SUMMARY
The extensive family of plant terpene synthases (TPSs) generally has a bi-domain structure, yet phylogenetic analyses consistently indicate that these evolved from larger diterpene synthases. In particular, that duplication of the diterpene synthase genes required for gibberellin phytohormone biosynthesis provided an early predecessor, whose loss of a ~220 amino acid “internal sequence element” (now recognized as the γ domain) gave rise to the precursor of modern mono- and sesqui-TPSs found in all higher plants. Intriguingly, TPSs are conserved by taxonomic relationships rather than function, demonstrating that such functional radiation has occurred both repeatedly and relatively recently, yet phylogenetic analyses assume that “internal/γ” domain loss represents a single evolutionary event. Here we provide evidence that such loss was not a singular event, but rather has occurred multiple times. Specifically, we provide an example of a bi-domain diterpene synthase, from Salvia miltiorrhiza, along with a sesquiterpene synthase from Triticum aestivum (wheat) that is not only closely related to diterpene synthases, but retains the ent-kaurene synthase activity relevant to the ancestral gibberellin metabolic function. Indeed, while the wheat sesquiterpene synthase clearly no longer contains the “internal/γ” domain, it is closely related to rice diterpene synthase genes that retain the ancestral tri-domain structure. Thus, these findings provide examples of key evolutionary intermediates underlying the bi-domain structure observed in the expansive plant TPS gene family, as well as indicating that “internal/γ” domain loss has independently occurred multiple times, highlighting the complex evolutionary history of this important enzymatic family.
INTRODUCTION
Extensive phylogenetic studies indicate that the expanded terpene synthase (TPS) gene family in plants are derived from the diterpene synthases found in all higher plants for the requisite production of the gibberellin phytohormones (Bohlmann et al. 1998, Trapp and Croteau 2001). However, most other plant TPSs are significantly shorter, with crystallographic analyses revealing the presence of two distinct domains (termed “α” and “β”), although only the C-terminalα domain is associated with the observed enzymatic activity (Christianson 2006). Thus, it has been hypothesized that there was an ancient loss of the corresponding sequence, which seems to represent loss of intervening exons rather than simple truncation, leading to its designation as an “internal sequence element” (Trapp and Croteau 2001). Recent crystallographic characterization of diterpene synthases have demonstrated that this “internal sequence element” forms a distinct “γ” domain (Köksal et al. 2011a, Köksal et al. 2011b)(Figure 1). However, the possibility that “internal/γ” domain loss may have independently occurred multiple times has not been considered, which may contribute to the current uncertainties regarding the origins of plant TPSs.
Figure 1. Diterpene synthase tri-domain structure.
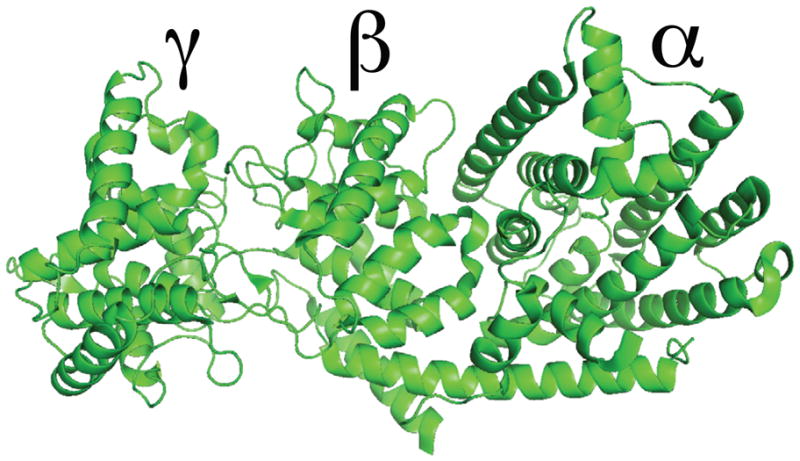
As exemplified by the crystal structure of taxadiene synthase from Taxus brevifolia (Köksal, et al. 2011b), with domain nomenclature as indicated.
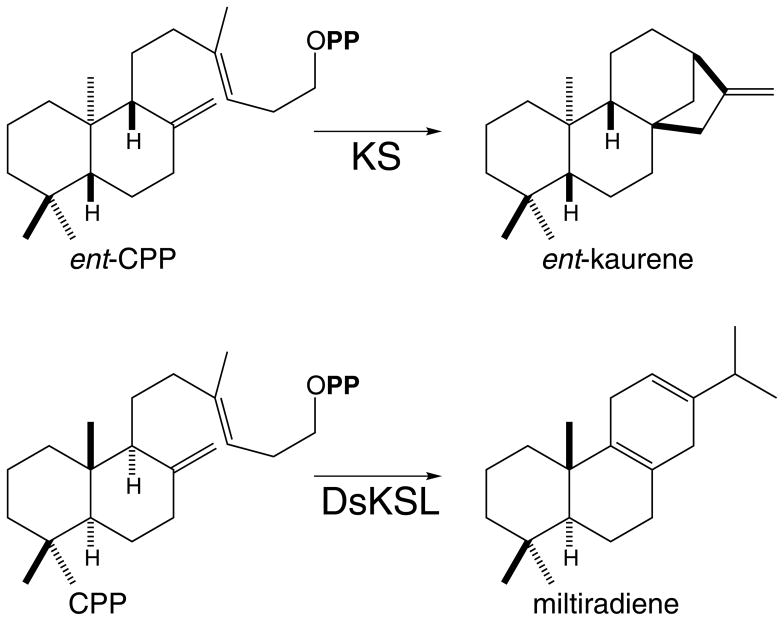
Gibberellin biosynthesis is initiated by dual cyclization reactions catalyzed by mechanistically distinct, but phylogenetically related diterpene synthases. These are ent-copalyl diphosphate (CPP) synthase (CPS), a class II diterpene cyclase whose action precedes that of ent-kaurene synthase (KS), which is an ent-CPP specific class I diterpene synthase whose catalytic activity is analogous to that of other TPS (Figure 2). The monofunctional CPS and KS observed in all higher plants seem to be derived by subfunctionalization of an ancestral bifunctional diterpene synthase (Keeling et al. 2010). In turn, this was likely formed by an ancient gene fusion between the two different classes of diterpene synthases (Morrone et al. 2009), with the class I activity associated with theα domain, and the class II activity associated with the βγ domains (Cao et al. 2010). Consistent with this hypothesis, based on correlated conservation of sequence and catalytic activity (Xu et al. 2004), and more recent crystallographic evidence (Köksal, et al. 2011a), it appears that the “internal/γ” domain plays a role in the class II cycloisomerization reaction. However, operating under the simplifying assumption that loss of the “internal/γ” domain occurred only once, phylogenetic analyses then indicate that bifunctional diterpene synthases arose from the monofunctional CPS and/or KS (Martin et al. 2004, Trapp and Croteau 2001).
