Abstract
The biogenesis of ER Exit Site/Golgi Junctions (EGJ) in bloodstream form African trypanosomes is investigated using tagged markers for ER Exit Sites, the Golgi, and the bilobe structure. The typical pattern is two EGJ in G1 phase (1 kinetoplast/1 nucleus, 1K1N) through S phase (2K1N), duplication to four EGJ in post-mitotic cells (2K2N), and segregation of two EGJ to each daughter. Lesser cell percentages have elevated EGJ copy numbers in all stages, and blocking cell cycle progression results in even higher copy numbers. EGJs are closely aligned with the flagellar attachment zone (FAZ) indicating nucleation on the FAZ-associated ER (FAZ:ER). Only the most posterior EGJ in each cell is in proximity to the bilobe, which is located at the base of the FAZ filament near the mouth of the flagellar pocket. These results indicate that EGJ replication in bloodstream trypanosomes is not tightly coupled to the cell cycle. Furthermore, segregation of EGJ is not obligately mediated by the bilobe, rather assembly of the EGJ on the FAZ:ER, which is coupled to the flagellar cytoskeleton, apparently ensures segregation with fidelity during cytokinesis. These findings differ markedly from procyclic form trypanosomes, and models highlighting these stage-specific differences in EGJ biogenesis are proposed.
Keywords: Trypanosome, ER exit site (ERES), Golgi, Cell Cycle, Centrin
INTRODUCTION
The Golgi apparatus is the central secretory organelle for modification and sorting of newly synthesized protein cargo in all eukaryotes (Mellman & Simons, 1992). In recent years single cell eukaryotes have provided novel insights into the basic process of Golgi duplication and segregation (He, 2007). There are two alternate models of Golgi replication during the cell cycle (Emr et al., 2009). In the ‘templated’ model the parental Golgi enlarges and via fission generates new daughter organelles. In the ‘de novo’ model new Golgi are generated from materials supplied directly by transport from ER exit sites (ERES). Examples of each can be found in parasitic protozoa - templated biogenesis in Toxoplasma gondii and de novo biogenesis, albeit in modified form, in the African trypanosome Trypanosoma brucei (He, 2007). Both these ancient protozoan lineages have small highly ordered subcellular architectures and many of the standard organelles, such as the Golgi, are present in single or reduced copy number. Consequently, it is critical that the Golgi be replicated and segregated with fidelity to ensure the viability of each daughter cell.
Trypanosomes are highly polarized cells with unique internal organization [see (Lacomble et al., 2008, Ralston et al., 2009)]. The elongated cell shape is maintained by a corset of subpellicular microtubules, and the single flagellum emerges from the posterior end via the flagellar pocket, the sole portal of endocytic/exocytic vesicular trafficking. The flagellum is nucleated by the basal body, which is in turn connected to the kinetoplast, the concatenated circular mitochondrial genome. Externally the flagellum adheres to the cell body as it extends in the anterior direction. Beneath the plasma membrane of this flagellar adherence zone (FAZ) is a quartet of anti-parallel microtubules and a unique FAZ filament that intercalate in the subpellicular corset. Interdigitating on the interior side of the microtubule quartet is an extension of the smooth ER call the FAZ-associated ER (FAZ:ER) (Vickerman, 1969a). Cell division in trypanosomes is a highly orchestrated process, with the first events being basal body duplication and initial outgrowth of the daughter flagellum and FAZ, followed by kinetoplast (K) and then nuclear (N) replication (Woodward & Gull, 1990). Once complete sets of organelles are present these are segregated longitudinally and cytokinesis generates two daughter cells. The ordered process of mitochondrial and nuclear genome replication allows any cell in an asynchronously replicating population to be positioned within the trypanosome cell cycle by visual inspection of organellar DNA content (1K1N→2K1N→2K2N→cytokinesis).
Golgi replication in T. brucei has only been studied in the procyclic insect form (PCF) (Field et al., 2000, He et al., 2004, Ho et al., 2006, Morriswood et al., 2009). Golgi duplication in this life cycle stage is a de novo process, although the new Golgi is primarily derived from the old Golgi, as opposed to the ER as in the ‘classic’ model. Each Golgi has an associated ERES and the two replicate synchronously. Initially in G1 cells (1K1N) there is a single Golgi in close proximity to the flagellar pocket. This Golgi duplicates early in the cell cycle, following basal body replication and initial outgrowth of the new flagellum, but prior to kinetoplast segregation. Thereafter two Golgi are present throughout the 2K1N and 2K2N stages respectively of S and G2/M phase cells. Additional late Golgi fragments with associated ERES have also been observed in post-mitotic PCF trypanosome, but these apparently disappear prior to cytokinesis. The significance of these ‘phantom’ structures is uncertain.
A novel centrin-containing structure, called the bilobe, is associated with PCF Golgi and is thought to regulate its replication (He et al., 2005, Selvapandiyan et al., 2007, Shi et al., 2008). In early G1 cells the anterior end of the bilobe is associated with the single Golgi, and formation of the new Golgi nucleates at the posterior end, suggesting that the bilobe is critical for correct Golgi duplication. Thereafter the bilobe splits and one bilobe remains with each Golgi through subsequent kinetoplast replication, mitosis and cytokinesis. In support of this model RNAi silencing of TbCen2 blocks Golgi duplication (He et al., 2005), as does silencing of another bilobe component, TbLRRP1 (Zhou et al., 2010). These studies also revealed the bilobe to be associated with the posterior end of the FAZ filament, at the mouth of the flagellar pocket, suggesting it plays additional roles in flagellar biogenesis and cell division (de Graffenried et al., 2008, Shi et al., 2008, Morriswood et al., 2009, Zhou et al., 2010). Thus the bilobe may be a multifunctional structure with roles in both Golgi duplication and as a cytoskeletal adaptor for Golgi segregation in insect stage trypanosomes.
Golgi biogenesis has not been investigated in the pathogenic bloodstream form (BSF) of T. brucei, but it might be expected to follow a similar process to that in PCF trypanosomes. Nevertheless, there are distinct stage-specific differences in trypanosome morphogenesis (Matthews, 2005), including flagellar biogenesis (Briggs et al., 2004), cytoskeleton (Hertz-Fowler et al., 2001, Vedrenne et al., 2002), relative positions of kinetoplasts and nuclei in pre-cytokinesis cells (Tyler et al., 2001), and post-nuclear subcellular architecture (Matthews et al., 1995). The latter feature may be most relevant in comparing secretory organelle biogenesis. In interphase PCF trypanosomes the flagellar pocket, with associated basal bodies and kinetoplast, is positioned approximately at the midpoint between the posterior end of the cell and the centrally located nucleus, while in BSF cells these organelles are fully posterior in location. Consequently there is an increased relative volume between the nucleus and flagellar pocket to accommodate secretory and endocytic organelles, which may in turn be related to the increased dependence of BSF trypanosomes on endocytosis (Langreth & Balber, 1975, Engstler et al., 2004). Given these well-documented stage-specific differences it seems likely that additional differences in Golgi replication and segregation may exist in BSF trypanosomes.
