Abstract
Human-associated microbiota is recognized to play vital roles in maintaining host health, and it is implicated in many disease states. While the initial surge in the profiling of these microbial communities was achieved with Sanger and next generation sequencing, many oligonucleotide microarrays have also been developed recently for this purpose. Containing probes complementary to small ribosomal subunit RNA gene sequences of community members, such phylogenetic arrays provide direct quantitative comparisons of microbiota composition among samples and between sample groups. Some of the developed microarrays including PhyloChip, Microbiota Array, and HITChip can simultaneously measure the presence and abundance of hundreds and thousands of phylotypes in a single sample. This review describes the currently available phylogenetic microarrays that can be used to analyse human microbiota, delineates the approaches for the optimization of microarray use, and provides examples of recent findings based on microarray interrogation of human-associated microbial communities.
Keywords: microbiota, microbial communities, microflora, phylogenetic, microarray, Microbiota Array, HITChip, PhyloChip
Introduction
Most microbes exist in nature not as populations of single species, but rather as multi-species assemblies – microbial communities. Such communities are found in ocean waters and terrestrial waterways, in soil, on the bark and leaves of plants, and on epithelial surfaces of animals (Kent & Triplett, 2002, Gao, et al., 2007, McLellan, et al., 2009, Olson & Kellogg, 2010, Grice & Segre, 2011). These microbial conglomerates play a central role in regulating the flow of energy and nutrients in the Earth’s ecosphere and are responsible for most of the biomass production in the ocean. Members of microbial communities process metabolites and energy as parts of a large network of interacting cells, where intermediary products of the metabolism of one species are often utilized by other members (Duncan, et al., 2004, Belenguer, et al., 2006, Flint, et al., 2008, De Vuyst & Leroy, 2011). This complex network of inter-species interactions also explains theh relatively young age of the field of microbial population ecology, since previously many of the community members could not be easily grown as pure cultures in a laboratory setting. The advent and development of molecular tools provided an opportunity to bypass the culturing requirements and study community composition, dynamics, and functionality directly.
Considerable progress has been made recently in the study of microbial populations associated with humans. Called the human microbiota, this microbial community is believed to contain at least 10 times the number of cells in the human body, with the cumulative microbial gene count estimated to be 100-fold larger than the human genome (Gill, et al., 2006). Renewed interest in the human microbiota is associated with the recognition of the important relationships these microbes form with our bodies. Human microbiota participate in the digestion of complex carbohydrates in the gut, in the protection of the host from pathogen invasion, in modulating proper immune function, and in the maintenance of epithelial homeostasis (Neish, 2009, Sekirov, et al., 2010). At the same time, perturbation of the normal microbiota have been shown to be associated with a number of diseases including dental plaque, bacterial vaginosis, psoriasis, atopic dermatitis, inflammatory bowel disease, obesity, and colon cancer (Larsen & Monif, 2001, Gao, et al., 2008, Sartor, 2008, Neish, 2009, Grice & Segre, 2011).
Different approaches can be taken to study complex microbial populations. We can look at the community membership where interrogation of small ribosomal subunit (SSU) RNA is used extensively to obtain the phylogenetic structure of the community (Suau, 2003, Sekirov, et al., 2010). We can profile community functionality by cataloguing the pool of functional genes and proteins present in the population through meta-genomic, meta-transcriptomic, and meta-proteomic approaches (Gill, et al., 2006, Klaassens, et al., 2007, Booijink, et al., 2010, Qin, et al., 2010, Arumugam, et al., 2011). We can also study the metabolic processes in microbial conglomerates by measuring levels of produced and consumed metabolites with metabolomic and metabonomic techniques (Marchesi, et al., 2007, de Graaf, et al., 2009, Martin, et al., 2010).
The majority of recent advances in our understanding of human microbiota structure and dynamic changes in disease were made through phylogenetic interrogation of SSU rRNA. Whereas many different techniques have been successfully employed to provide novel findings, next generation sequencing (NGS) and phylogenetic microarrays proved to be the most widely used. This review describes current advances in the use of phylogenetic microarrays to the study of human-associated microbiota.
