocel.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 60.Yang S, Lin HZ, Hwang J, Chacko VP, Diehl AM. Hepatic hyperplasia in noncirrhotic fatty livers: is obesity-related hepatic steatosis a premalignant condition? Cancer Res. 2001;61:5016–23. [PubMed] [Google Scholar]
- 61.Bugianesi E. Non-alcoholic steatohepatitis and cancer. Clin Liver Dis. 2007;11:191–207. x–xi. doi: 10.1016/j.cld.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Hill-Baskin AE, Markiewski MM, Buchner DA, Shao H, DeSantis D, Hsiao G, Subramaniam S, Berger NA, Croniger C, Lambris JD, Nadeau JH. Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum Mol Genet. 2009;18:2975–88. doi: 10.1093/hmg/ddp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yun M, Bang SH, Kim JW, Park JY, Kim KS, Lee JD. The importance of acetyl coenzyme A synthetase for 11C-acetate uptake and cell survival in hepatocellular carcinoma. J Nucl Med. 2009;50:1222–8. doi: 10.2967/jnumed.109.062703. [DOI] [PubMed] [Google Scholar]
- 64.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–8. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]