Previously my laboratory visualized the ERES and Golgi in BSF trypanosomes using epitope tagged TbSec23.2 (TbSec23.2:HA, a COPII coat component) and TbGT15 (TbGT15:Ty, a glycosyltransferase) as respective markers (Sevova & Bangs, 2009). These preliminary investigations revealed three key features: i) as in PCF trypanosomes, the Golgi is always in close proximity to an ERES (referred to hereafter as EGJ for ERES:Golgi Junction); ii) in G1 cells (1K1N) there are typically either one (~20%) or two (~80%) EGJs, in contrast to PCF cells which typically have 1 EGJ; and iii) in 1K1N cells with two EGJs the old EGJ is positioned forward close to the nucleus, and the less prominent and presumably newer EGJ is nearer the flagellar pocket, also as seen in PCF cells. This latter feature is much more overt in BSF trypanosomes due to the more posterior location of the flagellar pocket. At the time these results suggested that the ERES and Golgi replicate in concert early in the BSF cell cycle before the initiation of kinetoplast duplication, as is the case in PCF trypanosomes. However, increased Golgi number in BSF trypanosomes has been noted by others (Field et al., 2000), albeit non-quantitatively, and data presented below suggest that this interpretation is overly simplistic. This earlier work also demonstrated that the ERES nucleates on the FAZ:ER such that there is a transverse spatial organization (outside-to-inside) of flagellum, flagellar membrane, cell plasma membrane, FAZ, FAZ:ER, ERES, and Golgi [see Fig. 9 in (Sevova & Bangs, 2009)]. This organization places the Golgi in a distal (internal) position to the flagellar cytoskeleton, calling into question the role of the bilobe structure as a direct cytoskeletal adaptor for Golgi segregation. In addition, the greater distance between the old and new EGJs in BSF trypanosomes raises questions about the bilobe in regard to tethering old and new Golgi during replication and segregation.
RESULTS
Golgi Replication in BSF Trypanosomes
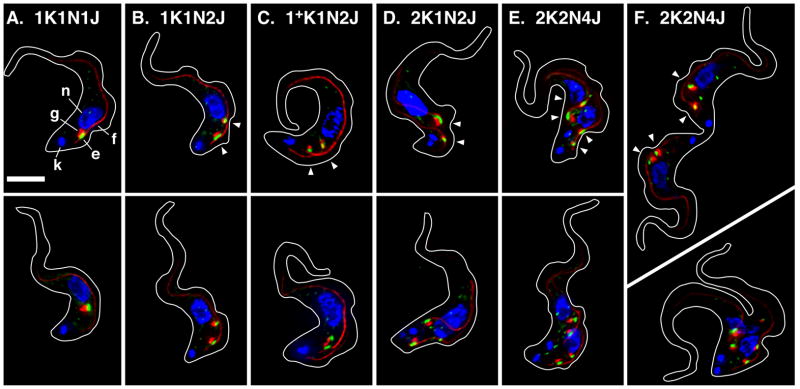
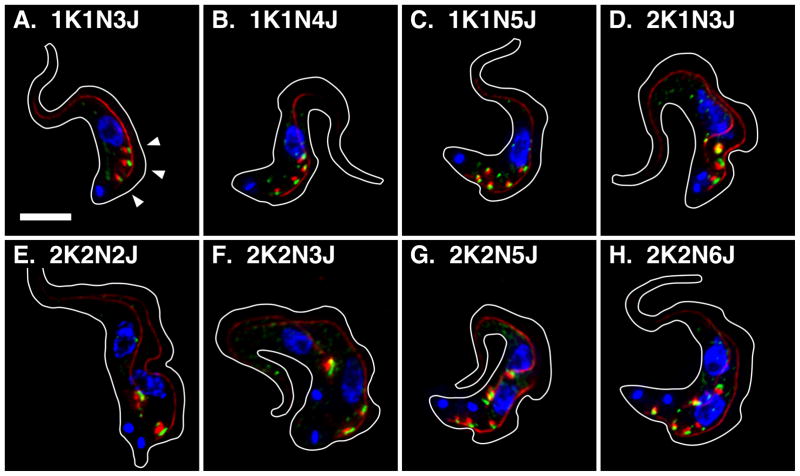
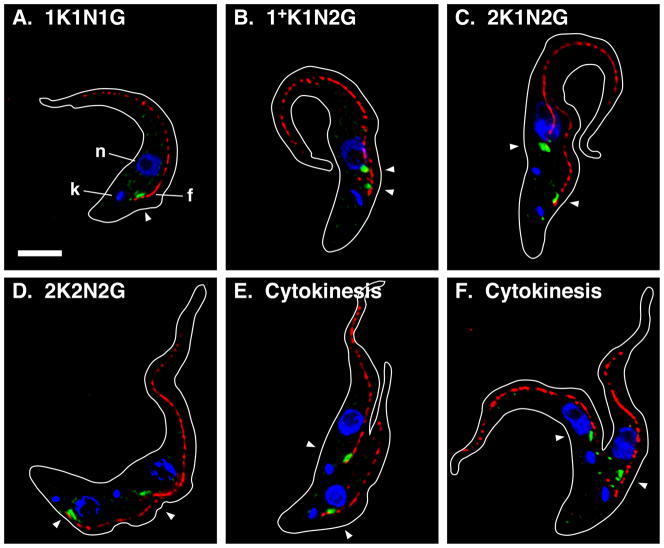
I have investigated EGJ distribution and copy number through the cell cycle of BSF trypanosomes using the dual tagged TbSec23.2HA/TbGT15Ty cell line (Sevova & Bangs, 2009). Log phase cells were stained for the EGJ and the FAZ filament, images of typical cells were acquired (Fig. 1) and EGJ copy number was scored as a function of cell cycle progression (Fig. 2A). In agreement with our previous results, ~20% of all 1K1N cells have one EGJ (Fig. 1A), ~75% have two EGJs (Fig. 1B), and the remainder have three or more. These EGJ are closely aligned with the single FAZ filament and remain so in 1+K1N2J cells as the new daughter FAZ emerges prior to kinetoplast scission (Fig 1C, 86.4% of 1+K1N2J cells, n = 2). In cells that have completed kinetoplast replication (2K1N) >65% still have two fully developed EGJs, but these are now independently aligned with the two daughter FAZ filaments (Fig. 1D, 94.5% of all 2K1N cells, n = 2). These results strongly suggest that the transition from 1K1N2J to 2K1N2J occurs by redistribution of one EGJ from the old FAZ to the new FAZ. Subsequently, as cells progress through mitosis (Fig. 1E; 2K2N) EGJ copy number doubles such that there are two independent EGJs aligned to each FAZ filament. This phenotype represents ~65% of all post-mitotic cells. Finally cytokinesis generates two 1K1N daughter cells each with two EGJs (Fig. 1F). Other atypical EGJ phenotypes were observed at all stages of the cell cycle, including 1K1N cells with three, four and five EGJs (Figs. 3A–C), 2K1N cells with three EGJs (Fig. 3D, this likely represents a normal intermediate in the transition from two to four EGJ), and 2K2N cells with two, three, five and six EGJs (Panels E–H). 2K2N cells with four asymmetrically distributed (1:3) EGJs were also observed (not shown). All of these organellar phenotypes represent lesser percentages of their respective stages of the cell cycle (Fig. 2) suggesting that the major pathway for EGJ replication and segregation is from recently divided daughter cells with two EGJ to post-mitotic cells with four EGJ. Subsequent cytokinesis generates two new daughter cells each with two EGJ. These data indicate that the critical stage for EGJ replication is during late S-phase leading up to mitosis (2K1N→2K2N transition).