Overview of currently available phylogenetic microarrays
While microarrays were originally developed (Schena, et al., 1995) and are widely used to monitor gene expression in both prokaryotic and eukaryotic cells, they are now also employed for comparative genomics, for DNA sequencing and single nucleotide polymorphism analysis, and for microbial detection (Wang, et al., 2002, Loy & Bodrossy, 2006). Several types of microarrays have been used to date to characterize the composition and function of microbial communities including community genome arrays, functional gene arrays, and phylogenetic microarrays (Zhou, 2003). Community genome arrays are constructed using whole genomic DNA isolated from strains in pure culture and allow detection of individual species and strains in simple and complex communities (Wu, et al., 2004). Functional gene arrays contain probes to genes encoding key enzymes involved in various biochemical processes, and are useful for monitoring physiological changes in microbial communities (Wu, et al., 2001, He, et al., 2007). An excellent example of a functional gene array is the GeoChip, which currently contains 84,000 oligonucleotide probes for genes involved in biogeochemical cycling of carbon, nitrogen, phosphorus, and sulfur, for genes involved in metal and antibiotic resistance, and for genes coding for bioremediation of organic compounds (Zhou, et al., 2010, Zhou, et al., 2011). Phylogenetic oligonucleotide arrays (phyloarrays) contain probes complementary to the small subunit (SSU) rRNA sequences and are thus well suited for the analysis of microbial community composition structure and variance. The choice of SSU rRNA for most phylogenetic studies is based on the ubiquity and sequence conservation of this molecule. Among different array types, POAs are currently the most popular due to the availability of a large set of near-full length rRNA sequences deposited in NCBI, EMBL, RDP, and Greengenes databases. Currently, over 480,000 total 16S rRNA sequences of human microbiota origin are catalogued in NCBI GenBank (with length ≥ 1000 bp). Based on these sequence databases, many different types of phylogenetic microarrays have been recently developed for the interrogation of human-associated microbiota as shown in Table 1.
Table 1.
Phylogenetic microarrays available to profile human microbiota
| Array name | Target community |
Sensitivity level * |
Platform | Detectable groups |
No. of all probes † |
No. of probes per group |
No. of each probe ‡ |
Reference |
|---|---|---|---|---|---|---|---|---|
| - | Intestinal biota | Species | Glass slide | 40 species | 120 Phylo. | 3 | 1 | (Wang, et al., 2004) |
| - | All bacteria | Species | Agilent | 1629 species | 10265 Tot. / 9121 Phylo. | N/A | 1 | (Palmer, et al., 2007) |
| PhyloChip** | All bacteria | Varied | Affymetrix | 842 subfamilies | 506944 Tot. / 297851 Phylo. | 11+ | 1 | (Brodie, et al., 2006) |
| HOMIM | Oral biota | Species | Glass slide | 272 species | 960 Tot. / 912 Phylo. | 1–2 | 2 | (Preza, et al., 2009) |
| Microbiota Array | Intestinal biota | Species | Affymetrix | 775 species | 16223 Tot. / 10976 Phylo. | 5–11 | 1 | (Paliy, et al., 2009) |
| HITChip | Intestinal biota | Species | Agilent | 1140 species | 4809 Tot. | 4–6 | 1 | (Rajilic-Stojanovic, et al., 2009) |
| AUS-HIT Chip | Intestinal biota | Varied | CustomArray | 500 species | 2242 Tot. / 2203 Phylo. | 1–2 | 3 | (Kang, et al., 2010) |
| - | Intestinal biota | Genus | Glass slide | 310 genera | 1461 Tot. / 1412 Phylo. | 1–48 | 2 | (Manges, et al., 2010) |
| V-Chip | Vaginal biota | Varied | Glass slide | 350 groups | 459 Phylo. | 1–3 | 1 | (Dols, et al., 2011) |
| OC chip | Oral biota | Varied | Glass slide | 350 groups | 420 Phylo. | 1–3 | 1 | (Crielaard, et al., 2011) |
N/A – information was not available
Species sensitivity is usually defined as ability to detect individual phylotypes defined at 98% similarity cutoff level; varied sensitivity describes arrays that contain probes for different taxonomic group levels from species to genera to families
Tot. – number of total probes on the array; Phylo. – number of probes targeting SSU sequences
Represents the number of different locations on the array each probe was placed on
The numbers are given for the version of the array (G2) that has been used most widely. An updated version of PhyloChip has been recently developed (Hazen, et al., 2010), the new format contains probes to 1,464 families and 10,993 sub-families of prokaryotes
The first phylogenetic array was developed by Guschin and colleagues in 1997. In their design, oligonucleotide probes complementary to the 16S rRNA sequences of selected genera of nitrifying bacteria were immobilized in gel pads on a glass slide (Guschin, et al., 1997). One of the first phyloarrays for human microbiota interrogation was described by Wang and co-workers, who placed 40-mer probes to 40 predominant gut bacteria onto epoxy slides. Using this array, researchers were able to detect between 25 and 37 species in human fecal samples (Wang, et al., 2004). The number of species and phylogenetic groups that can be detected by a single microarray has been expanded significantly in further studies. Palmer et al. queried the prokaryotic SSU rRNA database to develop oligonucleotide probes to over 1600 bacterial and archaeal species. The microarray was employed to profile fecal samples of 26 infants and showed that infant fecal microbiota displayed remarkable temporal and inter-personal variation (Palmer, et al., 2007). PhyloChip, an Affymetrix-based microarray, was also intended to cover all available prokaryotic 16S rRNA sequences and contains probes to operational taxonomic units (OTUs, also called phylotypes) representing thousands different subfamilies (Brodie, et al., 2006, Hazen, et al., 2010). This array has been used successfully in a number of studies looking not only at the human-associated microbiota (Cox, et al., 2010, Lemon, et al., 2010) but also profiling microbial communities in other environments (Wu, et al., 2010, Deangelis, et al., 2011, Mendes, et al., 2011). Another phylogenetic microarray based on Affymetrix photolithographic technology, Microbiota Array, was developed in our laboratory (Paliy, et al., 2009). This microarray was designed to be specific to bacteria residing in the human gastrointestinal tract, and contained multiple sets of probes to 775 different species. Microbiota Array has already been utilized to successfully profile distal gut populations of healthy adults and healthy children as well as children diagnosed with irritable bowel syndrome (IBS), inflammatory bowel disease (IBD), and obesity ((Agans, et al., 2011), (Rigsbee, et al., manuscript in submission), and (H. Kenche and O. Paliy, unpublished results)). Another microarray specific to human gut microbiota, HITChip, was developed in the group of Willem de Vos (Rajilic-Stojanovic, et al., 2009). This array can detect levels of 1140 different species and was employed in a number of recent studies (Biagi, et al., 2010, Booijink, et al., 2010, Van den Abbeele, et al., 2010).
