Abstract
MicroRNAs have recently been identified as important regulators of gene expression at the post-transcriptional level. While it has clearly been established that microRNAs influence the ontogeny of several immune cell lineages, the role of individual microRNAs during natural killer (NK) cell development has not been described. Here, we show that miR-181 expression levels have a profound impact on the development of human NK cells from CD34+ hematopoietic progenitor cells (HPC) and IFN-γ production in primary CD56+ NK cells. We also demonstrate that nemo-like kinase (NLK), an inhibitor of Notch signaling, is a target of miR-181 in NK cells, and knockdown of NLK mirrors the developmental effect of miR-181 over-expression. We conclude that miR-181 promotes NK cell development, at least in part, through the suppression of NLK, providing an important link between microRNAs and Notch signaling.
Introduction
MicroRNAs constitute a highly conserved family of small non-coding RNAs that regulate the translation and/or stability of mRNAs, thus influencing gene expression at the post-transcriptional level (1). MicroRNAs affect immune cell differentiation, inflammatory responses to infections and the development of immune-mediated disease (2). Several individual microRNAs with critical roles in the development of mouse T and B cell subsets have recently been described (3,4,5). While it is known that miR-223 regulates granzyme B translation during murine NK cell activation (6), and the NK cell compartment is reduced in mice with conditional knockouts of enzymes involved in the biogenesis of miRNAs (7), the contributions of individual microRNAs to NK cell development have not yet been reported.
miR-181a and miR-181b are the most well-studied members of the miR-181 family and are clustered together on two genomic locations on chromosomes 1 and 9. The mature transcripts for miR-181a and miR-181b are highly homologous with only three base pair differences distinguishing them (www.microrna.org). Given its established role in T and B cell development, we hypothesize that miR-181 may also regulate human NK cell development.
Materials and Methods
NK cell and CD34+ hematopoietic progenitor cell isolation
The use of all human tissue was approved by the Committee on the Use of Human Subjects in research at the University of Minnesota (Minneapolis, MN), and informed consent was obtained in accordance with the Declaration of Helsinki. NK cells were magnetically isolated from peripheral blood through negative selection (STEMCELL Technologies, Vancouver, BC). CD34+ hematopoietic progenitor cells (HPCs) were isolated from umbilical cord blood by double-column positive selection using anti-CD34 microbeads (Miltenyi Biotech, Auburn, CA).
Retroviral transductions and in vitro NK cell cultures
Scramble microRNA, full-length precursor miR-181a/b, miR-181a antisense knockdown, miR-181b antisense knockdown (System Biosciences, Mountain View, CA), siRNA scramble and NLK siRNA (Open Biosystems, Huntsville, AL) lentiviral vectors were used to transduce purified CD34+ cells. GFP+ cells were isolated by FACS and cultured for 28 days on the EL08-1D2 stromal line (9). The culture media and cytokines for NK cell differentiation are published (10). microRNA vectors were also used to transduce peripheral blood NK cells and NK92 cells. GFP+ cells were isolated by FACS. Peripheral NK cells were cultured for 14 days in the presence of 10 ng/ml IL-15 (R&D Systems, Minneapolis, MN). NK92 cells were cultured in alpha medium containing 12.5% fetal calf serum, 12.5% horse serum (HyClone Laboratories, Logan, UT), 0.1 mM β-mercaptoethanol, 100 U/mL penicillin, 100 U/mL streptomycin (Invitrogen, Carlsbad, CA), and 500 U/mL recombinant human IL-2 (Chiron, Emeryville, CA).
Functional assay for IFN-γ production in NK cells
Transduced peripheral blood NK cells were treated overnight with or without 1 ug/ml IL-12 and 10 ug/ml IL-18. Cells were then analyzed for the expression of GFP, CD56 and IFN-γ.
Flow cytometry and Abs used
Cell sorting was performed on the FACSAria (BD Biosciences, Franklin Lakes, NJ), and phenotypic analysis was performed on the FACSCalibur (BD Biosciences) or LSRII (BD Biosciences) using CellQuest Pro Software (BD Biosciences). The Abs used in this study were allophycocyanin (APC)-conjugated NCAM13.2 (CD56) and CD34, PE-conjugated CD56, NKG2A and CD16, APC-Cy7-conjugated CD117, FITC-conjugated CD94 (BD Biosciences), and Pacific Blue-conjugated IFN-γ (BioLegend, San Diego, CA).
