Abstract
Thirteen years after the demonstration of quantum dots as biological imaging agents, and nine years after the initial commercial introduction of bioconjugated quantum dots, the brightness and photostability of the quantum dots has enabled a range of investigations using single molecule tracking. These materials are being routinely utilized by a number of groups to track the dynamics of single molecules in reconstituted biophysical systems and on living cells, and are especially powerful for investigations of single molecules over long timescales with short exposure times and high pointing accuracy. New approaches are emerging where the quantum dots are used as “hard-sphere” probes for intracellular compartments. Innovations in quantum dot surface modification are poised to substantially expand the utility of these materials.
Introduction
Quantum dots are an alternative fluorescent material with unique spectral properties (broad excitation, narrow emission) and substantially improved photostability compared to conventional fluorescent molecules. These materials are based on inorganic semiconductor particles, typically 2—10 nm in size. Because the quantum dots are based on inorganic materials, they can be very bright, with extinction coefficients 10—50-fold higher than dye molecules of comparable emission spectra. The inorganic basis of the materials also accounts for the exceptional photostability, allowing observation of quantum dots over timescales that are more biologically limited than probe limited.
Since the initial publications in 1998, the use of semiconductor nanocrystals (also known as quantum dots, qdots or QDs) for biological detection has exploded [1,2]. As with many new technologies, there was a gap of several years from the initial reports to the appearance of robust biological applications using these new materials, due in part to the lack of availability of standardized materials, and due to the difficulties inherent in reproducibly synthesizing high-quality, luminescent, water-soluble quantum dots coupled to biological molecules. Nevertheless, bioconjugated quantum dots were made available commercially in 2002, following our publication demonstrating a robust chemistry based on use of an amphiphilic polymer to transfer the as-synthesized tri-octylphosphine oxide coated quantum dots to water and aqueous buffer systems [3]. These materials provided a relatively consistent starting point for biological researchers interested in the properties of the quantum dots, and were quickly used for a range of applications in single-particle tracking, both on living cells [4-7] and in reconstituted biophysical studies of single molecular motors [8-10].
The cornerstones of these related methodologies are the quantum dot brightness, which allows high signal-to-background detection and localization accuracy of <10 nm within a ~5 ms exposure, and the photostablity of the quantum dots, which allows tracking the position in 3D over prolonged periods. These properties enabled the observation of dynamic biological events that take place with rates up to ~100 s-1, over timescales from seconds to minutes.
Because the quantum dots are substantially larger than typical fluorescent molecules, the delivery across the plasma membrane to study intracellular protein dynamics remains a significant challenge. In this review, I will discuss some recent applications of QDs in reconstituted biophysical systems and the recent expansion of applications employing QDs for single-particle tracking on and in living cells. In addition, I will review some highlights of the recent literature on quantum dot surface chemistry and conjugation. Quantum dots are now being used in a broad array of applications, including flow cytometry, immunofluorescence, in-vivo imaging, and as direct and energy-transfer-based reporter systems, the review of which is beyond the scope of this contribution.