Figure 2. KS(L) reactions.
Reactions catalyzed by ent-kaurene synthases (e.g. from gibberellin phytohormone biosynthesis) and DsKSL.
The requirement for gibberellin production has provided a genetic reservoir of enzymatic genes that has given rise to a large super-family of diterpenoid natural products (Peters 2010). As part of our investigation of the biosynthesis of these “labdane-related” diterpenoids, particularly the KS-like (KSL) diterpene synthases that have arisen from gene duplication and divergence of KS, we have uncovered evidence demonstrating that “internal/γ” domain loss is a relatively facile event that has independently occurred multiple times. In particular, we recently identified such a KSL from Salvia miltiorrhiza (Gao et al. 2009), a Chinese medicinal herb also known as “dan shen”, and we designate this enzyme here DsKSL. Notably, while DsKSL is functionally analogous to other KSL, reacting stereospecifically with normal CPP, and catalyzing the production of miltiradiene/abieta-8,12-diene (Figure 2), it is considerably shorter than other KSL (as well as KS), and we show here this is due to lack of the usual “internal/γ” domain. Thus, DsKSL represents a bi-domain diterpene synthase, with a βα bi-domain structure analogous to mono- and sesqui-TPSs. Based on this observation, we reexamined other KSL in our collection, and found one from Triticum aestivum (wheat) that provides an example of recent “internal/γ” domain loss, as it is closely related to rice diterpene synthase genes that retain the ancestral γβα tri-domain structure. Indeed, this TaKSL5 not only exhibits strong sequence similarity to KS and other KSL across their commonβα domains, but also will still react with ent-CPP to produce significant amounts of ent-kaurene, although it appears to actually function as a sesquiterpene synthase, most readily reacting with (2E,6E)-farnesyl diphosphate to produce (E)-nerolidol.
RESULTS
DsKSL is a bi-domain terpene synthase
DsKSL is much shorter than other KSL, and amino acid (AA) sequence alignments suggested that this might reflect a bi-domain structure analogous to mono- and sesqui-TPSs (Figure 3). In particular, the homology between DsKSL and other KSL only begins after the “internal/γ” domain (i.e. DsKSL has a βα domain structure). Although DsKSL contains ~50 AA N-terminal to this point, plant diterpene synthases typically contain a poorly conserved N-terminal transit peptide directing the encoded pre-protein to the plastid. Thus, this first ~50 AA presumably represents the DsKSL plastid targeting sequence, which would be removed upon import, leaving the mature fully active enzyme in planta. To verify that this N-terminal sequence is not required for the observed activity, we carried out truncation studies. This demonstrated that a recombinant pseudo-mature construct, missing the first 54 AA, exhibited the same activity previously found with full-length DsKSL expressed as a fusion to glutathione S-transferase (Gao, et al. 2009). Specifically, using our previously described metabolic engineering system to co-express DsKSLd54 with a GGPP and various CPP synthases (Cyr et al. 2007), it was found to react stereospecifically with normal CPP to produce >95% miltiradiene. Further enzymatic analysis, using a purified 6×His tagged version of DsKSLd54 (Figure S1), yielded kinetic constants (KM = 6 ± 1μM and kcat = 4.3 ± 0.2 s−1) comparable to that exhibited by other diterpene synthases for their native substrates (Peters et al. 2000, Xu et al. 2007b), consistent with physiological relevance for this activity.
Figure 3. DsKSL lacks “internal/γ” domain.
Alignment of DsKSL with Solenaceae KSL homologs demonstrates homology only after the beginning of the β domain (indicated by *). Shown is alignment around N-terminal sequence of DsKSL.
DsKSL specifically acts as a diterpene synthase
Somewhat surprisingly, DsKSL is most closely related to two unusual TPS that are not diterpene synthases. Specifically, to a monoterpene synthase that reacts with the unusual cisoid C10 precursor neryl diphosphate (NPP) to produce largelyβ-phellandrene (Schilmiller et al. 2009), and a sesquiterpene synthase that is plastid-targeted and reacts with the unusual cisoid C15 precursor (2Z,6Z)-farnesyl diphosphate (FPP) to produce bergamotene and santalene (Sallaud et al. 2009), although both of these TPS also contain the “internal/γ” domain missing in DsKSL (Figure 3). To verify that DsKSL specifically acts as a diterpene synthase, we assessed its ability to react with other isoprenyl diphosphate substrates. In particular, we modified our metabolic engineering system to enable detection of sesquiterpene synthase activity. For activity with the general sesquiterpene precursor (2E,6E)-FPP this was simply accomplished by use of a previously described pMBIS plasmid, which carries such a trans-FPP synthase (Martin et al. 2003). In order to test activity with the unusual (2Z,6Z)-FPP, we cloned a synthetic version of the corresponding cis-FPP synthase into pACYC-Duet to enable co-expression with terpene synthases carried on pET derived vectors. Co-expressing DsKSLd54 with either type of FPP synthase only yielded the corresponding isomer of farnesol, the dephosphorylated primary alcohol resulting from the action of the endogenous phosphatases and found in control cultures, demonstrating that DsKSL does not possess any significant sesquiterpene synthase activity. This was verified by in vitro assays with purified DsKSL, which also was screened in vitro with the C10 geranyl diphosphate (GPP) and NPP, but it did not react with these monoterpene precursors either.