Several new phylogenetic array designs have recently been added to the growing list as shown in Table 1. As can be surmised from the table, different array designs vary in the platform used, in the lowest taxonomic level interrogated, and in the choice of targeted communities. For example, both PhyloChip and Microbiota Array were developed utilizing the Affymetrix platform, which enables arrays to contain multiple probes for each tested OTU and also provides means to curtail potential cross-hybridization through the use of mismatch probes. At the same time, HITChip and the array developed by Palmer and colleagues are based on the Agilent system, which allows more cost-efficient update of the array design (since no expensive lithographic masks are used) as well as the profiling of multiple samples on a single slide. When many arrays are needed, the use of standard glass slide platform remains the most economical method due to low costs of the glass slides; however, extensive tests are often required to validate the quality of printed slides.
Another important consideration distinguishing various microarray designs is the specificity level and the intended target community. Species (OTU, phylotype) level specificity provides the deepest interrogation that is close to that achieved by Sanger sequencing and is currently better than what next generation sequencing allows, and a number of phylogenetic arrays achieve this specificity (Table 1). Reviewing targeting breadth of the available arrays, both PhyloChip and the array developed by Palmer et al. were designed to contain probes to as many prokaryotic species as possible. Such a design allows the microarray to be used in a variety of studies focused on different types of microbial populations, and PhyloChip is an excellent example of this design strategy with recent reports of microarray interrogation of human stomach, intestine and mouth, watershed communities, tropical soil, and plant rhizosphere (Cox, et al., 2010, Lemon, et al., 2010, Wu, et al., 2010, Deangelis, et al., 2011, Mendes, et al., 2011). The drawback of such broad microarray design is an expected higher number of false positives due to the combination of large number of probes present and probe cross-hybridization; this can be mitigated to some extent by a rigorous probe selection process and stringent criteria for positive detection calls. In contrast, phylogenetic arrays specific to one particular community, such as Microbiota Array and HITChip developed for the interrogation of human gastro-intestinal biota, benefit from the reduced cross-hybridization potential, but can only be applied to the analysis of that particular community.
Other reports were made available recently that described microarrays based on non-traditional technologies. Candela et al. developed a DNA microarray based on the use of fragment ligation reaction coupled with the interrogation of the ligated products on a “detection” array (Candela, et al., 2010). In this approach, successful ligation of two adjacent oligonucleotides is dependent on the presence of the complementary target sequence; this method relies on high selectivity of ligase and therefore can distinguish single-nucleotide differences. The ligated products are quantified on a specially designed “universal” detection microarray that contains probes complementary to the oligo tag sequences (“zip-codes”) incorporated into the ligated oligonucleotides. The use of this universal array creates uniform hybridisation conditions for all zip-coded sequences, and the same array platform can be used with multiple ligation probe sets which provides great flexibility (Hultman, et al., 2008). One of the pilot ligation microarrays was made to quantify levels of 30 groups of human intestinal microbiota and was used to profile fecal samples of several young adults (Candela, et al., 2010). Another microarray platform, termed restriction site tagged microarray, was developed by Zabarovsky et al. (2003). In this method, a rare-cutting restriction enzyme is chosen, and a set of short tags (19 bases long) is developed that match sequences flanking recognition sites of that restriction endonuclease in the genome of a particular species. A collection of such tags for one species constitutes that species “passport”. After digesting genomic DNA with the chosen restriction enzyme, different species can be distinguished through DNA hybridisation to the custom microarray containing probes complementary to the restriction site flanking regions (Zabarovsky, et al., 2003). The use of such restriction enzyme passports allows efficient discrimination of even closely related strains of bacteria; however, this approach has not yet been used for the in-depth interrogation of human-associated microbiota. Indeed, since this method relies on the use of organism genome sequence in order to develop that species’ passport, it might not be well suited to profile microbial communities with many uncultured and yet unsequenced members. Finally, microarrays based on the interrogation of large subunit rRNA genes have also been recently developed (Mitterer, et al., 2004, Yoo, et al., 2009). For example, Yoo and colleagues designed a diagnostic microarray containing multiple probes to 23S ribosomal DNA and 16S–23S intergenic spacers of 39 pathogenic bacteria. The DNA microarray was shown to have 100% specificity (no false positives) and close to 100% sensitivity (almost no false negatives; both values were defined based on comparison to culture-based identification) (Yoo, et al., 2009).