Reconstituted biophysical systems—walking the line
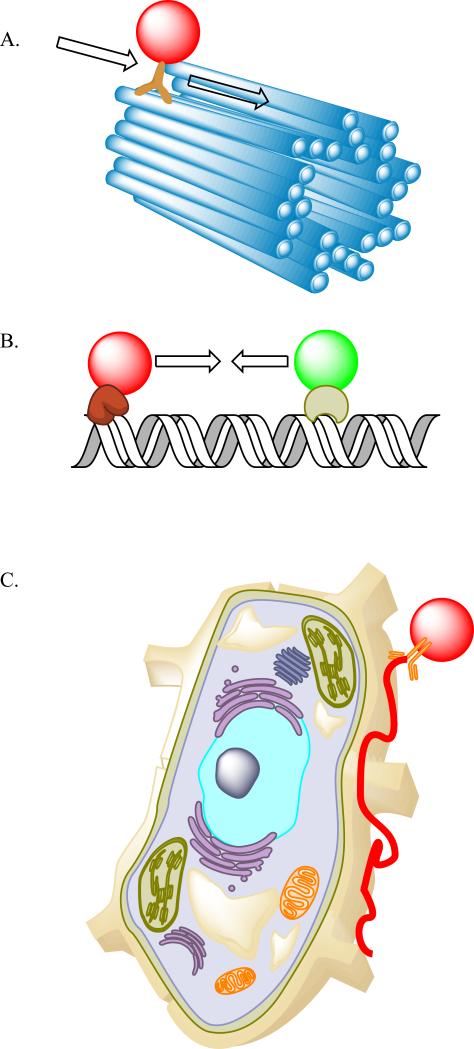
A number of studies have made use of the QD properties to investigate processive enzymes on immobilized tracks (Figure 1). The biophysical processes involved are well matched to the optical properties of the quantum dots, as many of these systems move with velocities near 1 μm/second, but with trajectories consisting of discrete steps (typically 30—80 nm steps). The quantum dots have become a routine tool in a number of labs to investigate the processivity of molecular motors on their pre-formed tracks. Some recent investigations have even used the quantum dots to study the influence of the motors on the tracks themselves. In addition to the study of typical molecular motors, recent work has demonstrated the utility of quantum dots for the study of DNA-Protein interactions in DNA replication and repair.
Figure 1. Single Particle Tracking in reconstituted biophysical systems and living cells.
A. The investigation of motor proteins involves quantum dot labeling of the motor domain, and subsequently measuring the trajectory of the QD labeled motor along the cytoskeletal track. B. DNA-Protein interactions can be probed with one or multiple QD labeled proteins, where the DNA is immobilized, and the interactions or disruptions of one labeled protein with another can be ascertained by dynamic imaging. C. Single receptors can be easily labeled and tracked at the surface of the cell or during internalization (not illustrated).
Motor proteins
One of the first motors studied using quantum dots was myosin V on actin. Early work by Warshaw and Trybus [10] demonstrated that N-terminal biotinylation of the head domains coupled with statistical labeling using a mixture of two spectrally distinct streptavidin quantum dot conjugates produced functional motors, reporting the position of the two heads relative to each other during myosin stepping. More recently, these investigators have coupled the step-size analysis with resistive load analysis using a tail-coupled laser trap to determine the correlations between the head-steps and tail-steps, providing additional evidence for the hand-over-hand mechanism of myosin V motion [11]. In addition, these investigators developed an approach that delivered labeled myosin Va motors into living cells through osmotic lysis of pinosomes, and visualized the random walk behavior of single motors in the cortical actin meshwork [12].
Nishikawa, Yanagida and colleagues demonstrated that fusion of a Halo-tag domain at the N-terminus of myosin VI allowed attachment of quantum dots to the head by a biotin-streptavidin quantum dot labeling approach. These motors displayed evidence of step-switching behavior, alternating between a hand-over-hand stepping motion and a shorter inchworm step [13].
Dahan's initial work with kinesin motors demonstrated that biotin-streptavidin mediated QD coupling could produce a functionally labeled motor, and proved that osmotic lysis of pinosomes could deliver the QD labeled motors into the cell [14]. These motors were not site-specifically labeled, so while the motors could be tracked within the cell, more quantitative biophysical analysis of the step size and motor dynamics was not possible. Recent studies from the collaboration between Warshaw and Trybus revealed that labeling of various kinesin-1 constructs by QDs using anti-His6 QD conjugates, streptavidin conjugates, or carboxylated quantum dots resulted in different dynamics in contact with microtubules. This analysis revealed a diffusive motion, where the kinesin remained microtubule bound, moving along the microtubule undirected, but without ATP hydrolysis. In addition, this study revealed that QD binding can serve as an active regulator, not just as a passive fluorescent label, depending on the mode of binding[15].
Two groups in particular have turned these problems upside down, and used the quantum dots to label the track (microtubules). This allows independent assessment of the influence of specific motor domains on their cellular routes. Coupled with sensitive 3D molecular localization, Nishizaka[16] and Diez[17] were able to visualize the rotation of single microtubules by immobilized motor domains.