Other bi-domain KSL – TaKSL5
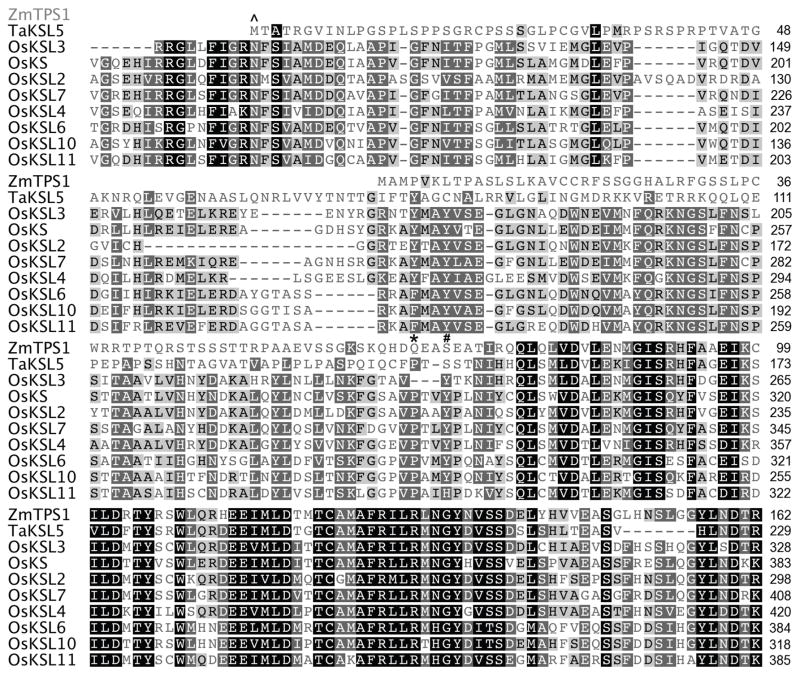
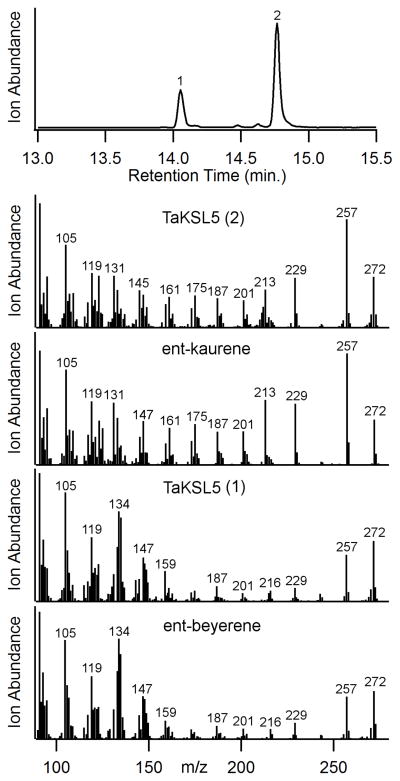
Intrigued by the bi-domain structure of DsKSL, we examined our collection of KSL genes and discovered another potential bi-domain diterpene synthase among those we are characterizing from wheat (T. aestivum). While complete results for the corresponding TaKSL gene family will be presented elsewhere, we report here characterization of this TaKSL5. Amino acid sequence alignment show that TaKSL5 is clearly missing the “internal/γ” domain, with clear homology after this point (i.e. in the βα domains) to other KSL, although it contains ~120 AA N-terminal to that point (Figure 4). Notably, while little activity was evident with the full-length gene product, removal of 121 AA resulted in a pseudo-mature construct that exhibited diterpene synthase activity in our metabolic engineering system. In particular, this TaKSL5d121 stereoselectively reacted with ent-CPP, and produced a mixture of ent-kaurene and ent-beyerene in a ~70:30 ratio (Figure 5).
Figure 4. TaKSL5 lacks “internal/γ” domain.
Alignment of TaKSL5 with other cereal KSLs demonstrates homology only after the beginning of the β domain (indicated by *). Notably, TaKSL5 appears to have lost complete exons, as the point at which homology becomes readily evident is an exon boundary in its closest homolog, OsKSL3 (indicated by #; see Figure S5).
Figure 5. TaKSL5 diterpene synthase activity.
GC-MS analysis of diterpene products from co-expression of TaKSL5d121 with ent-CPP synthase in E. coli. Shown are the chromatogram and the mass spectra of both products, with comparison to that of authentic standards for ent-kaurene and ent-beyerene (as indicated).
TaKSL5 as a sesquiterpene synthase
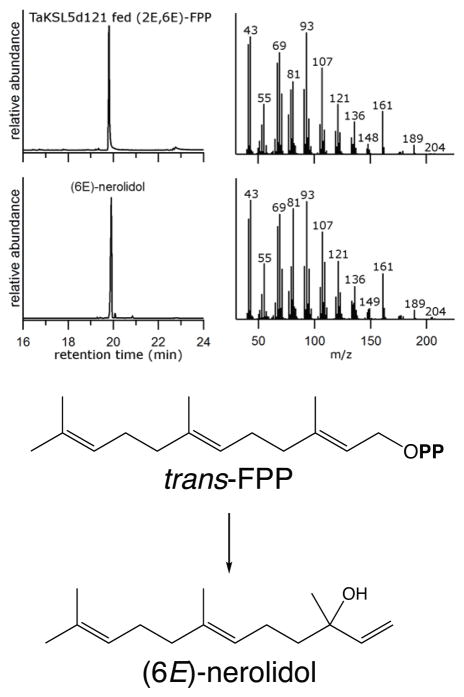
Intriguingly, TaKSL5 is closely related to a maize (Zea mays) sesquiterpene synthase (ZmTPS1), which reacts with trans-FPP and produces a mixture of (E)-β-farnesene, (E,E)-farnesol, and (E)-nerolidol (Schnee et al. 2002). Accordingly, we further co-expressed TaKSL5d121 with cis- and trans-FPP synthases, and found that it will react with trans- (but not cis-) FPP to produce (E)-nerolidol (Figure 6). Indeed, purified 6×His tagged TaKSL5d121 reacted much more readily with trans-FPP than ent-CPP in vitro, and it was possible to measure kinetics constants for TaKSL5d121 activity with trans-FPP (KM = 1.7 ± 0.2μM and kcat = 0.45 ± 0.01 s−1) that are consistent with physiological relevance for such sesquiterpene synthase activity (Figure S2). Given such broad reactivity, TaKSL5d121 also was screened in vitro against GPP and NPP, but neither was found to serve as a substrate.
Figure 6. TaKSL5 is a sesquiterpene synthase.
(a) GC-MS analysis of diterpene products from co-expression of TaKSL5d121 with cis-FPP synthase in E. coli. Shown is the chromatogram and the mass spectra, with comparison to that of an authentic standard (6E)-nerolidol. (b) Reaction catalyzed by TaKSL5 to produce (6E)-nerolidol from cis-FPP.