Quantitative RT-PCR
For microRNA qRT-PCR, cDNA was synthesized with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA), and primer/probe sets for miR-181a, miR-181b and RNU-24 were purchased from Applied Biosystems. qRT-PCR for Hes5 and NLK were performed with the SYBR Green reagent (Applied Biosystems). All primer/probe sets were purchased from Applied Biosystems except for NLK. NLK forward primer: 3’ CAGCCATATTTCCATCACC 5’ NLK reverse primer: 3’GACAACACCAAAGGCTCCAT 5’.
Western blot analysis
Western blots for NLK protein were performed with a rabbit anti-NLK polyclonal antibody (Millipore, Billerica, MA) at a 1:5000 dilution in 3% milk w/v overnight. A donkey ECL anti-rabbit IgG, horseradish peroxidase-linked whole antibody (GE Healthcare, Buckinghamshire, UK) was used for the secondary staining at a 1:500 dilution in 3% milk for 3 hours. Densitometric analysis was performed using an Alphaimager Gel Documentation system.
Results
miR-181 levels determine the efficiency of human NK cell development
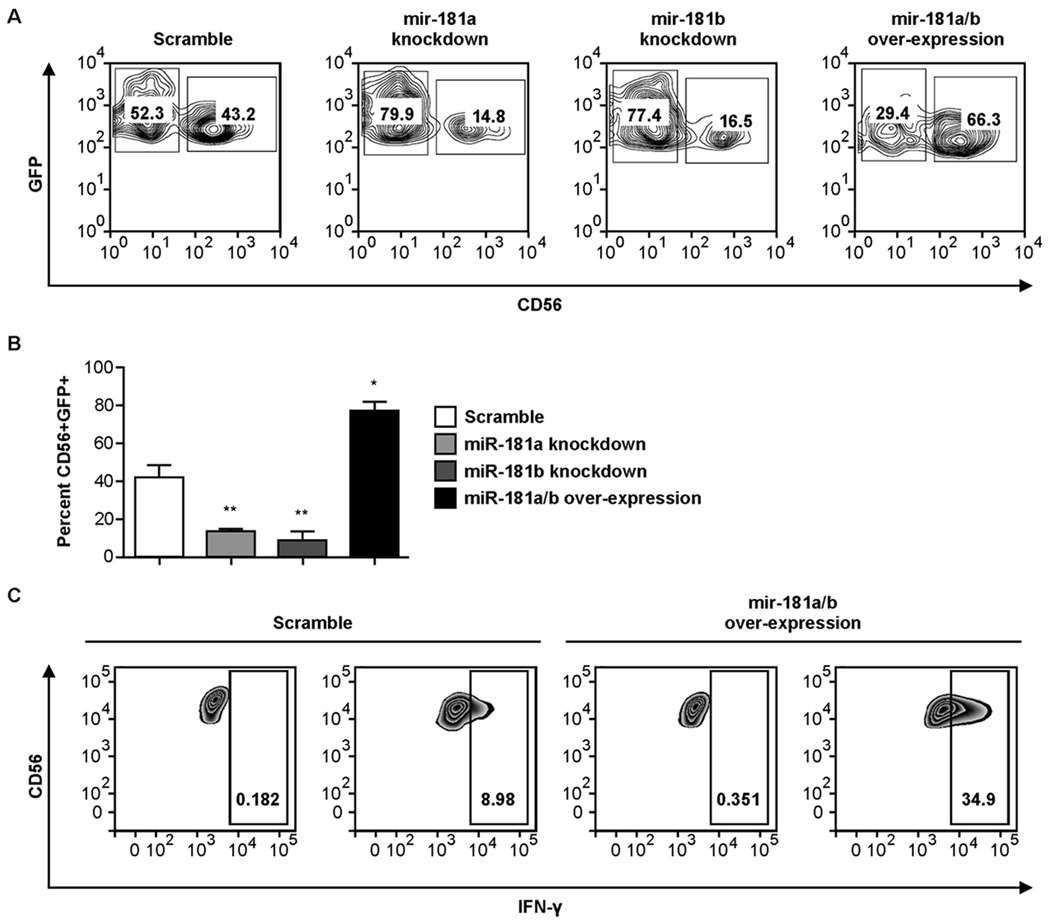
Validated scramble, miR-181a and miR-181b knockdown, and miR-181a/b over-expression lentiviral constructs were transduced separately into primary CD34+ hematopoietic progenitor cells (HPCs), which were subsequently differentiated along the NK cell lineage for 28 days. Knockdown of either miR-181a or miR-181b led to an ~3-fold decrease in the percentage of mature CD56+ NK cells. In contrast, over-expression of miR-181a/bresulted in a 2-fold increase in CD56+ NK cells at day 28 compared to scramble controls (Fig. 1A, 1B). A similar defect in NK cell development was seen when analyzing other markers of differentiated NK cells (NKG2A and CD16) in miR-181a and miR-181b knockdown cultures (Supplemental Fig. 1). These results were not due to differences in proliferation between cultures, as the total cell counts throughout development were comparable (Supplemental Fig 2B). To confirm that the results observed in Fig. 1 were not simply due to miR-181a or miR-181b affecting the expression of the CD56 protein, we transduced primary CD56+ cells from the peripheral blood of healthy donors with each expression construct and cultured the cells for 14 days. Knockdown or over-expression of miR-181 had no impact on CD56 expression, proliferation or survival in these cells (Supplemental Fig. 2A). Collectively, these results suggest that the expression of miR-181a and miR-181b are important for the differentiation of precursors through the NK cell lineage.