Figure 1. ERES:Golgi junction morphology through the cell cycle in BSF trypanosomes.
Panels A–E. Typical images for the predominant EGJ phenotypes as determined in Fig. 2A. Panel F. Images of cells in cytokinesis. Cultured log phase (<5 × 105/ml) TbSec23.2HA/TbGT15Ty BSF cells were fixed/permeabilized and stained with rabbit anti-HA, monoclonal anti-Ty, and monoclonal anti-FAZ filament. Secondary staining is green for TbSec23.2HA, and red for TbGT15Ty and the FAZ filament. Cells were stained with DAPI (blue) to reveal kinetoplast (k) and nuclei (n). The ERES (e, TbSec23.2HA), Golgi (g, TbGT15Ty), and FAZ filament (f) are indicated in Panel A (top) only. All images are merged three channel summed stack projections of deconvolved z-series. Outlines of individual cells were traced from matched DIC images. All cells are oriented with the anterior end up. Kinetoplast (K), nuclear (N), and EGJ (J) copy numbers are indicated. Arrowheads indicate EGJs (top row only). Bar indicates 5 μm (shown in Panel A only).
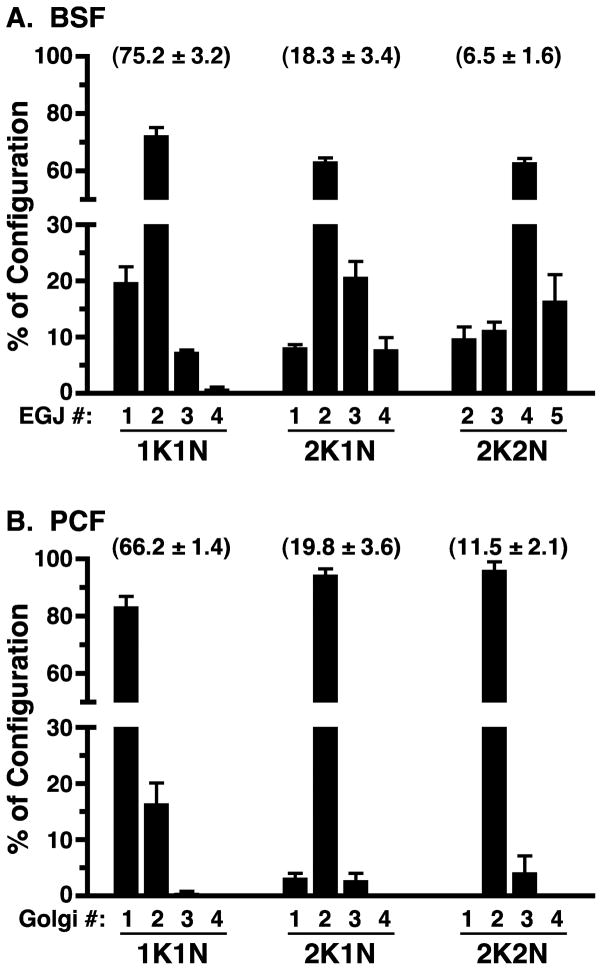
Figure 2. Quantification of EGJ or Golgi copy number through the trypanosome cell cycle.
Log phase BSF TbSec23.2HA/TbGT15Ty cells were stained for the EGJ, and log phase PCF cells were stained for the Golgi, as described in Figs. 1 & 5, respectively. BSF EGJ (Panel A) or PCF Golgi (Panel B) copy numbers were determined with >500 BSF or >300 PCF cells scored in in each of three independent experiments. For BSF cells only anti-HA+/anti-Ty+ structures aligned with the FAZ filament were scored. For PCF cells only anti-TbGRASP-positive structures aligned with the FAZ filament were scored. Data are presented as percent of each kinetoplast/nuclear configuration (1K1N, 2K1N, 2K2N; mean ± s.e.m.) comprised by each EGJ or Golgi copy number. Numbers in parentheses indicate the percent of total cells in each stage of the cell cycle (mean ± s.d.).
Figure 3. Atypical EGJ copy numbers in BSF trypanosomes.
TbSec23.2HA/TbGT15Ty BSF cells were fixed, stained, and imaged exactly as described in Fig. 1. All images are merged three channel summed stack projections of deconvolved z-series. Organellar copy numbers are indicated. Arrowheads indicate EGJs and bar indicates 5 μM (Panel A only).
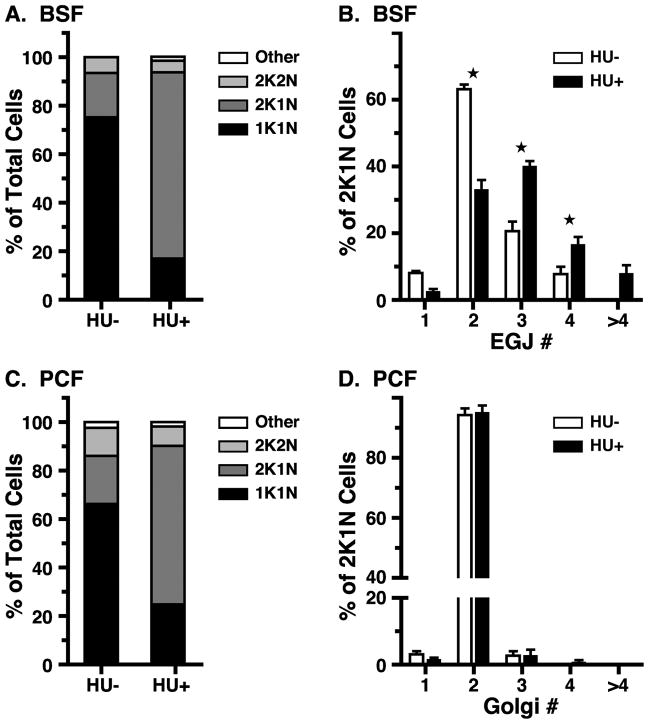
The presence of atypical EGJ copy numbers at all stages of kinetoplast and nuclear replication suggests that EGJ replication is not tightly coupled to the cell cycle. BSF trypanosomes can be arrested predominantly at the 2K1N stage by hydroxyurea treatment (Forsythe et al., 2009), and this strategy was used to investigate cell cycle control of EGJ replication. Log phase BSF cells were exposed to hydroxyurea, following which organellar phenotypes were determined (Fig. 4A). Consistent with Forsythe et al. (2009), the 2K1N configuration increased dramatically (18.3 ± 1.9% to 76.7 ± 0.5%) with a compensatory decrease in 1K1N cells (75.2 ± 1.8% to 17.0 ± 2.3%), indicating a cell cycle block in S phase. Simultaneously, the dominant EGJ copy number in the 2K1N configuration increased from predominantly two (~65%) in asynchronously replicating populations to a broader distribution of 2 to ≥4 in arrested cells (Fig. 4B). These results indicate that EGJ replication continues even when cycling is blocked. This lack of tight coupling, in conjunction with asymmetric segregation into daughter cells, can account for all of the less prevalent atypical EGJ copy numbers seen in asynchronously replicating cells.