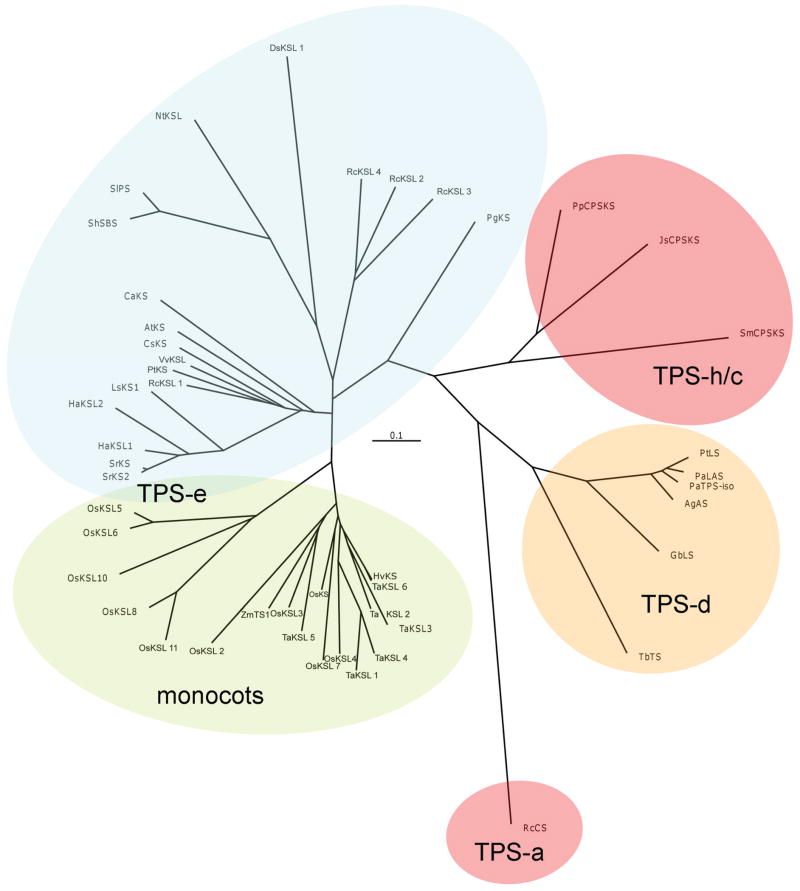
Molecular phylogenetic analysis
Diterpene synthases can be found in several of the defined TPS sub-families (Chen et al. 2011). Extensive phylogenetic analysis utilizing neighbor-joining (Figure 7), as well as parsimony (Figure S3) and maximum likelihood algorithms (Figure S4), all demonstrate that both DsKSL and TaKSL5 clearly fall within the KS(L) containing TSP-e sub-family. More specifically, these analyses suggest that the “internal/γ” domain deletion event leading to DsKSL occurred after the divergence of the Solanaceae and Lamiaceae dicot families. In particular, due to the presence of this domain in the closely related and co-clustered β-phellandrene and bergamotene/santalene synthases (SlPS and ShSBS, respectively), which are from Solanaceae species, relative to the Lamiaceae derived DsKSL. Thus, rather than representing the ancient “internal/γ” domain loss event postulated by previous TPS phylogenetic analyses, the βα bi-domain structure of DsKSL seems to reflect a separate, more recent such evolutionary event. However, particularly given its phylogenetic isolation, it also is possible that DsKSL represents an evolutionary relict wherein its placement in the TPS-e/f sub-family reflects conservation of function with other KS(L) i.e. as CPP-specific class I diterpene synthases.
Figure 7. Phylogenetic relationships of diterpene synthases.
Unrooted tree constructed via the neighbor-joining algorithm, including essentially only class I di-TPSs (along with a few closely related other TPSs defined in the text), and with previously defined TPS sub-family assignments indicated (Chen, et al. 2011).
On the other hand, TaKSL5 clearly falls within the monocot KS(L) cluster. Specifically, TaKSL5 clusters with KSL from rice (Oryza sativa), most closely OsKSL3 and the maize nerolidol synthase ZmTPS1 (Figure 7). While OsKSL2 and OsKSL3 appear to be pseudo-genes encoding inactive diterpene synthases (Xu et al. 2007a), both also clearly contain the “internal/γ” domain (Figure 4). However, the presence of ZmTPS1 in this phylogenetic cluster, even in nucleotide sequence based analyses, does not enable straightforward assignment of when during cereal plant family diversification, the domain loss event leading to TaKSL5 may have occurred. For example, the observed sequence similarity between TaKSL5 and ZmTPS1 may reflect functional conservation of both as nerolidol synthases, despite the relatively large evolutionary distance between maize and wheat, particularly relative to the closer relationship of wheat and rice. Nevertheless, the “internal/γ” domain loss event leading to the βα bi-domain TaKSL5 and ZmTPS1 does seem to have occurred at least following the split between monocots and dicots.
DISCUSSION
Previous phylogenetic analysis of the plant TPS super-family has left the origins of the commonly observed bi-domain TPS structure somewhat opaque. In particular, while it seems clear that plant TPSs arose from a larger diterpene synthase involved in primary (i.e., gibberellin) biosynthesis, it has been assumed that loss of the “internal/γ” domain was a unique evolutionary event. However, the possibility that the corresponding “internal/γ” domain loss has occurred multiple times was indicated by standard amino acid sequence alignment analysis, although it was ignored, leading to phylogenetic trees indicating that bifunctional diterpene synthases arose from the monofunctional CPS and/or KS (Trapp and Croteau 2001). Subsequent phylogenetic analysis including substantially more sequences also was consistent with multiple independent domain loss events, but did not question this assumption, leaving the evolutionary origins of the TPS super-family unclear (Martin, et al. 2004). More recent analysis has suggested that such “internal/γ” domain loss seems to have occurred independently in gymnosperms relative to angiosperms, but did not further address the question of how much more frequently this may have occurred (Chen, et al. 2011).
Here we provide evidence indicating that “internal/γ” domain loss in diterpene synthases has in fact independently occurred multiple times. Specifically, characterization of DsKSL has demonstrated that this is a bi-domain diterpene synthase that retains specificity for normal CPP, much like the other members of the TPS-e sub-family with which it clusters, despite clearly having lost the “internal/γ” domain in what appears to be a relatively recent evolutionary event. In addition, TaKSL5 is even more closely related to KS and other KSL, yet also is a bi-domain TPS. Indeed, while TaKSL5 most likely functions as a sesquiterpene (nerolidol) synthase, it also retains the ent-CPP substrate specificity and ent-kaurene product output characteristic of KS, similarly consistent with relatively recent loss of the “internal/γ” domain from a KS(L) precursor. Intriguingly, wheat contains a very closely related paralog to TaKSL5, which appears to be a polyploidy derived homoeolog that similarly no longer contains the “internal/γ” domain, and that differs largely in the upstream N-terminal sequence. This may reflect changes in splicing, which might further lead to alteration in final sub-cellular localization of the encoded peptide, consistent with the observed change in function from di- to sesqui-TPS activity.
The βα bi-domain TaKSL5 is most closely related to the corresponding regions of the tri-domain OsKSL3 (Figures 3 & 6), which is a pseudo-gene containing an inactivating frame-shift mutation. While previous attempts to discern the original activity of OsKSL3 by correction of the frame-shift mutation did not result in appreciable activity in vitro (Xu, et al. 2007a), expression of this corrected OsKSL3 in our metabolic engineering system demonstrated its ability to convert ent-CPP into ent-kaurene. This functional conservation between TaKSL5 and OsKSL3 is further consistent with relatively recent divergence. In addition, while activity was similarly not observed with the more distantly related full-length OsKSL2, it could be truncated to yield a bi-domain TPS that also exhibited activity with ent-CPP (albeit with only trace production of an unknown diterpene). While this might be taken as evidence that such “internal/γ” domain loss simply occurs as a result of random drift, it should be noted such truncation was not more widely applicable, as analogous truncation of the most closely related wheat TPS, TaKSL2, led to an inactive enzyme.