DNA-Protein interactions
Two groups in particular have extended the use of QDs in reconstituted biophysical analysis in a fruitful new direction: the study of DNA-protein interactions. These groups used innovative approaches to orient and capture DNA, either as hydrodynamically stretched curtains[18-20] which can be extended and released on-demand, or as hydrodynamically stretched tightropes[21], where the DNA extends between two beads, providing a free-floating double-stranded DNA that can be accessed by labeled proteins. Both approaches allow for facile labeling of the DNA structures with intercalating dyes, simultaneous with the localization of QD-tagged proteins. This has provided new insights into the interactions of these proteins with their DNA substrates. These studies have been applied to DNA repair complexes [20-22], DNA translocases [19] and nucleosome-DNA assemblies [18]. In these experiments, the long-term stability of the quantum dots allows detailed measurements of the sequential timing of protein interactions with the DNA and DNA-bound proteins at particular sites.
Cellular single particle tracking
The unique stability and brightness of the quantum dots, coupled with the relatively large size, has proven to be a natural fit for tracking proteins and molecules at the cell surface. Because the viscosity of the membrane is substantially higher than that of the media, the diffusional slowing induced by a quantum dot is insignificant even for measurements of lipid diffusion. These experiments have used a variety of labeling and targeting approaches to deliver QDs to the cellular target including metabolic biotinylation, biotin labeling of a physiological ligand, antibody sandwich labeling and genetic targeting of avidin constructs. This experimental framework has been applied to a range of biological targets, such as GPI-anchored proteins [23], AMPA-receptors [24], voltage gated ion channels such as BK [25], complex receptor assemblies such as the high-affinity IgE receptor [26], and receptor tyrosine kinases, such as the insulin receptor [27]. While most of these approaches involve analysis of several trajectories, typically in two-dimensions, a new approach by Wells et al. that uses a 4-position sensor to lock-in to a particle position based on maximal intensity has allowed 3D tracking of a single quantum dot through surface labeling, diffusion, and subsequent internalization throughout the volume of the cell [28].
The size of the quantum dots has sometimes posed problems in these tracking experiments, for example by limiting access to synaptic sites as reported by Groc and colleagues [5]. However, a trio of papers have used the hard-sphere properties of quantum dots to assess exclusion cutoffs in biological systems, establishing size-cutoffs for Kiss-and-Run vesicle fusion [29], organization of nuclear chromatin [30], and the porosity of the actin meshwork in rapidly migrating cells [31]. Clearly, combining the hard-sphere properties of QDs with single-particle tracking methods could emerge as a powerful approach to address questions such as the influence of cellular sieving on motors, or on the sorting of receptors in living, crowded systems.
Innovations in quantum dot chemistry
The commercially available quantum dots, which make up the bulk of materials being used for biological investigations, are based on minor modifications of the amphiphilic polymer chemistry reported in 2002 [3]. These materials are relatively large, with hydrodynamic diameters near 20—30 nm, limiting the utility of these materials to problems where the size is either a benefit or a limited liability. A number of groups have developed alternative approaches to reduce the quantum dot size in solution, as outlined in Figure 2. There have been a large number of innovations in quantum dot chemistry. These are highlighted as a representative, rather than a comprehensive list.