Figure 4. Effect of cell cycle arrest on EGJ copy number.
Panels A & B. Log phase BSF TbSec23.2HA/TbGT15Ty cells were treated with hydroxyurea (HU+, 10 μg/ml, 6 hr), and then stained for the EGJ as in Fig. 1. Panels C & D. Log phase PCF cells were treated with hydroxyurea (HU+, 13.2 μg/ml, ~16 hr), and then stained for the Golgi as in Fig. 5. Panels A & C. DAPI stained cells (200–300 per experiment) were scored for kinetoplast and nuclear configuration. Data are presented as percent of total cells (mean, n = 3). Panels B & D. HU-treated 2K1N (>300) and control cells were scored for EGJ or Golgi copy number as indicated, and as described in Fig. 2. Data are presented as percent of all 2K1N cells (mean ± s.e.m., n = 3). All data for control cells (HU-, Panels C & D) are re-plotted from Fig. 2. Stars indicate statistically significant differences (p<0.05) between matched data sets by paired t-test.
Golgi Replication in PCF Trypanosomes
BSF trypanosomes present a very different profile of Golgi replication than that reported for PCF parasites (Field et al., 2000, He et al., 2004). To confirm these differences we repeated our analyses with logarithmically growing PCF cells. The TbGT15Ty epitope tag did not perform reliably in transgenic PCF trypanosomes, so the Golgi was visualized by immunostaining for the Golgi matrix protein TbGRASP (He et al., 2004). Quantification of Golgi copy number and representative images are presented in Fig. 2B and Fig. 5, respectively. Typically a single Golgi was found in 1K1N cells (>80%), always in close proximity to the posterior base of the FAZ filament where the flagellum emerges from the flagellar pocket (Fig 5A). In contrast, 2K1N cells overwhelmingly (>90%) contained two Golgi, one aligned with the base of the old anterior FAZ filament, and one likewise aligned with the base of the new posterior FAZ filament (Fig. 5C). Thereafter, two Golgi were maintained through mitosis and cytokinesis (Fig 5D–F). Close inspection also indicated that many of the ~20% of 1K1N cells that contained two Golgi had enlarged unsegregated kinetoplasts (Fig. 5B, 1+K1N), consistent with Golgi duplication preceding kinetoplast replication, and in tight concordance with outgrowth of the new flagellum. These data indicate that the critical point for Golgi replication in PCF trypanosomes is early in S-phase at the 1K1N→2K1N boundary.
Figure 5. Golgi morphology through the cell cycle in PCF trypanosomes.
Typical images for the predominant PCF Golgi phenotypes as determined in Fig. 2B. Cultured log phase PCF cells (<5×106/ml) were fixed/permeabilized and stained with rabbit anti-TbGRASP (green) and monoclonal anti-FAZ filament (red). Cells were also stained with DAPI. Kinetoplast (k), nucleus (n), and FAZ filament (f) are indicated (Panel A only). Golgi are indicated by arrowheads. All images are merged three channel summed stack projections of deconvolved z-series. Outlines of individual cells were traced from matched DIC images. All cells are oriented with the anterior end up. Panels A–D. Kinetoplast (K), nuclear (N), and Golgi (G) copy numbers are indicated. A cell with partially replicated kinetoplast (1+K) is presented in Panel B. Panels E & F. Cells in early and late cytokinesis, respectively. Additional ‘Golgi’ spots in Panel F may represent ‘phantom’ Golgi observed by (He et al., 2004). Note also that the FAZ filament consistently presents a more ‘discontinuous’ profile than seen in BSF trypanosomes. Bar indicates 5 μm (shown in Panel A only).
This pattern of Golgi duplication, which is in general agreement with the prior work of others (Field et al., 2000, He et al., 2004, de Graffenried et al., 2008)1, contrasts dramatically with the observed pattern for BSF trypanosomes. In particular, the strict maintenance of two Golgi in 2K1N and 2K2N cells suggests that Golgi duplication is tightly coupled to the cell cycle. To investigate this possibility log phase PCF trypanosomes were subjected to HU treatment and the effect on Golgi copy number quantified. As reported previously (Chowdhury et al., 2008), and similar to BSF trypanosomes, HU treatment of PCF cells resulted in dramatic arrest at the 2K1N configuration (Fig. 4C, >65%). However, no change in Golgi copy number was seen in these arrested cells (Fig. 4D), indicating that Golgi duplication is indeed tightly coupled to the PCF cell cycle.
The Bilobe in BSF Trypanosomes
These results show that the EGJ in BSF trypanosomes, regardless of how many are present in a given cell, is always closely aligned with the FAZ filament, consistent with assembly on the FAZ:ER as previously demonstrated (Sevova & Bangs, 2009). Of necessity these EGJs are distributed longitudinally along the FAZ filament and only one can be proximal to the posterior end. This contrasts sharply with PCF trypanosomes where EGJ copy number is tightly regulated and each EGJ is always closely associated with the base of the FAZ filament. In addition, the bilobe structure, which is closely associated with each Golgi in PCF cells, also localizes specifically to the posterior end of the FAZ filament (He et al., 2005, Selvapandiyan et al., 2007, Shi et al., 2008). A similar bilobe structure does exist in BSF trypanosomes in close proximity to the flagellum as it emerges from the flagellar pocket (Morriswood et al., 2009), but its association with the FAZ filament through the BSF cell cycle has not been defined, nor has its relationship to the EGJ.