Optimizing the use of phylogenetic microarrays
Phylogenetic microarrays are one of a number of currently available molecular approaches for the interrogation of complex microbial communities. We provide a short comparison of the most widely used methods in Table 2; a more detailed comparison of these and other technologies is available in a comprehensive review of gut microbiota (Sekirov, et al., 2010). The main advantages of phylogenetic microarrays compared with other methodologies include (i) ability to profile one sample at a time, which is useful in clinical studies and as a diagnostic tool; (ii) quantitative nature of the acquired data allowing direct comparison of levels of each OTU between samples; (iii) short processing and data acquisition times (only two days from sample to data using Microbiota Array); and (iv) currently lower costs compared with NGS, especially if we take into account that microarrays typically provide greater overall level of coverage of sample mixture (Xu, et al., 2011). The main limitation of microarrays is their inability to reveal novel species in any samples, since the arrays can only detect those sequences for which they contain probes. In addition, the design, use, and analysis of microarrays are technically demanding and require extensive testing, validation, and optimization (Hashsham, et al., 2004). For example, cross-hybridization of DNA fragments to multiple probes needs to be controlled and adjusted for to avoid artificially over-estimating microbiota diversity (Rigsbee, et al., 2011).
Table 2.
Molecular methods for the analysis of human microbiota
| Phylogenetic microarrays |
NGS | Sanger sequencing |
FISH | qPCR | |
|---|---|---|---|---|---|
| Taxonomic resolution | Good | Moderate | Very good | Moderate | Moderate |
| Throughput capability | High | High | Moderate | Low | Low |
| Quantitative | Yes | Semi | No | Yes | Yes |
| Sensitivity | High | High | Moderate | Low | Very high |
| Cost | Moderate to High | High | Very high | Low | Moderate |
| Main advantage | High-throughput quantitation | Novel sequence discovery | Taxonomic identification | Cell visualisation | Sensitivity |
NGS – next generation sequencing; FISH – fluorescent in situ hybridization; qPCR – quantitative real-time PCR
A number of approaches have been developed in our laboratory and by others to improve the robustness of the estimates provided by phylogenetic microarrays. The work in our group has focused on three aspects. We have developed a mathematical model of 16S rRNA gene amplification (an experimental strategy employed in most studies to selectively enrich DNA samples with 16S rRNA genes) linked to phylogenetic microarray detection based on the Microbiota Array design. The model aimed to determine optima for the amount of starting genomic DNA material and for the number of amplification cycles to be used in PCR reaction in order to achieve detection of the maximum fraction of community members and at the same time maintain good accuracy of quantitative abundance measurements (Paliy & Foy, 2011). The model showed that the optimum experimental conditions included a combination of small amount of starting genomic DNA (up to 50 ng) and moderate number of PCR amplification cycles (15– 20). We also developed two adjustment algorithms intended to improve the concordance between the actual bacterial numbers in the community and the distribution of measured DNA hybridization signals. The first algorithm accounts for the predicted cross-hybridization of 16S rRNA gene fragments among different species. It models the measured total signal as a combination of the true signal (binding of the complementary fragment to its target) and cross-hybridization signal due to erroneous hybridizations. Levels of cross-hybridization signals were estimated from microarray validation experiments and were incorporated into the algorithm to calculate the true signal (Rigsbee, et al., 2011). The goal of the second algorithm was to adjust the normalized microarray signal by an estimated number of 16S rRNA gene copies per species genome. Since different species of bacteria are known to possess a wide range of ribosomal RNA-encoding gene copies per genome (1–15, see (Lee, et al., 2009)), measured total signal for each species is not only an indication of that species abundance but also of the number of 16S rRNA genes that species possesses. The use of the copy number adjustment algorithm allowed a better estimate of the actual species abundance in each sample (Rigsbee, et al., 2011). Improvements in experimental and analytical procedures for other arrays were also described (Hamady, et al., 2010, Salonen, et al., 2010, Schatz, et al., 2010, Holmes, et al., 2011), and general optimizations of the phylogenetic microarray design and use are also available (Peplies, et al., 2003, Letowski, et al., 2004, Avarre, et al., 2007). It has also been a good practice in most studies to confirm microarray findings with other molecular techniques. Good concordance of the microarray results was found for comparisons to NGS (Claesson, et al., 2009, Manges, et al., 2010, van den Bogert, et al., 2011), qPCR (Paliy, et al., 2009, Kang, et al., 2010, Agans, et al., 2011), and FISH (Rajilic-Stojanovic, et al., 2009).