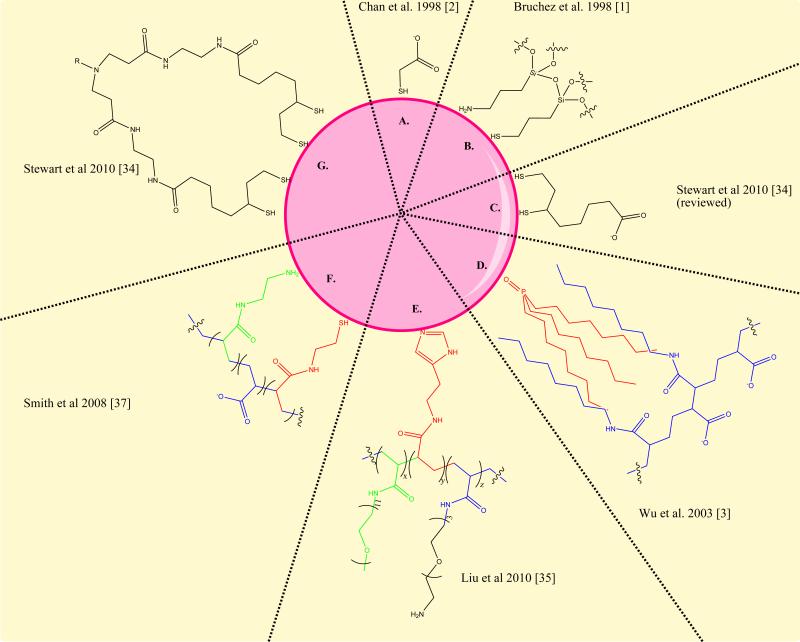
Figure 2. Variations of QD surface chemistry.
Over the past thirteen years, quantum dot surface chemistry has progressed substantially. Some of the initial coatings (A, B, C) were of limited stability and reproducibility. The amphiphilic polymer stabilization (D) developed in 2002, was implemented in the commercially available materials from Invitrogen. More recent chemistries developed to improve the stability and size of quantum dots use multiple attachment groups in RAFT synthesized polymers (E), modified polyacrylic acid (F) or discrete multivalent ligands (G). These final 3 alternatives result in a roughly 50% decrease in the overall hydrodynamic diameter of the aqueous quantum dots.
These chemical improvements have focused on reducing the size and increasing the chemical stability of the water-soluble quantum dots. Generally, the approach that has proven most fruitful at addressing these issues is the use of multivalent ligands. Multivalent thiols [32-34], imidazoles [35] and combinations of amines and thiols [36] have been used to produce materials with long-term stability, retained luminescence, and smaller sizes (roughly half the size). Introduction of PEG, hydroxyl, and sulfobetaine capping groups have produced quantum dots with reduced non-specific binding [35,37,38]. One innovative chemical approach introduced photoconvertible molecules into an amphiphilic stabilizer, producing quantum dots that can be robustly and reversibly photo-activated [39].
To simplify the use of QDs in biological experiments, a number of groups have developed straightforward conjugation and targeting strategies. The commercial streptavidin conjugates (Invitrogen) have 2—80 binding sites per quantum dot [40]. Because this multivalency can have significant biological influence, approaches that simplify conjugation or that robustly prepare mono-functional conjugates have been pursued. The available suite of coupling chemistries used to prepare various quantum dot conjugates now includes bioorthogonal chemistry [41], metal-affinity binding [42], intein-mediated coupling [43,44], scFv binding [45] and split protein complementation [46]. This flexibility allows quantum dots to be more seamlessly integrated into conventional cell-biology measurements.
Conclusions
Quantum dots represent an unusual addition to the toolkit for biological detection. Unlike conventional fluorescent dyes, the engineering aspects of QDs will allow significant improvements to accumulate over time, as innovations developed in motivated academic and commercial labs find their way into widely available materials and garner broader adoption. The use of QDs in biophysical studies and single particle tracking has provided a critical initial application where the benefits outweigh the liabilities. In addition, the investigators advancing these methods have clearly articulated the limits and advantages of the materials. Collaborations with the chemists developing the materials are giving rise to alternative modifications of QDs with improved properties for next-generation experiments. We can hope that the material improvements we are seeing in the literature are quickly translated into robust products to ensure that a broader community is able to use the advantages of QD-based detection. Personally, I am glad to see that quantum dots are now rarely referred to as a “novel” fluorophore in papers where they are used to address important biological problems.
Highlights.
Quantum dots are bright, stable probes with many uses in single particle tracking.
QD labeled molecular motors have established important biophysical mechanisms.
QDs have been used recently in studies of DNA-protein interactions.
QDs remain a standard tool in single particle tracking on living cells.
Innovations in QD chemistry have improved the biophysics and usability as probes.