To investigate these issues TbCen2, a component of both the bilobe and basal bodies (He et al., 2005), was HA tagged and constitutively over-expressed in the BSF trypanosomes, allowing TbCen2HA localization relative to the FAZ filament at all stages of the cell cycle. TbCen2HA was seen as an elongated structure prominently associated with the base the FAZ filament in 1K1N cells (Fig. S1A). Faint staining was also seen along the flagellum. This is likely a true localization and not due to over-expression as the pan-specific centrin antibody 20H5, which cross reacts with TbCen2, also stains the trypanosomal flagellum (He et al., 2005, Morriswood et al., 2009). A similar pattern of prominent colocalization with the base of each FAZ filament was seen in all other stages of the cell cycle, including cytokinesis (Fig. S1B–D). These data confirm that a TbCen2-positive bilobe structure exists in BSF trypansomes, and demonstrate that it is closely associated with the posterior end of the FAZ filament as shown previously in PCF cells.
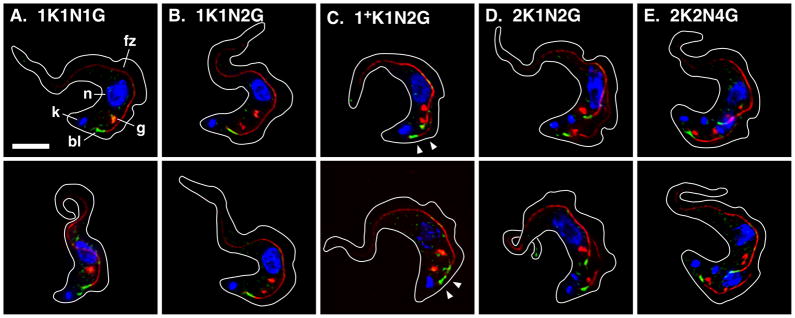
To investigate the spatial relationship to the Golgi TbCen4, another bilobe component (Shi et al., 2008), was in situ epitope tagged with YFP in the TbGT15Ty background. This marker combination allowed simultaneous imaging of both structures in formaldehyde-fixed/detergent-permeabilized cells. Early in the cell cycle (1K1N) all Golgi, whether present in single or double copy, were typically positioned forward of the bilobe along the FAZ filament (Figs. 6A & 6B). As seen for the entire EGJ (Fig. 1C), these Golgi remained associated with the old FAZ filament after duplication of the bilobe and early outgrowth of the new FAZ filament (Figs. 6C, 1+K1N). Upon completion of flagellar and kinetoplast replication Golgi appeared on the new FAZ filament, but again these were positioned forward of the bilobe (Figs. 6D, 2K1N). Only in post-mitotic cells (2K2N) did the most hindmost Golgi appear in proximity to the bilobe, but this was more overt on the posterior daughter FAZ (Figs. 6E), and was variable from cell to cell (not shown). These results confirm that there is no obligate association of Golgi with the bilobe in BSF trypanosomes, and that Golgi and bilobe duplication are independent events.
Figure 6. Spatial relationship of the bilobe and Golgi in BSF trypanosomes.
Cultured log phase (<5 × 105/ml) TbGT15Ty/TbCen4-YFP BSF cells were fixed/permeabilized and stained with rabbit anti-GFP (green), monoclonal anti-Ty (red), and monoclonal anti-FAZ filament (red). Cells were stained with DAPI (blue) to reveal kinetoplast (k) and nuclei (n). The bilobe (bl, TbCen4-YFP), Golgi (g, TbGT15Ty), and FAZ filament (f) are indicated in Panel A (top) only. Representative images of cells with the predominant Golgi copy number at all stages of the cell cycle are presented. All images are merged three channel summed stack projections of deconvolved z-series. Outlines of individual cells were traced from matched DIC images. The posterior newly replicating bilobe/FAZ filaments in cells that have not completed kinetoplast duplication are indicated (Panel C, 1+K, arrowheads). Kinetoplast (K), nuclear (N), and Golgi (G) copy numbers are indicated. Bar indicates 5 μm (shown in Panel A only).
DISCUSSION
These results, in conjunction with published studies of other laboratories, allow for detailed models for biogenesis of the ERES:Golgi junction in two stages of the trypanosome life cycle (Fig. 7). These models take into account well documented stage-specific morphological differences: i) connection at the flagellar tip during outgrowth of the daughter flagellum in PCF cells (Briggs et al., 2004); ii) relative positioning of daughter nuclei and kinetoplasts in post-mitotic cells (Tyler et al., 2001); and iii) more posterior location of the flagellar pocket in BSF trypanosomes (Matthews et al., 1995). In interphase (1K1N) PCF cells EGJ copy number is commonly one (~80%). EGJ duplication occurs early in S-phase following initial outgrowth of the daughter flagellum and prior to kinetoplast scission (1+K1N). This EGJ copy number is then tightly maintained until cytokinesis distributes a single EGJ to each daughter cell. In contrast, interphase BSF cells typically have two EGJ (~75%), one of which is redistributed to the new FAZ early in S-phase. Thereafter de novo EGJ duplication occurs on both FAZ filaments in late S-phase as 2K1N cells progress through nuclear replication and mitosis. This typically results in post-mitotic cells with four EGJs, two of which are aligned with each FAZ filament and which are segregated to each daughter cell during cytokinesis. However, EGJ duplication apparently can occur at any time in BSF cells leading to atypical EGJ copy numbers at all stages of the cell cycle.
Figure 7. Stage specific models for EGJ biogenesis in trypanosomes.
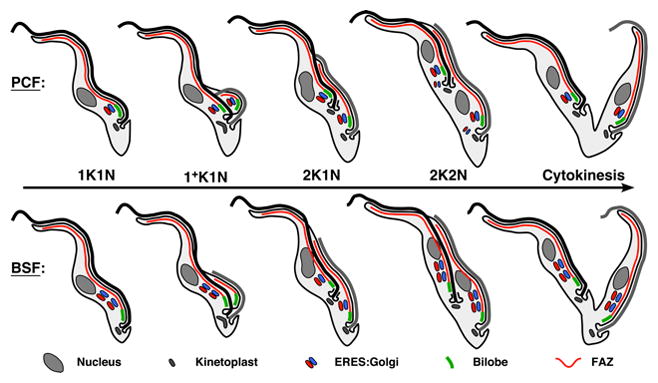
Diagrams of EGJ duplication through the trypanosome cell cycle. The PCF model (top) is based partly on data presented herein and primarily on the published data of others (Field et al., 2000, He et al., 2005, Selvapandiyan et al., 2007, de Graffenried et al., 2008, Shi et al., 2008, Morriswood et al., 2009). The BSF model (bottom) is based primarily on data presented herein, but also on (Morriswood et al., 2009). Key features of each model are discussed in the text. In replicating cells the old flagellum is black and the new flagellum is gray. Note that in 2K2N PCF cells smaller phantom EGJ are transiently present that are not aligned with the FAZ filament or bilobe. Also note that while the EGJ is known to be in close association with the bilobe in PCF cells (top), the precise degree of association of the hindmost EGJ with the bilobe is not certain in BSF cells (bottom).