More critically, the “internal/γ” domain loss events that led to DsKSL and TaKSL5 seem to have occurred separately from each other, and appear to be substantially more recent than the ancient such event (i.e. very early in angiosperm evolution), which has been suggested by previous phylogenetic analyses to lead to the majority of mono- and sesqui-TPSs i.e. those found in the TPS-a, -b, and -g sub-families (Bohlmann, et al. 1998, Chen, et al. 2011, Martin, et al. 2004, Trapp and Croteau 2001). Nevertheless, there is similarity between the “internal/γ” domain loss events leading to DsKSL and TaKSL5, which seems to have both occurred with the 3′ end of the “deletion” falling within a particular intron, such that their shared homology begins precisely at the beginning of an exon (Figures 3 & S5). By contrast, it has been suggested that the more ancient “internal/γ” domain loss event occurred within the preceding exon (Trapp and Croteau 2001).
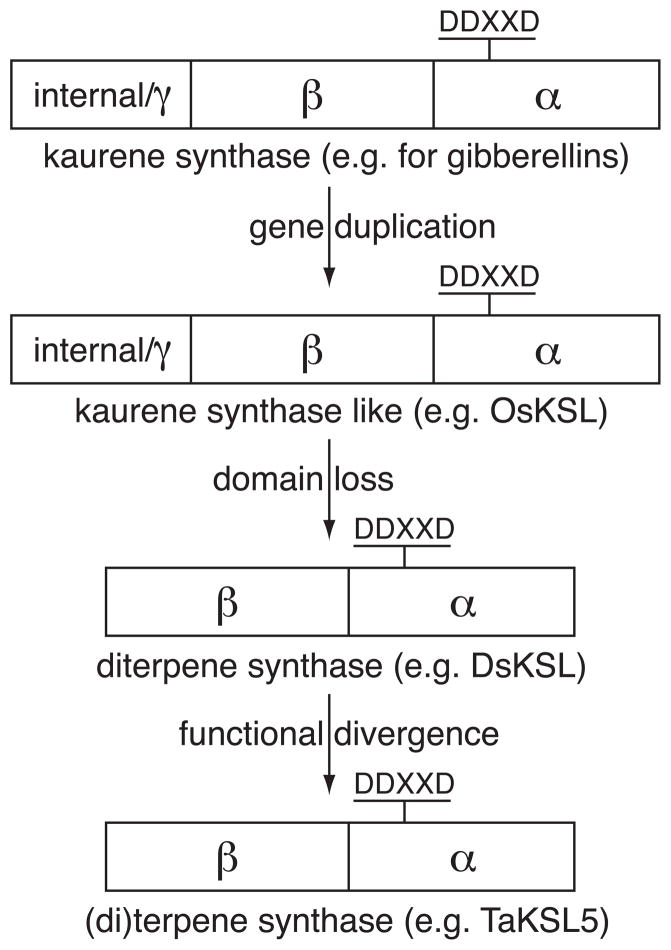
Regardless of exact details, given the observations reported here indicating that “internal/γ” domain loss has occurred repeatedly in diterpene synthases, specifically KSL, it seems appropriate to revisit the proposed origins for plant TPSs. Specifically, we hypothesize that other TPSs arose most directly from KSL (Figure 8). This would be consistent with earlier suggestions that the ancestral bifunctional diterpene synthase underwent subfunctionalization to yield the monofunctional CPS and KS, with independent diversification of the ancestral gene to yield the bifunctional diterpene synthases observed in gymnosperms (Keeling, et al. 2010). However, it is more detailed in specifying that the other TPSs were generated by a sequential process, with gene duplication of KS leading to formation of a KSL no longer required for gibberellin biosynthesis, which then could undergo “internal/γ” domain loss. Presumably, such a drastic alteration in protein structure is not compatible with the strict requirements for enzymatic specificity and/or regulatory mechanisms associated with gibberellin biosynthesis, explaining why KS are consistently observed to contain the “internal/γ” domain. Indeed, the findings reported here provide examples of the putative intermediates in such a process. In particular, DsKSL is a bi-domain diterpene synthase, representing the initial step of “internal/γ” domain loss with retention of function, while TaKSL5 represents the next step in being a sesquiterpene synthase that retains notable sequence and functional conservation with KS(L). In any case, and perhaps most importantly, our data strongly indicate that loss of this “internal/γ” domain has independently occurred multiple times, which is consistent with previous phylogenetic relationships derived from sequence analysis (Bohlmann, et al. 1998, Martin, et al. 2004, Trapp and Croteau 2001), but highlights the evolutionary complexity of plant terpene synthase gene family expansion.
Figure 8. Proposed process for the repeated evolution of bi-domain terpene synthases.
Gene duplication and divergence of the ent-kaurene synthase (KS) required for gibberellin biosynthesis leads to KS-like (KSL) diterpene synthases, whose function can diverge. Subsequent loss of the “internal/γ” domain results in bi-domain KSLs such as DsKSL. Further functional divergence then enables reaction with other isoprenyl diphosphate substrates, presumably via intermediate states still able to catalyze the ancestral diterpene synthase reaction such as TaKSL5.
EXPERIMENTAL PROCEDURES
Cloning and truncation of DsKSL and TaKSL5
Due to difficulties in manipulation of the native gene sequence, a full-length version of DsKSL was synthesized with codon-optimization for expression in Escherichia coli (Genscript USA Inc., Piscataway, NJ). To remove the putative 5′ transit peptide sequence, as predicted by the ChloroP 1.1 Server (Emanuelsson et al. 1999), as well as based on AA sequence alignment (Figures 2, 3 and S6) and previous experience with truncating rice KSL (Xu, et al. 2007a), a series of truncations were made by PCR (d22, d46, d54, d72, and d84), and subsequently cloned into the Gateway system via the pENTR/Shine-Delgarno/D-TOPO directional cloning kit (Invitrogen, Carlsbad, California). Likewise, two closely related TaKSL, designated TaKSL5-1 and TaKSL5-2 were cloned and multiple predicted transit sequence truncation site constructs were developed in the same manner for both, specifically for TaKSL5-2 (d34, d94, d121, and d131) and its putative homoeolog TaKSL5-1 (d34, d73, d100, and d112). TaKSL5-2d121 was found to have optimal activity relative to all other constructs, and TaKSL5-2 has been designated TaKSL5 here. These constructs were verified by complete sequencing and then transferred via directional recombination into pDEST14, pDEST15, and pDEST17 expression vectors using LR clonase (Invitrogen).