Application of phylogenetic microarrays to human microbiota analysis
Many different studies have been successfully performed utilizing phylogenetic microarrays for the interrogation of human-associated microbiota in health and disease. In this section we present examples of the use of three different microarrays for such high-throughput analysis.
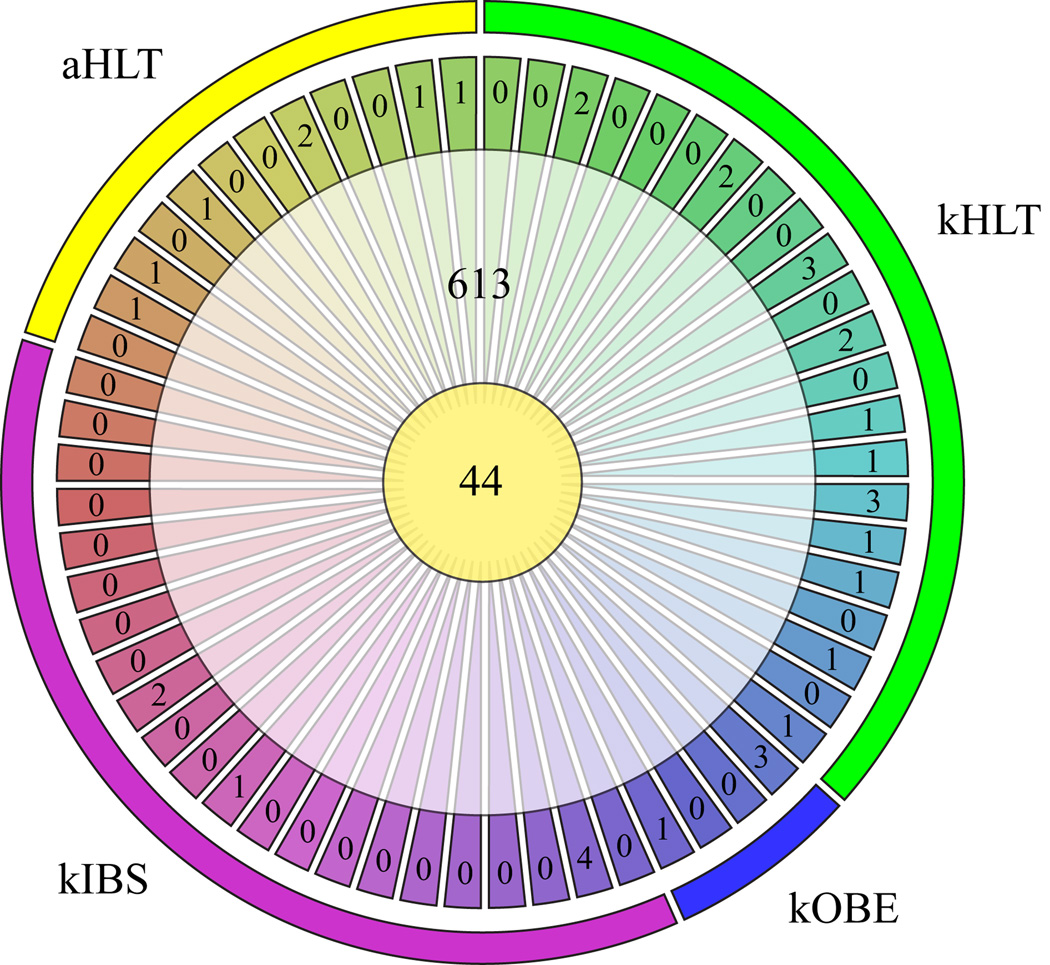
Since each particular microarray measures the hybridization level of every interrogated sequence, it provides quantitative signal for each examined OTU in every sample. This feature allows not only direct comparisons of OTU abundancesbetween samples, but it also provides an opportunity to assess if a particular OTU is detected in all or most samples. A set of such OTUs can be considered to form a core microbiome of species present in every community of particular human microbiota, which can potentially be attributed to an important role of these OTUs in inter-species and host-microbial interactions. Using Microbiota Array, we have recently profiled 60 samples of human fecal microbiota in healthy adults and adolescents and in adolescents diagnosed with obesity and diarrhea-predominant IBS ((Agans, et al., 2011) and (Rigsbee, et al., manuscript in submission)). We have now defined a robust core of 44 microbial phylotypes that were reliably detected in at least 59 samples (Figure 1). We allowed the core bacterial species to be missing from one sample because consideration of individual microbiomes revealed cases where members of a particular genus were completely absent in one individual. An example was provided by a patient with IBS who had no members of genus Faecalibacterium detected in her fecal microbiota (Rigsbee, et al., manuscript in submission); this genus constituted between 4% and 15% of total bacterial abundance in all other samples. Among core species were members of the genera Anaerostipes, Bacteroides, Blautia, Coprococcus, Dorea, Eubacterium, Faecalibacterium, Peptostreptococcus, Roseburia, and Streptococcus. Most of the gut microbiota phylotypes belonged to the so-called “shared” group, which we defined as those present in multiple but not all samples. As can be observed from Figure 1, many individual microbiomes contained unique species that were detected only in that particular sample. Phylogenetic microarrays have also been used to obtain core microbiomes in other studies (Rajilic-Stojanovic, et al., 2009, Jalanka-Tuovinen, et al., 2011); for example, a core of 75 OTUs was detected in lung microbiota among eight patients diagnosed with chronic obstructive pulmonary disease (Huang, et al., 2010).