These models illustrate both common and stage-specific features of EGJ replication in trypanosomes. The most obvious commonality is the close partnering of each ERES with a Golgi apparatus. This undoubtedly reflects the biogenesis of these paired organelles, which has been shown by time-lapse microscopy to be tightly coupled spatiotemporally in PCF cells (Ho et al., 2006). The quantitative data presented here indicate that this is likely true in the BSF stage as well. Also in common is the close alignment of the EGJ with the FAZ filament. Our previous work indicated that the EGJ nucleates on the FAZ-associated ER in BSF trypanosomes (Sevova & Bangs), and the close alignment of the Golgi with the FAZ filament seen here strongly suggests that this is also the case in PCF trypanosomes.
Beyond the timing of replication, the most obvious EGJ differences in PCF vs. BSF trypanosomes are coupling to the cell cycle and localization relative to the bilobe - features that may be related. In PCF cells a single EGJ is closely associated with each bilobe throughout the cell cycle, no other stable EGJs are observed and EGJ replication is tightly regulated. When cell division is arrested duplication stops completely. Knockdown of some bilobe components (He et al., 2005, Zhou et al., 2010) blocks both Golgi duplication and cell growth, but bilobe and Golgi replication can be uncoupled by ablation of TbPLK (polo-like kinase) (de Graffenried et al., 2008), a regulator of the cell cycle (Kumar & Wang, 2006, Hammerton et al., 2007). The bilobe is at the base of the FAZ filament in close proximity to the mouth of the flagellar pocket. Structurally this is the most complex region of the cell, with basal bodies, kinetoplast, flagellar collar, and flagellum all in close and complex relationships (Bonhivers et al., 2008, Lacomble et al., 2008). Collectively, these considerations suggest that the bilobe may be a multifunctional complex in PCF trypanosomes with roles in templating EGJ duplication and as a cytoskeletal adaptor to ensure its segregation during cytokinesis (see below for an alternative view). The apparently obligate nature of these roles provides an explanation for the tight coupling of EGJ replication to cell cycle progression, which is also dependent on the bilobe.
In contrast, coupling of EGJ replication to the cell cycle, and the physical relationship between the EGJ and the bilobe, seem to be much looser in BSF trypanosomes, calling into question a role for the bilobe in regulating EGJ duplication and subsequent segregation during cytokinesis. Three observations support this view: i) only the most posterior EGJ is proximal to the bilobe and this association does not appear to be particularly close; ii) in contrast to PCF trypanosomes, there is no connection of adjacent EGJs by the bilobe suggesting no role in EGJ duplication; and iii) images of cells in cytokinesis indicate that a bilobe association is not obligate for EGJ segregation. Perhaps the hindmost EGJ in each BSF daughter cell does segregate by association with the bilobe, thereby insuring that at least one EGJ is delivered to each daughter cell. However, all other EGJs apparently segregate by virtue of their nucleation on the FAZ:ER and consequent close association with the core FAZ cytoskeleton. Thus the FAZ:ER via its own poorly understood association with the flagellar cytoskeleton may be all that is required for coupling EGJ segregation to cytokinesis. However, this does not preclude a role for the bilobe in EGJ redistribution to the newly developing FAZ as BSF cells progress through kinetoplast and flagellar replication. Some mechanism must exist for redistributing these EGJs and the bilobe could participate in this process. Establishing such a role may require direct simultaneous imaging of the EGJ and bilobe by live cell imaging in immobilized cells, something that is not currently possible with BSF trypanosomes (Price et al., 2010).
Collectively these data argue against a direct role for the bilobe in EGJ replication and segregation in BSF trypanosomes, and this in turn suggests an alternative view of EGJ duplication and segregation in PCF parasites. The close association of the EGJ and the bilobe in PCF cells may be only coincidental due to the compressed volume between the nucleus and the flagellar pocket in this stage of the life cycle. In this scenario the effect of ablating bilobe components on EGJ replication would be an indirect consequence of blocking cell cycle progression. For instance, silencing TbLRRP1 in PCF trypanosomes blocks formation of the new FAZ in daughter cells (Zhou et al., 2010), which in turn could indirectly mediate the subsequent block in Golgi duplication. Thus, the function of the bilobe in the trypanosome cell cycle may be related primarily to FAZ replication. In this regard it is worth noting that some bilobe markers behave like cytoskeletal components, being resistant to detergent and high salt treatments (Morriswood et al., 2009, Zhou et al., 2010). Whether these apparent differences in bilobe functionality in PCF versus BSF trypanosomes are real or just perceived cannot be said at the present time. Nevertheless, it is clear that the bilobe plays an important role in the complex process of cell division in PCF cells, and this is likely to be true in BSF parasites as well. How the bilobe contributes to EGJ replication in BSF cells, if at all, will require silencing of specific bilobe components as have been performed in PCF trypanosomes. However, the data presented here suggest that EGJ replication in BSF trypanosomes will continue unabated when bilobe function is impaired.
Two final issues merit comment. First, ‘phantom’ EGJs arise late in the cell cycle in PCF trypanosomes (He et al., 2004, He et al., 2005) (Fig. 7). These transient structures are not closely aligned with the FAZ:ER or the bilobe (Zhou et al., 2010), and likely derive from inappropriate nucleation on internal portions of the ER not associated with the FAZ. This suggests that EGJ biogenesis may also be promiscuous in PCF trypanosomes, but only those EGJ closely associated with the FAZ, via the FAZ:ER and/or the bilobe, are stably maintained through cytokinesis. Thus, in BSF trypanosomes, with more space along the post-nuclear FAZ:ER to accommodate additional EGJs, these structures may segregate efficiently to each daughter cell, thereby accounting for the higher EGJ copy number throughout the cell cycle. Second, all of these data and speculations point toward a central role for the FAZ:ER in EGJ biogenesis, with or without assistance from the bilobe. This unusual ER structure, with its close association with the flagellar cytoskeleton, has been known since the earliest electron microscopy studies on trypanosomes (Vickerman, 1969b, Vickerman, 1969a), yet how it is formed and maintained is a complete mystery - one worthy of a more modern molecular investigation.
MATERIALS & METHODS
Maintenance of Trypanosomes
Lister 427 strain bloodstream (BSF, MITat1.2 expressing VSG221) and procyclic (PCF) form Trypanosoma brucei were grown in HMI-9 and Cunningham’s media respectively as described in (Sevova & Bangs, 2009, Tazeh et al., 2009). The in situ epitope-tagged BSF TbGT15Ty cell line is described in (Sutterwala et al., 2008), and an equivalent PCF cell line was generated for these studies (data not shown). The dual in situ epitope-tagged TbSec23.2HA/TbGT15Ty BSF cell line is described in (Sevova & Bangs, 2009). The epitope tags in all these cell lines were introduced by homologous chromosomal recombination to minimize problems frequently associated with over expression. For hydroxyurea (HU) treatment BSF cultures were seeded at 5×104 cells/ml, and at 18 hr (~4×105 cells/ml) HU was added to 10 μg/ml (151 μM). After 6 hours cells were processed for immunofluorescence. Similarly, log phase PCF cultures were seeded at 1×106 cells/ml with 13.2μg/ml (200 μM) HU and were processed for immunofluorescence at ~16 hours.