Analysis of DsKSL and TaKSL5 activity
The activity of the pDEST14/sDsKSL deletion constructs and the pDEST15/TaKSL5 deletion constructs were tested by expression in our previously described metabolic engineering system (Cyr, et al. 2007, Morrone et al. 2010). Briefly, this enabled co-expression of DsKSL with both a GGPP synthase and a CPP synthase (either one producing normal, ent-, or syn-CPP, separately). This system was further modified to enable analysis of sesquiterpene synthase activity by either use of a previously described pMBIS construct that includes a trans-FPP synthase (Martin, et al. 2003), or incorporation of the previously described cis-FPP synthase (Sallaud, et al. 2009). The latter was accomplished by synthesis of a codon-optimized (for expression in E. coli) gene construct (Genscript USA Inc., Piscataway, NJ), which was cloned into the Gateway system via the pENTR/Shine-Delgarno/D-TOPO directional cloning kit and verified by complete sequencing prior to transfer via directional recombination into a previously described DEST cassette containing pACYC-Duet (Novagen, Madison, WI) vector for co-expression (Morrone, et al. 2010). Hexane extracts from the resulting recombinant cultures were analyzed on a Varian (Palo Alto, CA) 3900 GC with Saturn 2100 ion trap mass spectrometer (MS) in electron ionization (70 eV) mode. Separation of the injected sample (1 μl) was achieved with a H2 flow rate of 2 mL/min with a temperature program of 3-min at 50 °C, followed by a gradient from 50 to 300 °C at 14 °C/min with a 3-min hold at 300 °C.
Expression and purification of sDsKSLd54 and TaKSL5d121
For protein purification pDEST17/sDsKSLd54 and pDEST17/TaKSL5d121 constructs (which provide a 6×His tag for purification) were transformed into chemically competent C41 cells (Lucigen Corp., Middleton, WI) and cultured overnight on NZY plates. Culture tubes containing 5 mL of NZY media with 50μg/mL carbenicillin were inoculated with 3–5 colonies from the over-night transformation plates, grown at 37 °C to an optical density of 0.5, and used to inoculate a 2800-mL firnbach flask containing 1 L of NZY and 50 μg/mL carbenicillin. These cultures were grown in an incubator/shaker at 37°C to a density of 0.7, then the temperature dropped to 16 °C. One hour later 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) was added to induce expression of both transformed plasmids, and fermentation continued overnight at 16 °C. Subsequently, the cultures were transferred to two 500 mL tubes and centrifuged for 20 min at 6,370× g. Cell pellets were re-suspended in 10 mL of protein lysis buffer (10 mM Tris-HCl, pH 6.8, 10 mM MgCl2, 1 mM dithiothreitol, 10% glycerol) and sonicated for 10 s three times on ice and transferred to 50 mL tubes and centrifuged for 20 min at 8,000× g at 4 °C to pellet cell debris. The cleared lysate was transferred to 2 mL of pre-washed nickel-NTA bead (Qiagen, Valencia, CA) slurry and incubated for 1 h at 4 °C. Thereafter, the nickel-NTA beads were rinsed three times with 20 mL of binding buffer (50 mM NaHPO4, 300 mM NaCl, 10 mM MgCl2, 10 mM imidazole). The sDsKSLd54 was then eluted by addition of 3 mL of elution buffer (50 mM NaHPO4, 300 mM NaCl, 10 mM MgCl2, 150 mM imidazole) to the bead bed twice. Eluted proteins were subjected to dialysis (50 mM NaHPO4, 10% glycerol, 10 mM MgCl2) three times for 1 h in a 25-kDa molecular weight cutoff membrane (Spectrum Laboratory Products, Inc., Gardena, CA) to remove the imidazole. The same protocol was used to purify the His-tagged TaKSLd121 protein. The purified proteins were quantified with a spectrophotometer using A280 and the calculated extinction coefficients for the corresponding peptides, and used immediately for enzymatic assays.
Biosynthesis of normal and ent-CPP
To produce substrate for sDsKSLd54 kinetics studies, normal CPP was synthesized enzymatically using a mutant version of abietadiene synthase that contains an inactive class I active site (rAgAS:D621A)(Peters et al. 2001). The previously characterized copalyl diphosphate synthase from Arabidopsis (AtCPSd84) was purified for synthesis of ent-CPP substrate, required for TaKSL5d121 assays. A 6×His tagged form of the mutant recombinant AgAS and AtCPSd84 were purified in the same manner as described above for sDsKSLd54 and TaKSL5d121. For 1 mL reaction assays, 150 nM of purified enzyme was added to reaction buffer consisting of 100 mM HEPES pH 7.2, 50 uL glycerol, 100 mM KCl, 1 mg alpha-casein, 100 μM MgCl2, 5 mM dithiothreitol (DTT) and 200μM (E,E,E)-geranylgeranyl diphosphate (GGPP) substrate (Isoprenoids LC, Tampa, Florida). Reactions were allowed to incubate 12 h at 22 °C in the dark. To confirm 100% conversion of GGPP to normal-CPP, samples of the reactions were de-phosphorylated with calf intestinal alkaline phosphatase (New England Biolabs, Mass.), extracted with equal volume HPLC-grade hexanes (Sigma, St. Louis, MO), filtered, concentrated, and product turnover quantified using gas chromatography with flame ionization detection (GC-FID 6890N)(Agilent, Palo Alto, CA) with an HP-5 column, for comparison to authentic standards. Separation of the injected sample (3 μl) was achieved with a H2 flow rate of 2 mL/min with a temperature program of 3-min at 40 °C, followed by a gradient from 40 to 320 °C at 20 °C/min with a 3-min hold at 320 °C.
DsKSLd54 kinetic assays
Initial assays that were carried out to verify enzymatic activity were measured in the linear response range, including time-course experiments and varying enzyme concentration. Additional time course assays were performed to provide preliminary data to optimize the time parameter for substrate concentration based kinetic assays. Ultimately, a concentration of 270 nM was chosen for kinetic assays with a 3-min reaction time at 30 °C. Individual 1-mL assays were carried using a buffer containing 50 μM NaHPO4, 100 mM KCl, 10 mM MgCl2, 10% glycerol, and 2 mM DTT. A concentration range from 0.5–20 μM CPP substrate was used. At the end of the 3-min reactions the enzyme was deactivated by incubating the reaction vial at 80 °C for 1 min, followed by quenching in ice. Assays were extracted with hexanes and the organic solvent removed, evaporated to completion, dissolved in 50 μL of hexanes containing an internal standard (geranylgeraniol), and quantified by GC-FID analysis. In particular, with 3 μL injections run with a H2 flow rate of 2 mL/min using a temperature program of 3-min at 40 °C, followed by a gradient from 40 to 320 °C at 20 °C/min with a 3-min hold at 320 °C. Assays were performed in triplicate and averaged for each represented data point. The plotted data was fit using Kaleidagraph (Synergy Software, Reading, PA).