Acknowledgements
MPB prepared this manuscript with the support of National Institutes of Health grants 5R01GM086237 and the National Technology Center for Networks and Pathways grant 7U54RR022241.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*Of Special Interest
**Of Outstanding Interest
- 1.Bruchez M, Jr., Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281(5385):2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 2.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281(5385):2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Immunofluorescent labeling of cancer marker her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21(1):41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 4.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302(5644):442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 5.Groc L, Heine M, Cognet L, Brickley K, Stephenson FA, Lounis B, Choquet D. Differential activity-dependent regulation of the lateral mobilities of ampa and nmda receptors. Nat Neurosci. 2004;7(7):695–696. doi: 10.1038/nn1270. [DOI] [PubMed] [Google Scholar]
- 6.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Quantum dot ligands provide new insights into erbb/her receptor-mediated signal transduction. Nat Biotechnol. 2004;22(2):198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 7.Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. One at a time, live tracking of ngf axonal transport using quantum dots. Proc Natl Acad Sci U S A. 2007;104(34):13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toprak E, Enderlein J, Syed S, McKinney SA, Petschek RG, Ha T, Goldman YE, Selvin PR. Defocused orientation and position imaging (dopi) of myosin v. Proc Natl Acad Sci U S A. 2006;103(17):6495–6499. doi: 10.1073/pnas.0507134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126(2):335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warshaw DM, Kennedy GG, Work SS, Krementsova EB, Beck S, Trybus KM. Differential labeling of myosin v heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J. 2005;88(5):L30–32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu H, Kennedy GG, Warshaw DM, Trybus KM. Simultaneous observation of tail and head movements of myosin v during processive motion. J Biol Chem. 2010;285(53):42068–42074. doi: 10.1074/jbc.M110.180265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12 *.Nelson SR, Ali MY, Trybus KM, Warshaw DM. Random walk of processive, quantum dot-labeled myosin va molecules within the actin cortex of cos-7 cells. Biophys J. 2009;97(2):509–518. doi: 10.1016/j.bpj.2009.04.052. [A useful protocol for delivering qdot conjugates into the cytoplasm of living cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishikawa S, Arimoto I, Ikezaki K, Sugawa M, Ueno H, Komori T, Iwane AH, Yanagida T. Switch between large hand-over-hand and small inchworm-like steps in myosin vi. Cell. 2010;142(6):879–888. doi: 10.1016/j.cell.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 14.Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Tracking individual kinesin motors in living cells using single quantum-dot imaging. Nano Lett. 2006;6(7):1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 15.Lu H, Ali MY, Bookwalter CS, Warshaw DM, Trybus KM. Diffusive movement of processive kinesin-1 on microtubules. Traffic. 2009;10(10):1429–1438. doi: 10.1111/j.1600-0854.2009.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16 **.Yajima J, Mizutani K, Nishizaka T. A torque component present in mitotic kinesin eg5 revealed by three-dimensional tracking. Nat Struct Mol Biol. 2008;15(10):1119–1121. doi: 10.1038/nsmb.1491. [An elegant demonstration of 3-d tracking of quantum dots to trace out rotation of microtubules under motor activity.] [DOI] [PubMed] [Google Scholar]
- 17 **.Nitzsche B, Ruhnow F, Diez S. Quantum-dot-assisted characterization of microtubule rotations during cargo transport. Nat Nanotechnol. 2008;3(9):552–556. doi: 10.1038/nnano.2008.216. [An interesting 3-d tracking approach, where the target is using microtubule sliding as a nanotechnology assembly/transport system.] [DOI] [PubMed] [Google Scholar]