Bilobe epitope tagging constructs
An HA-tagged TbCen2 construct was prepared using pXS6neo:HA3x (details available on request), a derivative of the constitutive expression vector pXS5 (Alexander et al., 2002). First, the entire TbCen2 orf (Tb927.8.1080, codons 1–196) was amplified from T. brucei strain 427 genomic DNA and inserted upstream of the aldolase intergenic region using flanking 5′ HindIII and 3′ EcoRI sites, thereby placing in frame a 3xHA tag (1x: YPYDVPDYA) at the C-terminus. Second, the TbCen2 3′ UTR (nts 1–520 relative to the stop codon) was amplified by PCR and inserted downstream of the neor cassette using flanking 5′ PacI and 3′ SacI sites. All cloning steps were confirmed by DNA sequencing. This construct was initially designed for in situ epitope tagging of the native TbCen2 gene, but failed for this purpose. Consequently, for ectopic over-expression of TbCen2HA by the pXS6 rRNA promotor the entire plasmid was linearized with NotI, transfected into the BSF TbGT15Ty cell line by electroporation (Sevova & Bangs, 2009), and double tagged TbGT15Ty/TbCen2HA clonal cell lines were derived under G418 selection. Expression of TbCen2HA was confirmed by western blotting with anti-HA (data not shown). An in frame C-terminal fusion of YFP with native TbCen4 gene was achieved by in situ epitope tagging. The Topo:TbCen4-YFP plasmid (a generous gift of Dr. Cynthia He) was restricted with PacI/NsiI, electroporated into the TbSec23.2HA/TbGT15Ty BSF cell line, and triply tagged clonal cell lines were selected with blastocidin.
Antibodies
Rabbit anti-HA (Sigma, St. Louis MO) and mouse anti-Ty (UAB Hybridoma Facility, Birmingham, AL) were used to detect epitope tags. Rabbit anti-GRASP (He et al., 2004) and mouse mAb L3B2 anti-FAZ (Kohl et al., 1999) were generously provided by Graham Warren (Max F. Perutz Laboratories, Vienna) and Keith Gull (University of Oxford), respectively. Goat Alexa Fluor secondary antibodies were from Molecular Probes Inc. (Eugene, OR). Rabbit anti-GFP was from Invitrogen (Carlsbad, CA).
Immunofluorescence Microscopy
Unless stated otherwise, all immunostaining was performed as previously described with minor alterations (Sevova & Bangs, 2009). Briefly, following settling onto Superfrost slides (Sigma, BSF) or plain slides (PCF), cells were sequentially treated (30 min, RT) with PBS/2% formaldehyde (EM grade), PBS/0.5% NP-40, and PBS/10% normal goat serum (NGS)/0.1% NP-40 (blocking buffer). Alternatively, for detection of epitope tagged TbCen2 with anti-HA, cells were fixed and permeabilized with methanol/acetone as described previously (Alexander et al., 2002) and then treated with blocking bufffer. In each case fixed/permeabilized cells were stained with primary antibodies diluted in blocking buffer, washed with PBS/1% NGS, and stained with the appropriate Alexa Fluor secondary antibodies (1/200) and DAPI (500 ng/mL) in blocking buffer. After washing, slides were mounted with PBS:Glycerol (1:1). Serial image stacks (0.2 micron Z-increment) were collected with capture times from 100–500 msec (100x PlanApo, oil immersion, 1.4 na) on a motorized Zeiss Axioplan IIi equipped with a rear-mounted excitation filter wheel, a triple pass (DAPI/FITC/Texas Red) emission cube, differential interference contrast (DIC) optics, and a Orca AG CCD camera (Hamamatsu, Bridgewater, NJ). All images were collected with OpenLabs 5.0 software (Improvision Inc., Lexington, MA) and individual channel stacks were deconvolved by a constrained iterative algorithm, pseudocolored, and merged using Velocity 5.0 software (Improvision Inc). The xyz pixel precision of this arrangement has been validated in (Sevova & Bangs, 2009) (see Figure S1 therein).
Acknowledgments
The author is grateful to lab members past and present, in particular Dr. Eli Theel (née Sevova) for conceptual contributions to the early phase of this work, and to Kevin (Bubba) Schwartz and Jason (Stan) Silverman for assistance in preparing the tagged TbCen cell lines. Likewise, I thank Professor Tony Sinai (University of Kentucky) for thoughtful discussions. I also acknowledge Drs. Graham Warren (Max F. Perutz Laboratories, Vienna) and Keith Gull (Oxford University) for generous gifts of antibodies, and Cynthia He (National University of Singapore) for the Topo:TbCen4-YFP tagging vector. This work supported by United States Public Health Service Grant R01 AI35739 to JDB.
Footnotes
de Graffenried et al. (2008) noted somewhat higher percentages of cells with >2 Golgi at all stages of the PCF cell cycle. This variance, which is a matter of degree only, likely stems from the criteria used here of scoring only GRASP-positive structures in close alignment with the FAZ filament, thereby eliminating ‘phantom’ Golgi from consideration (see Fig. 2 legend).