Figure 1. The expression levels of miR-181a and miR-181b determine the efficiency of human NK cell development from CD34+ progenitors and IFN-γ production by primary CD56+ peripheral blood NK cells.
(A) Purified CD34+ cells were transduced with lentiviral vectors over-expressing scramble microRNA, miR-181a antisense knockdown RNA, miR-181b antisense knockdown RNA or full-length precursor miR-181a/b RNA and cultured for 28 days in NK cell differentiation conditions. Representative FACS plots of CD56 expression in GFP+ cells are shown. (B) Cumulative data showing CD56 expression in GFP+ cells at day 28 after each set of transductions. Results are shown as the mean CD56 expression of 4 donors in two separate experiments with 2 donors in each experiment with error bars representing SEM. Statistically significant differences from the scramble control were determined by one-way ANOVA (*p<0.05, **p<0.01). (C) GFP-gated FACS plots of intracellular IFN-γ production in CD56+ peripheral blood NK cells transduced with lentiviral vectors over-expressing scramble microRNA or full-length precursor miR-181a/b RNA and stimulated IL-12/18. Plots are representative of the analysis of three healthy donors.
Over-expression of miR-181a/b enhances IFN-γ production in primary CD56+ NK cells
The observation that over-expression of miR-181a/b promotes NK cell development led us to hypothesize that miR-181a/b could also enhance NK cell function. To test this hypothesis, we transduced primary peripheral blood CD56+ NK cells with the scramble control or the miR-181a/b over-expression vector. Over-expression of miR-181a/b significantly enhanced IFN-γ production by NK cells in response to cytokine stimulation compared to scramble controls (58 ± 12% SEM vs. 13 ± 2% SEM, p = 0.003) (Fig. 1C). In this setting the enhanced function is analogous to the enhanced activation seen in T-cells where miR-181 targets phosphatases (11).