Figure 1. Core fecal microbiome defined with Microbiota Array.
The figure displays the distribution of detected OTUs among 60 samples of human fecal microbiota obtained from four groups of participants. Outmost bands illustrate sample designation to four different groups. Individual outer segments show phylotypes unique to each analyzed sample; inner circle represents core species detected in at least 59 samples of each group; shared set (middle donut) enumerates OTUs detected in more than 1 but less than 59 samples of the group. aHLT – healthy adults, kHLT – healthy adolescent children (kids), kOBE – obese children, kIBS – children diagnosed with IBS.
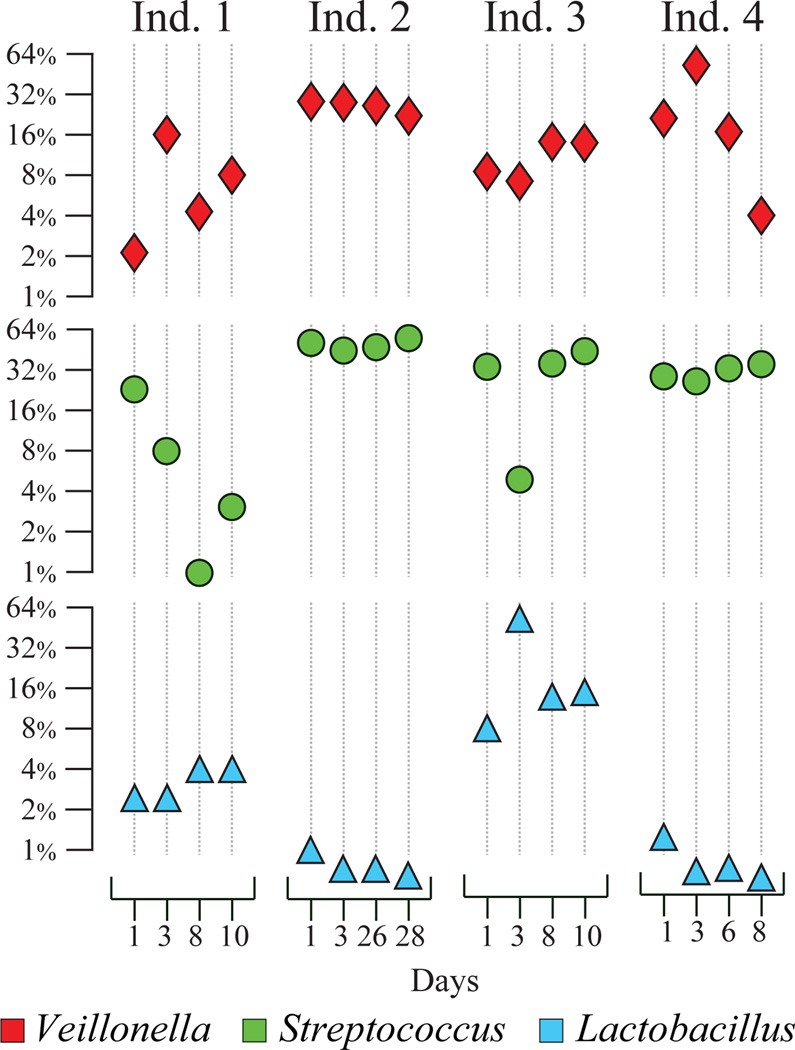
The diversity and temporal stability of microbiota in ileum of patients with ileostomy was studied with HITChip (Booijink, et al., 2010). Ileal contents had high amounts of Streptococcus, Veillonella, and Lactobacillus (Figure 2). Overall, the ileal microbiota was less complex than that typically observed in the distal gut. Profiling ileal biota over a period of 28 days indicated that microbial communities were not only sufficiently different among participants, but were also unstable in the same individual even within one day, since substantial differences were detected in microbial profiles between morning and evening samples collected on the same day (Booijink, et al., 2010). Such temporal differences are in contrast with previous reports indicating relative stability of human distal gut microbiota over long periods of time (Zoetendal, et al., 1998, Costello, et al., 2009, Rajilic-Stojanovic, et al., 2009, Claesson, et al., 2011, Jalanka-Tuovinen, et al., 2011), which was related by the authors to the more significant fluctuations of lumenal contents in the small intestine compared with those of the colon (Booijink, et al., 2010).