- 18 **.Visnapuu ML, Greene EC. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat Struct Mol Biol. 2009;16(10):1056–1062. doi: 10.1038/nsmb.1655. [A powerful framework for robust investigations of DNA-protein interactions at a single molecule level.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19 **.Finkelstein IJ, Visnapuu ML, Greene EC. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature. 2010;468(7326):983–987. doi: 10.1038/nature09561. [A very interesting study using collisions between two proteins on a single DNA, illustrating relative energetics and priority of bound species.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17(8):932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21 **.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37(5):702–713. doi: 10.1016/j.molcel.2010.02.003. [A complementary method to the DNA curtain approach where the DNA is stretched across two beads, allowing free access from all directions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Tessmer I, Croteau DL, Erie DA, Van Houten B. Functional characterization and atomic force microscopy of a DNA repair protein conjugated to a quantum dot. Nano Lett. 2008;8(6):1631–1637. doi: 10.1021/nl080316l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinaud F, Michalet X, Iyer G, Margeat E, Moore HP, Weiss S. Dynamic partitioning of a glycosyl-phosphatidylinositol-anchored protein in glycosphingolipid-rich microdomains imaged by single-quantum dot tracking. Traffic. 2009;10(6):691–712. doi: 10.1111/j.1600-0854.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opazo P, Labrecque S, Tigaret CM, Frouin A, Wiseman PW, De Koninck P, Choquet D. Camkii triggers the diffusional trapping of surface ampars through phosphorylation of stargazin. Neuron. 2010;67(2):239–252. doi: 10.1016/j.neuron.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Won S, Kim HD, Kim JY, Lee BC, Chang S, Park CS. Movements of individual bkca channels in live cell membrane monitored by site-specific labeling using quantum dots. Biophys J. 2010;99(9):2853–2862. doi: 10.1016/j.bpj.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews NL, Pfeiffer JR, Martinez AM, Haaland DM, Davis RW, Kawakami T, Oliver JM, Wilson BS, Lidke DS. Small, mobile fcepsilonri receptor aggregates are signaling competent. Immunity. 2009;31(3):469–479. doi: 10.1016/j.immuni.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giudice J, Leskow FC, Arndt-Jovin DJ, Jovin TM, Jares-Erijman EA. Differential endocytosis and signaling dynamics of insulin receptor variants ir-a and ir-b. J Cell Sci. 2011;124(Pt 5):801–811. doi: 10.1242/jcs.076869. [DOI] [PubMed] [Google Scholar]
- 28 **.Wells NP, Lessard GA, Goodwin PM, Phipps ME, Cutler PJ, Lidke DS, Wilson BS, Werner JH. Time-resolved three-dimensional molecular tracking in live cells. Nano Lett. 2010;10(11):4732–4737. doi: 10.1021/nl103247v. [The brightness and stability of the quantum dots allow robust 3-d tracking of single molecules on the surface of the cell and during the internalization using a feedback loop system with 4 detectors for position.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Li Y, Tsien RW. The dynamic control of kiss-and-run and vesicular reuse probed with single nanoparticles. Science. 2009;323(5920):1448–1453. doi: 10.1126/science.1167373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. Embo J. 2009;28(24):3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31 *.Keren K, Yam PT, Kinkhabwala A, Mogilner A, Theriot JA. Intracellular fluid flow in rapidly moving cells. Nat Cell Biol. 2009;11(10):1219–1224. doi: 10.1038/ncb1965. [A series of QDs with different PEG lengths and hydrodynamic diameters revealed the porosity of the cortical actin network in keratocytes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dif A, Boulmedais F, Pinot M, Roullier V, Baudy-Floc'h M, Coquelle FM, Clarke S, Neveu P, Vignaux F, Le Borgne R, Dahan M, et al. Small and stable peptidic pegylated quantum dots to target polyhistidine-tagged proteins with controlled stoichiometry. J Am Chem Soc. 2009;131(41):14738–14746. doi: 10.1021/ja902743u. [DOI] [PubMed] [Google Scholar]