miR-181a and miR-181b transcript levels increase during NK cell development
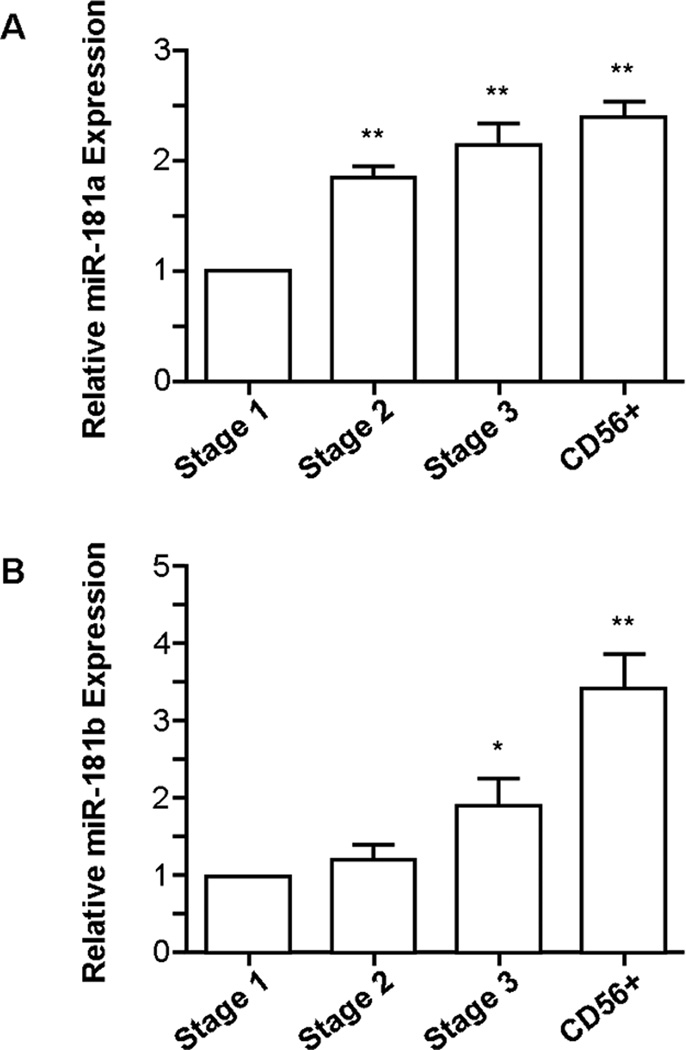
We isolated stage 1 NK cell progenitors (CD34+CD117−CD94−CD56−), stage 2 progenitors (CD34+CD117+CD94−CD56−), stage 3 progenitors (CD34−CD117+CD94−CD56lo/−) and lineage-committed CD56+ NK cells from umbilical cord blood (12). Consistent with the observation that high expression levels of miR-181a or miR-181b are associated with efficient NK cell development (Fig. 1A, 1B), we observed a steady increase in miR-181a (Fig. 2A) and miR-181b (Fig. 2B) mature transcript levels throughout normal NK cell development, providing physiologic relevance to our findings.
Figure 2. The expression levels of miR-181a and miR-181b increase throughout NK cell development.
qRT-PCR analysis of (A) miR-181a and (B) miR-181b levels throughout NK cell development (n=4). Results are shown as the mean expression values normalized against stage 1. Error bars represent SEM, and statistical significance was determined by one-way ANOVA (*p<0.05, **p<0.01).
Nemo-like kinase (NLK) is a miR-181 target in NK cells
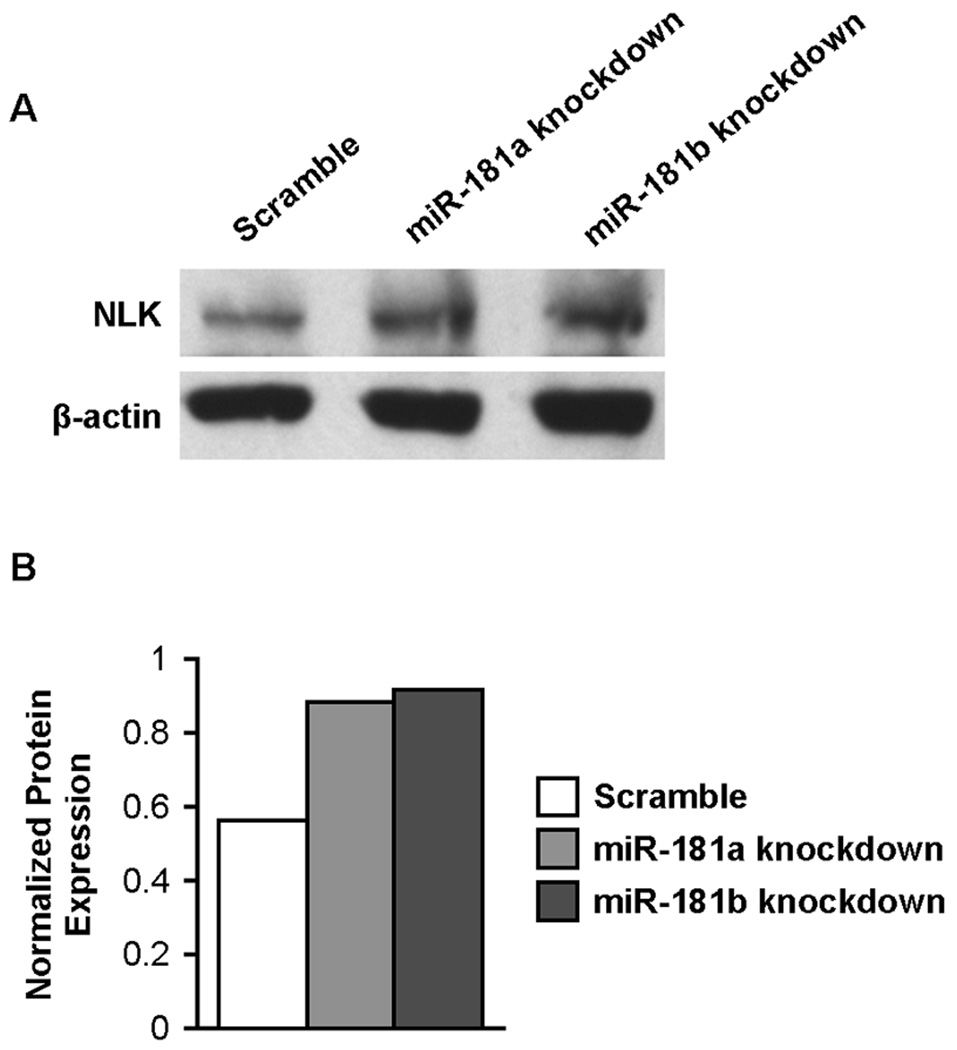
A combined database search using miRGator (http://genome.ewha.ac.kr/miRGator/miRGator.html) yielded 951 unique, annotated targets shared by miR-181a and miR-181b. We searched this list for genes with potential relevance to NK cell development and identified NLK, which has a 