Figure 2. Relative contribution of three bacterial genera to the ileal microbial communities in four patients with ileostomy as measured by HITChip.
Each column represents a specific day as shown, each set of four columns corresponds to all samples collected from a single patient. Y axes (log2 scale) show relative abundance values of each genus. Figure is based on the data from (Booijink, et al., 2010).
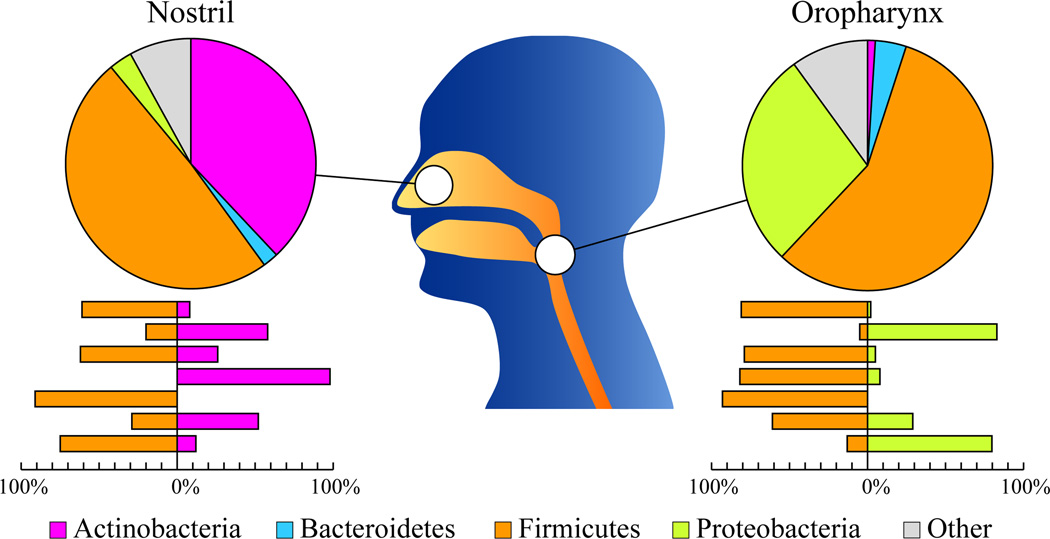
PhyloChip microarray was employed by Lemon et al. (2010) to examine bacterial microbiota of the nostril and oropharynx in seven healthy adults. Diversity and stability of oropharynx microbiota were higher than those of the nostril microbes. Four phyla – Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes – accounted for the majority of the detected bacteria (Figure 3). Interestingly, while Firmicutes and Actinobacteria predominated in the nostril, Firmicutes and Proteobacteria abounded in the oropharynx. Nostril microbiota was thus more similar to that found on the skin, whereas oropharynx communities resembled that of the saliva. While Firmicutes were the most prevalent phylum in both regions, distinct families dominated numerically in each site. Moreover, a striking inverse relationship was observed in the relative abundances of the Firmicutes and another prevalent phylum in each sample as shown in Figure 3 (Lemon, et al., 2010).
Figure 3. Relative distributions of bacterial phyla in the nostril and oropharyngeal samples as detected by PhyloChip.
Pie charts show relative contribution of each phylum (average among seven profiled individuals) to overall microbiota abundance in each region. Bar graphs below each pie chart show relative amount (% total) of the two most abundant phyla in each individual sample. The figure is adapted from data in (Lemon, et al., 2010) with permission.
Outlook
Utilizing phylogenetic microarrays, NGS and Sanger sequencing, initial large-scale studies of human microbiota focused on the compositional analysis of this complex microbial community, and we now have relatively good understanding of which community members are present at different sites and how the community structure fluctuates over time. Phylogenetic microarrays are now also actively used to obtain quantitative data on the changes experienced by human-associated microbial communities in different diseases of the gut, skin, airways, and vaginal canal. The examples provided in the section above illustrate a wide diversity of projects successfully employing phylogenetic microarrays for the analysis of human-associated microbial communities, and they highlight a variety of questions that can be answered with this technology. Phylogenetic arrays were also used to study gut microbiota development in infants (Palmer, et al., 2007, Cox, et al., 2010); altered fecal microbiota in patients with IBD (Kang, et al., 2010), IBS (Kajander, et al., 2008) and Clostridium difficile infection (Manges, et al., 2010); oral microbiota in children (Crielaard, et al., 2011), adults (Olson, et al., 2011) and in the elderly (Preza, et al., 2009); differences in airway microbiota of pediatric and adult cystic fibrosis patients (Cox, et al., 2010); and microbiota associated with bacterial vaginosis (Dols, et al., 2011). One potential barrier to the wider use of phyloarrays by many researchers is the complexity of the de novo array design and the substantial effort required to validate and test array performance. Even so, a panel of the phylogenetic microarrays already available (Table 1) makes it possible to analyze most human-associated microbial communities.