References
- Alexander DL, Schwartz KJ, Balber AE, Bangs JD. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J Cell Sci. 2002;115:3255–3263. doi: 10.1242/jcs.115.16.3253. [DOI] [PubMed] [Google Scholar]
- Bonhivers M, Nowacki A, Landrein N, Robinson DR. Biogenesis of the trypanosome endo-exocytic organelle is cytoskeleton mediated. PLoS Biol. 2008;6(5):e105. doi: 10.1371/journal.pbio.0060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs LJ, McKean PG, Baines A, Moreira-Leite F, Davidge J, Vaughan S, Gull K. The flagella connector of Trypanosoma brucei: an unusual mobile transmembrane junction. J. Cell Sci. 117: 1641–1651. 2004 doi: 10.1242/jcs.00995. [DOI] [PubMed] [Google Scholar]
- Chowdhury AR, Zhao Z, Englund PT. Effect of hydroxyurea on procyclic Trypanosoma brucei: an unconventional mechanism of achieiving synchronous growth. Euk Cell. 2008;7:425–428. doi: 10.1128/EC.00369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graffenried CL, Ho HH, Warren G. Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J Cell Biol. 2008;181:431–438. doi: 10.1083/jcb.200708082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini A, Malhotra V, Marsh BJ, Nakano A, Pfeffer S, Rabouille C, Rothman JE, Warren G, Wieland FT. Journeys through the Golgi - taking stock in a new era. J Cell Biol. 2009;187:449–453. doi: 10.1083/jcb.200909011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstler M, Thilo L, Weise F, Grünfelder CG, Schwarz H, Boshart M, Overath P. Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J Cell Sci. 2004;117:1105–1115. doi: 10.1242/jcs.00938. [DOI] [PubMed] [Google Scholar]
- Field H, Sherwin T, Smith AC, Gull K, Field MC. Cell-cycle and develpmental regulation of TbRAB31 localisation, a GTP-locked Rab protein from Trypanosoma brucei. Mol Biochem Parasitol. 2000;106:21–35. doi: 10.1016/s0166-6851(99)00192-9. [DOI] [PubMed] [Google Scholar]
- Forsythe GR, McCulluch R, Hammerton TC. Hydroxyurea-induced synchronization of bloodstream stage Trypanosoma brucei. Mol Biochem Parasitol. 2009;164:131–136. doi: 10.1016/j.molbiopara.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerton TC, Kramer S, Tetley L, Boshart M, Mottram JC. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol Microbiol. 2007;65:1229–1248. doi: 10.1111/j.1365-2958.2007.05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CY, Ho HH, Malsam J, Chalouni C, West CM, Ullu E, Toomre D, Warren G. Golgi duplication in Trypanosoma brucei. J Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CY, Pypaert M, Warren G. Golgi duplication in Trypanosoma brucei requires centrin2. Science. 2005;310:1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
- He CY. Golgi biogenesis in simple eukaryotes. Cell Microbiol. 2007;9:566–572. doi: 10.1111/j.1462-5822.2006.00874.x. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Ersfeld K, Gull K. CAP5.5, a life-cycle-regulated, cytoskeleton-associated protein is a member of a novel family of calpain-related proteins in Trypanosoma brucei. Mol Biochem Parasitol. 2001;116:25–34. doi: 10.1016/s0166-6851(01)00296-1. [DOI] [PubMed] [Google Scholar]
- Ho HH, He CY, de Graffenried CL, Murrells LJ, Warren G. Ordered assembly of the duplicating Golgi in Trypanosoma brucei. Proc Natl Acad Sci USA. 2006;103:7676–7681. doi: 10.1073/pnas.0602595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L, Sherwin T, Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Euk Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- Kumar P, Wang CC. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Euk Cell. 2006;5:92–102. doi: 10.1128/EC.5.1.92-102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by elctron microscope tomography. J Cell Sci. 2008;122:1081–1090. doi: 10.1242/jcs.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langreth SG, Balber AE. Protein uptake and digestion in bloodstream and culture forms of Trypanosoma brucei. J Protozool. 1975;22:40–53. doi: 10.1111/j.1550-7408.1975.tb00943.x. [DOI] [PubMed] [Google Scholar]
- Matthews KR, Sherwin T, Gull K. Mitochondrial genome repositioning during the differentiation of the African trypanosome between life cycle forms is microtubule mediated. J Cell Sci. 1995:108. doi: 10.1242/jcs.108.6.2231. [DOI] [PubMed] [Google Scholar]
- Matthews KR. The developmental cell biology of Trypanosoma brucei. J Cell Sci. 2005;118:283–290. doi: 10.1242/jcs.01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Simons K. The Golgi complex: in vitro, veritas? Cell. 1992;68:829–840. doi: 10.1016/0092-8674(92)90027-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B, He CY, Sealey-Cardona M, Yelnik JT, Pypaert M, Warren G. The bilobe structure of Trypanosoma brucei contains a MORN-repeat protein. Mol Biochem Parasitol. 2009;167:95–103. doi: 10.1016/j.molbiopara.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Price HP, MacLean L, Marrison J, O’Toole PJ, SDJ Validation of a new method for immobilizing kinetoplastid parasites for cell imaging. Mol Biochem Parasitol. 2010;169:66–69. doi: 10.1016/j.molbiopara.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston KS, Kabututu ZP, Melehani JH, Oberholzer M, Hill KL. The Trypanosoma brucei flagellum: moving parasites in new directions. Annu Rev Microbiol. 2009;63:335–362. doi: 10.1146/annurev.micro.091208.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvapandiyan A, Kumar P, Morris JC, Salisbury JL, Wang CC, Nakhasi HL. Centrin 1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol Biol Cell. 2007;18:3290–3301. doi: 10.1091/mbc.E07-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevova ES, Bangs JD. Streamlined architecture and GPI-dependent trafficking in the early secretory pathway of African trypanosomes. Mol Biol Cell. 2009;20:4739–4750. doi: 10.1091/mbc.E09-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Franklin JB, Yelnik JT, Ebersberger I, Warren G, He CY. Centrin4 coordinates cell and nuclear division in T. brucei. J Cell Sci. 2008;121:3062–3070. doi: 10.1242/jcs.030643. [DOI] [PubMed] [Google Scholar]
- Sutterwala SS, Hsu FF, Sevova ES, Schwartz KJ, Zhang K, Key P, Turk J, Beverley SM, Bangs JD. Developmentally regulated sphingolipid synthesis in African trypanosomes. Mol Microbiol. 2008;70:281–296. doi: 10.1111/j.1365-2958.2008.06393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazeh NN, Silverman JS, Schwartz KJ, Sevova ES, Sutterwala SS, Bangs JD. The role of AP-1 in developmentally regulated post-Golgi trafficking in Trypanosoma brucei. Euk Cell. 2009;8:1352–1361. doi: 10.1128/EC.00156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler KM, Matthews KR, Gull K. Anisomorphic cell division by African trypanosomes. Protist. 2001;152:367–378. doi: 10.1078/1434-4610-00074. [DOI] [PubMed] [Google Scholar]
- Vedrenne E, Probst M, Schneider A, Renggli SECK, Baltz T. Two related subpellicular cytoskeleton-associated proteins in Trypanosoma brucei stabilize microtubules. Mol Biol Cell. 2002;13:1058–1070. doi: 10.1091/mbc.01-06-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman K. The fine structure of Trypanosoma congolense in its bloodstream phase. J Protozool. 1969a;16:54–69. doi: 10.1111/j.1550-7408.1969.tb02233.x. [DOI] [PubMed] [Google Scholar]
- Vickerman K. On the surface coat and flagellar adhesion in trypanosomes. J Cell Sci. 1969b;5:163. doi: 10.1242/jcs.5.1.163. [DOI] [PubMed] [Google Scholar]
- Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Gheiratmand L, Chen Y, Lim TK, Zhang J, Li S, Xia N, Liu B, Lin Q, He CY. A comparative proteomic analysis reveals a new bi-lobe protein required for bilobe duplication and cell division in Trypanosoma brucei. PLoS one. 2010;5:e9660. doi: 10.1371/journal.pone.0009660. [DOI] [PMC free article] [PubMed] [Google Scholar]