3’ UTR site with strong alignment scores for predicted miR-181a and miR-181b binding. NLK is an evolutionarily conserved protein kinase that negatively regulates Notch-dependent transcriptional activation by decreasing the formation of its ternary complex (13). Because we and others have shown that Notch signaling supports human NK cell development (14, 15), we hypothesized that miR-181a and miR-181b promote NK cell development by down-regulating a negative regulator of Notch signaling. To establish that NLK is a target of miR-181 in NK cells, we knocked down miR-181a and miR-181b individually in NK92 cells and measured NLK protein expression by Western blot. Knockdown of either miR-181a or miR-181b led to a substantial increase in NLK protein levels (Fig. 3A, 3B), supporting a role for miR-181 in the negative regulation of NLK expression in NK cells. miR-181 has previously been implicated in the regulation of NLK expression in hepatic cells (16).
Figure 3. Knockdown of miR-181a and miR-181b increase NLK protein levels in NK cells.
(A) Western blots of NLK and β-actin protein in sort-purified NK92 cells transduced with lentiviral vectors over-expressing scramble microRNA, miR-181a antisense knockdown RNA or miR-181b antisense knockdown RNA. (B) Densitometric analysis of NLK protein expression normalized against β-actin protein levels for each lane. Results are representative of two independent experiments.