TaKSL5d121 assays
Substrate screening assays and kinetic assays were carried out using 620 nM purified TaKSL5d121, using the same assay as described above for DsKSLd54, except with a 30-min incubation time for screening various substrates (3-min for kinetic assays), followed by extraction with 2 mL of pentane. TaKSL5d121 was screened against all available diphosphate substrates, including NPP, GPP, (2E,6E)-FPP, GGPP (Isoprenoids LC, Tampa, Florida), and ent-CPP, all at a concentration of 100 μM. Pentane extracts were concentrated to 50 μL and initially injected on a Varian (Palo Alto, CA) Saturn 3900 GC-MS as stated above and for kinetics on an Agilent (Palo Alto, CA) 6890N GC-FID for quantification. In particular, with 3 μL injections run with a H2 flow rate of 2 mL/min using a temperature program of 3-min at 70 °C, followed by a gradient from 70 to 240 °C at 6 °C/min with a 3-min hold at 240 °C.
Phylogenetic analyses
The diterpene and other TPSs sequences used here are as follows (GenBank accession): DsKSL (EF635966), ZmTPS1 (NP_001105097), ShSBS (FJ194970), SlPS (FJ797957), and TaKSL5 (AB597961); the other wheat KS(L) TaKSL1 (AB597957), TaKSL2 (AB597958), TaKSL3 (AB597959), TaKSL4 (AB597960), and TaKSL6 (AB597962); the rice KS(L) OsKS (BAE72099)(Xu, et al. 2007a), pseudo-genes OsKSL2 (DQ823350) and OsKSL3 (DQ823351), syn-pimaradiene synthase OsKSL4 (AB126934)(Wilderman et al. 2004), ent-pimaradiene synthase OsKSL5 (DQ823352)(Kanno et al. 2006), ent-isokaurene synthase OsKSL6 (DQ823353), ent-cassadiene synthase OsKSL7 (DQ823354)(Cho et al. 2004), syn-stemarene synthase OsKSL8 (AB118056)(Nemoto et al. 2004), ent-sandaracopimaradiene synthase OsKSL10 (DQ823355)(Otomo et al. 2004), and syn-stemodene synthase OsKSL11 (DQ100373)(Morrone et al. 2006); HvKSL (AY551436) from barley (Hordeum vulgare); uncharacterized KS(L) from Ricinus communis (castor bean) RcKSL1 (XP_002533694), RcKSL2 (XM_002525795), RcKSL3 (XM_002525790), and RcKSL4 (XM_002525796), along with the known casbene synthase RcCS (XM_002513288)(Mau and West 1994); putative KS(L) from sunflower (Helianthus annuus) HaKSL1 (CBL42917), HaKSL2 (CBL42916), grape (Vitis vinifera) VvKSL (XM_002264969), tobacco (Nicotiana tabacum) NtKSL (AY528645), cucumber (Cucumis sativus) CsKS (AB045310), coffee (Coffea arabica) CaKS (FJ409845) and poplar (Populus trichocarpa) PtKS (XM_002311250); the known ent-kaurene synthases AtKS (AF034774) from Arabidopsis thaliana (Yamaguchi et al. 1998), SrKS (AAD34295) and SrKS2 (AAD34294) from Stevia rebaudiana (Richman et al. 1999), LsKS1 (BAB12441) from Lactuca sativa (Sawada et al. 2008), and PgKS (ACY25275) from Picea glauca (Keeling, et al. 2010); the abietadiene synthases AgAS (AGU50768) from Abies grandis (Stofer Vogel et al. 1996), GbLS (Q947C4) from Ginkgo biloba (Schepmann et al. 2001), and PaLS (AY473621) from Picea abies, as well as paralogous isopimaradiene synthase PaTPS-iso (Q675L5)(Martin, et al. 2004); the taxadiene synthase TbTS (TBU48796) from Taxus brevifolia (Wildung and Croteau 1996); an uncharacterized bifunctional diterpene synthase from the lycophyte Selegnella moellendorffii, SmCPS/KS (XP_002960350); and the bifunctional ent-kaurene synthases JsCPS/KS (BAJ39816) from the liverwort Jungermannia subulata (Kawaide et al. 2011), and PpCPS/KS (BAF61135) from the bryophyte Physcomitrella patens (Hayashi et al. 2006).
The relevant amino acid sequences were imported into CLC Bio Sequence Viewer 6 (CLC Bio, Cambridge, Mass.) and aligned (ClustalW) using gap settings of gap open cost=10 and gap extension cost=1. For phylogenetic analysis all sequences were truncated at the N-termini of the universally conserved β domain. The amino acid (AA) start position for these truncations were as follows: DsKSL (AA80), TaKSL1 (AA317), TaKSL2 (AA339), TaKSL3 (AA337), TaKSL4 (AA318), TaKSL5-1 (AA126), TaKSL6 (AA345), ZmTPS1 (AA73), OsKS (AA294), OsKSL2 (AA126), OsKSL3 (AA239), OsKSL4 (AA331), OsKSL5 (AA293), OsKSL6 (AA295), OsKSL7 (AA319), OsKSL8 (AA294), OsKSL10 (AA229), OsKSL11 (AA296), HvKS (AA342), RcKSL1 (AA226), RcKSL2 (AA276), RcKSL3 (AA312), RcKSL4 (AA285), PtKS (AA238), HaKSL1 (AA274), HaKSL2 (AA277), SrKS (AA277), SrKS2 (AA277), LsKS1 (AA285), CsKS (AA270), AtKS (AA270), CaKS (AA285), ShSBS (AA272), NtKSL (AA236), VvKSL (AA287), SlPS (AA273), PtKS (AA238), PtLS (AA333), AgAS (AA352), PaTPS-iso (AA350), PaLAS (AA342), TbTS (AA351), GbLS (AA354), RcCS (AA110), PgKS (AA250), JsCPS/KS (AA373), PpCPS/KS (AA367), SmCPS/KS (AA227).
Phylogenetic trees and bootstrap values (100 replicates) were calculated utilizing Phylogenetic Analysis Using Parsimony (PAUP)(Rogers and Swofford 1998), PhyML 3.0 (maximum likelihood)(Guindon et al. 2010), and the Neighbor-joining algorithm (Saitou and Nei 1987). This was performed using the PAUP 4.0 analysis software (Sinauer Associates, Sunderland, MA), Geneious Pro 5.1.6 (Biomatters, Auckland, New Zealand), and CLC Bio installed on a quad-core CPU desktop computer with 4 GB of memory running Windows 7 64-bit operating system. The presented phylogenetic trees were plotted using TreeView (Page 2002) and Geneious Pro 5.1.6.