- 33.Clarke S, Pinaud F, Beutel O, You C, Piehler J, Dahan M. Covalent monofunctionalization of peptide-coated quantum dots for single-molecule assays. Nano Lett. 2010;10(6):2147–2154. doi: 10.1021/nl100825n. [DOI] [PubMed] [Google Scholar]
- 34.Stewart MH, Susumu K, Mei BC, Medintz IL, Delehanty JB, Blanco-Canosa JB, Dawson PE, Mattoussi H. Multidentate poly(ethylene glycol) ligands provide colloidal stability to semiconductor and metallic nanocrystals in extreme conditions. J Am Chem Soc. 2010;132(28):9804–9813. doi: 10.1021/ja102898d. [DOI] [PubMed] [Google Scholar]
- 35.Liu W, Greytak AB, Lee J, Wong CR, Park J, Marshall LF, Jiang W, Curtin PN, Ting AY, Nocera DG, Fukumura D, et al. Compact biocompatible quantum dots via raft-mediated synthesis of imidazole-based random copolymer ligand. J Am Chem Soc. 2010;132(2):472–483. doi: 10.1021/ja908137d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith AM, Nie S. Minimizing the hydrodynamic size of quantum dots with multifunctional multidentate polymer ligands. J Am Chem Soc. 2008;130(34):11278–11279. doi: 10.1021/ja804306c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kairdolf BA, Mancini MC, Smith AM, Nie S. Minimizing nonspecific cellular binding of quantum dots with hydroxyl-derivatized surface coatings. Anal Chem. 2008;80(8):3029–3034. doi: 10.1021/ac800068q. [DOI] [PubMed] [Google Scholar]
- 38.Muro E, Pons T, Lequeux N, Fragola A, Sanson N, Lenkei Z, Dubertret B. Small and stable sulfobetaine zwitterionic quantum dots for functional live-cell imaging. J Am Chem Soc. 2010;132(13):4556–4557. doi: 10.1021/ja1005493. [DOI] [PubMed] [Google Scholar]
- 39.Diaz SA, Menendez GO, Etchehon MH, Giordano L, Jovin TM, Jares-Erijman EA. Photoswitchable water-soluble quantum dots: Pcfret based on amphiphilic photochromic polymer coating. ACS Nano. 2011;5(4):2795–2805. doi: 10.1021/nn103243c. [DOI] [PubMed] [Google Scholar]
- 40.Mittal R, Bruchez MP. Biotin-4-fluorescein based fluorescence quenching assay for determination of biotin binding capacity of streptavidin conjugated quantum dots. Bioconjug Chem. 2011;22(3):362–368. doi: 10.1021/bc100321c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han HS, Devaraj NK, Lee J, Hilderbrand SA, Weissleder R, Bawendi MG. Development of a bioorthogonal and highly efficient conjugation method for quantum dots using tetrazine-norbornene cycloaddition. J Am Chem Soc. 2010;132(23):7838–7839. doi: 10.1021/ja101677r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roullier V, Clarke S, You C, Pinaud F, Gouzer GG, Schaible D, Marchi-Artzner V, Piehler J, Dahan M. High-affinity labeling and tracking of individual histidine-tagged proteins in live cells using ni2+ tris-nitrilotriacetic acid quantum dot conjugates. Nano Lett. 2009;9(3):1228–1234. doi: 10.1021/nl9001298. [DOI] [PubMed] [Google Scholar]
- 43 *.Xia Z, Xing Y, So MK, Koh AL, Sinclair R, Rao J. Multiplex detection of protease activity with quantum dot nanosensors prepared by intein-mediated specific bioconjugation. Anal Chem. 2008;80(22):8649–8655. doi: 10.1021/ac801562f. [Intein mediated protein conjugation demonstrated with QD conjugates. This could be a very versatile conjugation method that would allow facile preparation of recombinant protein conjugates with controlled stoichiometry.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charalambous A, Andreou M, Skourides PA. Intein-mediated site-specific conjugation of quantum dots to proteins in vivo. J Nanobiotechnology. 2009;7(9) doi: 10.1186/1477-3155-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iyer G, Michalet X, Chang YP, Pinaud FF, Matyas SE, Payne G, Weiss S. High affinity scfv-hapten pair as a tool for quantum dot labeling and tracking of single proteins in live cells. Nano Lett. 2008;8(12):4618–4623. doi: 10.1021/nl8032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinaud F, Dahan M. Pnas plus: Targeting and imaging single biomolecules in living cells by complementation-activated light microscopy with split-fluorescent proteins. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1101929108. [DOI] [PMC free article] [PubMed] [Google Scholar]