Knockdown of NLK enhances human NK cell development
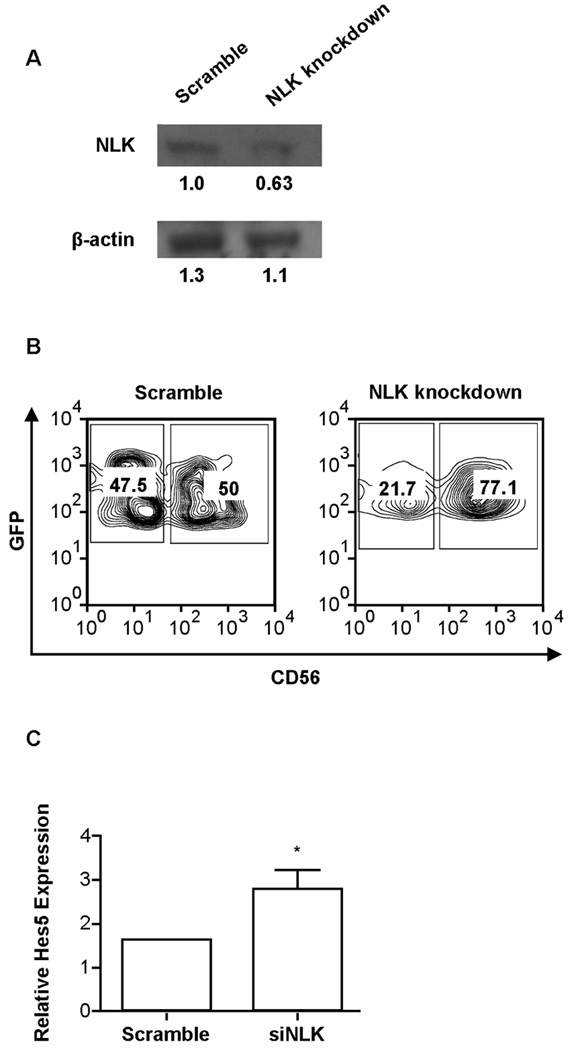
A corollary of our hypothesis that miR-181a and miR-181b promote NK cell development by targeting NLK, is that knockdown of NLK will enhance NK cell development. Following this reasoning, we used RNA interference technology to successfully knockdown NLK expression, as determined by Western blot analysis (Fig. 4A). CD34+ HPCs were transduced with either scramble siRNA or NLK siRNAlentiviral vectors and differentiated along the NK cell lineage for 28 days. Knockdown of NLK led to an increase in NK cell development compared to scramble controls as measured by the percentage of GFP+ cells that acquired CD56 in culture (74.9% ± 7.6 SEM vs. 48.1% ± 5.4 SEM, p = 0.008) (Fig. 4B), mirroring the phenotype observed with over-expression of miR-181a or miR-181b (Fig. 1A, 1B).
Figure 4. Knockdown of NLK promotes NK cell development and is associated with an increase in Hes5 expression.
(A) Western blots of NLK and β-actin protein in sort-purified HEK293T cells transduced with scramble and siNLKlentiviral vectors. Values shown below each band represent denstometric analysis of protein expression. (B) Purified CD34+ cells were transduced with lentiviral vectors over-expressing scramble siRNA or NLK siRNA and cultured in NK cell differentiation conditions for 28 days. Representative FACS plots from GFP+ cells are shown. (C) qRT-PCR analysis of Hes5 expression in NK cells from scramble siRNA and NLK siRNA in GFP-isolated CD56+ cells from day 28 cultures (n=3). Error bars represent SEM, and statistical significance was determined by one-way ANOVA (*p<0.05).
Knockdown of NLK correlates with an increase in the expression of the Notch target Hes5
To answer the question of whether NLK knockdown enhances signaling through the Notch pathway, we analyzed Hes5 transcript levels in developing NK cells expressing the siRNA targeting NLK that were sorted out of culture at day 28. Compared to scramble controls, we observed ~3-fold more Hes5 transcript in NK cells harboring the siNLK expression vector (Fig. 3F). These results support the conclusion that miR-181 influences NK development by regulating the Notch pathway.
Discussion
The development of hematopoietic progenitor cells into distinct blood lineages is controlled by a complex set of molecular events that must be coordinated to regulate proliferation, commitment and maturation simultaneously. The recent discovery of microRNAs adds a new layer to this complexity, as several microRNAs have unique expression patterns in cells of the innate and adaptive immune systems (2). Ectopic expression of miR-181a in mouse progenitor cells leads to a substantial increase in the B cell compartment (8), and high levels of miR-181a are necessary for progression through the double-positive stage of thymocyte development (17). Here, we show that miR-181 family members are also important for the development of NK cells. Therefore, miR-181 contributes to the development of all three lymphoid lineages.