Supplementary Material
Acknowledgments
The work reported here was supported by grants from the Key Project of Chinese National Programs for Fundamental Research and Development (973 Program: 2006CB-504700) and Chinese National Programs for High Technology Research and Development (863 Program: 2007AA02Z104) to L.H., Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to T.T., and a grant from the NIH to R.J.P. (GM076324).
References
- Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y, Mann FM, Huang C, Mukkamala D, Mudock MP, Mead ME, Prisic S, Wang K, Lin KY, Chang TK, Peters RJ, Oldfield E. Diterpene cyclases and the nature of the isoprene fold. Proteins. 2010;78:2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- Cho EM, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, Oikawa H, Toshima H, Shibuya N, Nojiri H, Omori T, Nishiyama M, Yamane H. Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J. 2004;37:1–8. doi: 10.1046/j.1365-313x.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ. A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc. 2007;129:6684–6685. doi: 10.1021/ja071158n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 2006;580:6175–6181. doi: 10.1016/j.febslet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T. Characterization of a rice gene family encoding type-A diterpene cyclases. Biosci Biotechnol Biochem. 2006;70:1702–1710. doi: 10.1271/bbb.60044. [DOI] [PubMed] [Google Scholar]
- Kawaide H, Hayashi K, Kawanabe R, Sakigi Y, Matsuo A, Natsume M, Nozaki H. Identification of the single amino acid involved in quenching the ent-kauranyl cation by a water molecule in ent-kaurene synthase of Physcomitrella patens. FEBS J. 2011;278:123–133. doi: 10.1111/j.1742-4658.2010.07938.x. [DOI] [PubMed] [Google Scholar]
- Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S, Bohlmann J. Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol. 2010;152:1197–1208. doi: 10.1104/pp.109.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Structure of Copalyl Diphosphate Synthase from Arabidopsis thaliana, a Protonation-Dependent Diterpene Cyclase. Nat Chem Biol. 2011a;7:431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature. 2011b;469:116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Faldt J, Bohlmann J. Functional characterization of nine norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 2004;135:1908–1927. doi: 10.1104/pp.104.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Mau CJD, West CA. Cloning of casbene synthase cDNA: Evidence for conserved structural features among terpenoid cyclases in plants. Proc Natl Acad Sci U S A. 1994;91:8497–8501. doi: 10.1073/pnas.91.18.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ. Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 2009;583:475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- Morrone D, Jin Y, Xu M, Choi SY, Coates RM, Peters RJ. An unexpected diterpene cyclase from rice: Functional identification of a stemodene synthase. Arch Biochem Biophys. 2006;448:133–140. doi: 10.1016/j.abb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ. Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol. 2010;85:1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Cho EM, Okada A, Okada K, Otomo K, Kanno Y, Toyomasu T, Mitsuhashi W, Sassa T, Minami E, Shibuya N, Nishiyama M, Nojiri H, Yamane H. Stemar-13-ene synthase, a diterpene cyclase involved in the biosynthesis of the phytoalexin oryzalexin S in rice. FEBS Lett. 2004;571:182–186. doi: 10.1016/j.febslet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Otomo K, Kanno Y, Motegi A, Kenmoku H, Yamane H, Mitsuhashi W, Oikawa H, Toshima H, Itoh H, Matsuoka M, Sassa T, Toyomasu T. Diterpene cyclases responsible for the biosynthesis of phytoalexins, momilactones A, B, and oryzalexins A-F in rice. Biosci Biotechnol Biochem. 2004;68:2001–2006. doi: 10.1271/bbb.68.2001. [DOI] [PubMed] [Google Scholar]
- Page RD. Visualizing Phylogenetic Trees Using TreeView. John Wiley & Sons, Inc; 2002. [DOI] [PubMed] [Google Scholar]
- Peters RJ. Two rings in them all: The labdane-related diterpenoids. Nat Prod Rep. 2010;27:1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ, Flory JE, Jetter R, Ravn MM, Lee HJ, Coates RM, Croteau RB. Abietadiene synthase from grand fir (Abies grandis): Characterization and mechanism of action of the "pseudomature" recombinant enzyme. Biochemistry. 2000;39:15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- Peters RJ, Ravn MM, Coates RM, Croteau RB. Bifunctional abietadiene synthase: free diffusive transfer of the (+)-copalyl diphosphate intermediate between two distinct active sites. J Am Chem Soc. 2001;123:8974–8978. doi: 10.1021/ja010670k. [DOI] [PubMed] [Google Scholar]
- Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE. Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J. 1999;19:411–421. doi: 10.1046/j.1365-313x.1999.00531.x. [DOI] [PubMed] [Google Scholar]
- Rogers JS, Swofford DL. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Syst Biol. 1998;47:77–89. doi: 10.1080/106351598261049. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sallaud C, Rontein D, Onillon S, Jabes F, Duffe P, Giacalone C, Thoraval S, Escoffier C, Herbette G, Leonhardt N, Causse M, Tissier A. A novel pathway for sesquiterpene biosynthesis from (Z,Z)-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell. 2009;21:301–317. doi: 10.1105/tpc.107.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Katsumata T, Kitamura J, Kawaide H, Nakajima M, Asami T, Nakaminami K, Kurahashi T, Mitsuhashi W, Inoue Y, Toyomasu T. Germination of photoblastic lettuce seeds is regulated via the control of endogenous physiologically active gibberellin content, rather than of gibberellin responsiveness. J Exp Bot. 2008;59:3383–3393. doi: 10.1093/jxb/ern192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepmann HG, Pang J, Matsuda SP. Cloning and characterization of Ginkgo biloba levopimaradiene synthase, which catalyzes the first committed step in ginkolide biosynthesis. Arch Biochem Biophys. 2001;392:263–269. doi: 10.1006/abbi.2001.2438. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci U S A. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Kollner TG, Gershenzon J, Degenhardt J. The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-beta-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol. 2002;130:2049–2060. doi: 10.1104/pp.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stofer Vogel B, Wildung MR, Vogel G, Croteau R. Abietadiene synthase from grand fir (Abies grandis) J Biol Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- Trapp SC, Croteau RB. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildung MR, Croteau RB. A cDNA clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J Biol Chem. 1996;271:9201–9204. doi: 10.1074/jbc.271.16.9201. [DOI] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ. Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 2004;39:309–318. doi: 10.1111/j.1365-313X.2004.02137.x. [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya N, Coates RM, Peters RJ. Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry. 2007a;68:312–326. doi: 10.1016/j.phytochem.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Peters RJ. Following evolution’s lead to a single residue switch for diterpene synthase product outcome. Proc Natl Acad Sci USA. 2007b;104:7397–7401. doi: 10.1073/pnas.0611454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Sun T, Kawaide H, Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998;116:1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.