Our data suggests that miR-181 promotes the development of human NK cells, at least in part, through down-regulation of NLK, which has the effect of increasing Notch activity. We have previously shown that Notch signaling is important in NK cell development (15), but miR-181 targeting of NLK points to a cell-intrinsic effect in the modulation of Notch signaling. Cell-intrinsic modulation of Notch signaling is necessary as earlier stages of NK cell development require Notch signals, but these signals are detrimental to development at later stages. Given the availability of Notch ligands in the bone marrow compartment during development, the maturing NK cells must be capable of shutting down Notch signals in a cell-intrinsic manner. In addition, we show that expression of miR-181 also enhances IFN-γ production in NK cells, possibly through regulation of the Notch pathway. The production of IFN-γ is a tightly controlled process that appears to involve multiple microRNAs including miR-29 (18).
NLK also functions within the Wnt signaling pathway to antagonize canonical Wnt/β-Catenin signaling (19), and pharmacological inhibition of this pathway inhibits NK cells development in vitro (20). We analyzed several genes that are validated targets of the Wnt pathway (21) in NK92 cells with knockdown or over-expression of miR-181 and observed a moderate effect on their expression levels that did not reach statistical significance in all cases (Supplemental Fig. 2C), which warrants further study.
This is the first report to demonstrate the importance of an individual microRNA during NK cell development. It is becoming increasingly clear that NK cell development is tied to a complex process of NK cell commitment, acquisition of NK cell receptors and ultimately the acquisition of function that will be needed to protect against infection and cancer. Our data shows that miR-181 directly regulates the developmental process at least in part through interaction with Notch pathways.
Supplementary Material
Acknowledgements
This work was supported by NIH AI50656, NIH HL55417 and P01 CA111412.
Footnotes
The authors have no financial conflicts.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 3.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehniger TA, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DTG, Lanier LL. Distinct Requirements of MicroRNAs in NK Cell Activation, Survival, and Function. J. Immunol. 2010;185:3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 9.Oostendorp RA, Harvey KN, Kusadasi N, de Bruijn MF, Saris C, Ploemacher RE, Medvinsky AL, Dzierzak EA. Stromal lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood. 2002;99:1183–1189. doi: 10.1182/blood.v99.4.1183. [DOI] [PubMed] [Google Scholar]
- 10.Cichocki F, Miller JS. In vitro development of human Killer-Immunoglobulin Receptor-positive NK cells. Methods Mol. Biol. 2010;612:15–26. doi: 10.1007/978-1-60761-362-6_2. [DOI] [PubMed] [Google Scholar]
- 11.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA. Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishitani T, Hirao T, Suzuki M, Isoda M, Ishitani S, Harigaya K, Matsumoto K, Itoh M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat. Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- 14.Beck RC, Padival M, Yeh D, Ralson J, Cooke KR, Lowe JB. The Notch ligands Jagged2, Delta1, and Delta4 induce differentiation and expansion of functional human NK cells from CD34(+) cord blood hematopoietic progenitor cells. Biol. Blood Marrow Transplant. 2009;15:1026–1037. doi: 10.1016/j.bbmt.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachanova V, McCullar V, Lenvik T, Wangen R, Peterson KA, Ankarlo DE, Panoskaltsis-Mortari A, Wagner JE, Miller JS. Activated notch supports development of cytokine producing NK cells which are hyporesponsive and fail to acquire NK cell effector functions. Biol. Blood Marrow Transplant. 2009;15:183–194. doi: 10.1016/j.bbmt.2008.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji J, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu G, Min H, Yue S, Chen CZ. Pre-miRNA loop nucleotides control the distinct activities of miR-181a-1 and miR-181c in early T cell development. PLoS One. 2008 doi: 10.1371/journal.pone.0003592. [Epub Oct. 31]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma F, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-g. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 19.Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman N, Shibuya H, Moon RT, Ninomiya-Tsuji J, Matsumoto K. The TAK1-NLK Mitogen-Activated Protein Kinase Cascade Functions in the Wnt-5a/Ca2+ Pathway To Antagonize Wnt/β-Catenin Signaling. Mol. Cell. Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grzywacz B, Kataria N, Kataria N, Blazar BR, Miller JS, Verneris MR. Natural killer-cell differentiation by myeloid progenitors. Blood. 2011;117:3548–3558. doi: 10.1182/blood-2010-04-281394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staal FJ, Weerkamp F, Baert MR, van den Burg CM, van Noort M, de Haas EF, van Dongen JJ. Wnt target genes identified by DNA micro arrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J. Immunol. 2004;172:1099–1108. doi: 10.4049/jimmunol.172.2.1099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.