The combination of moderate costs and quantification capability of phyloarrays make them an attractive option as a choice of high-throughput method for current and future studies of human-associated microbiota composition. Especially appealing is a simultaneous application of phylogenetic microarrays and next generation or Sanger sequencing to the analysis of the same microbial population (Crielaard, et al., 2011, van den Bogert, et al., 2011). While SSU rRNA gene sequencing provides the ability to identify novel members of such communities, microarrays can be used to quantitatively compare phylotype abundances among samples and between sample groups.
While recent studies have employed microarrays and sequencing to answer “Who is there?” type questions, the future directions of microbiota research will likely involve the use of a combination of novel molecular tools to (i) obtain a large-scale view of the interactions among microbiota members and between microbiota and human host, and to (ii) link microbiota function and activity to different diseases. In this integrative approach, microarray analysis can be supplemented with other high-throughput systems biology methods including metabonomics, meta-transcriptomics, and meta-proteomics (Klaassens, et al., 2007, Booijink, et al., 2010, Martin, et al., 2010). Combining these techniques would allow us to simultaneously profile community composition (phylogenetic microarrays, SSU RNA gene sequencing), overall gene content (meta-genomics), and gene (meta-transcriptomics) and protein (meta-proteomics) expression, and we will be able to link these datasets to the metabolite levels measured in the same fecal or lumenal samples (metabonomics). Only such truly integrative strategy can provide satisfactory understanding of the complex interplay among microbiota members and between microbiota and human host in both health and disease.
There are also several potential options for future improvements in phylogenetic microarray design and use. Availability of a large set of genome sequences of human-associated microbiota members through Human Microbiome Project (Peterson, et al., 2009) and metaHIT initiative (Qin, et al., 2010) opens an opportunity to design phylogenetic detection arrays based on functionally conserved genes such as groEL, rpoB, gyrA, and tuf (Loy & Bodrossy, 2006). Some phylogenetic microarrays such as Microbiota Array can already measure the levels of not only SSU rRNA genes but can also profile the abundances of SSU rRNAs itself, which provide estimates of the metabolic activity of community members (Rigsbee, et al., 2011). Moreover, the arrays can be eventually expanded to include probes to microbial functional genes; this would allow us to combine on one array community structure interrogation based on phylogenetic probes with community function description based on the availability and abundance of metabolic genes (Louis & Flint, 2007). At the same time, phylogenetic microarrays can be used as microbial diagnostic arrays in clinical settings, where their ability to provide species level detection of hundreds of human microbiota members in a short period of time can aid in disease diagnosis and the choice of best available treatment (Loy & Bodrossy, 2006).
Acknowledgments
We are grateful to Willem de Vos, Erwin Zoetendal, Seungha Kang, Bart Keijser, Frank Schuren, Amee Manges, Michael Markey, and Susan Lynch for valuable comments on the manuscript. The work in Paliy laboratory is supported by the National Institutes of Health grants AT003423 and HD065575.
References
- Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77:404–412. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarre JC, de Lajudie P, Bena G. Hybridization of genomic DNA to microarrays: a challenge for the analysis of environmental samples. J Microbiol Methods. 2007;69:242–248. doi: 10.1016/j.mimet.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booijink CC, Boekhorst J, Zoetendal EG, Smidt H, Kleerebezem M, de Vos WM. Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl Environ Microbiol. 2010;76:5533–5540. doi: 10.1128/AEM.00502-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booijink CC, El-Aidy S, Rajilic-Stojanovic M, et al. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- Brodie EL, Desantis TZ, Joyner DC, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M, Consolandi C, Severgnini M, et al. High taxonomic level fingerprint of the human intestinal microbiota by ligase detection reaction--universal array approach. BMC Microbiol. 2010;10:116. doi: 10.1186/1471-2180-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, O'Sullivan O, Wang Q, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MJ, Huang YJ, Fujimura KE, et al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One. 2010;5:e8745. doi: 10.1371/journal.pone.0008745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MJ, Allgaier M, Taylor B, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf AA, Maathuis A, de Waard P, Deutz NE, Dijkema C, de Vos WM, Venema K. Profiling human gut bacterial metabolism and its kinetics using [U-13C]glucose and NMR. NMR Biomed. 2009;23:2–12. doi: 10.1002/nbm.1418. [DOI] [PubMed] [Google Scholar]
- De Vuyst L, Leroy F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol. 2011 doi: 10.1016/j.ijfoodmicro.2011.03.003. [in press] [DOI] [PubMed] [Google Scholar]
- Deangelis KM, Allgaier M, Chavarria Y, et al. Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS One. 2011;6:e19306. doi: 10.1371/journal.pone.0019306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dols JA, Smit PW, Kort R, et al. Microarray-based identification of clinically relevant vaginal bacteria in relation to bacterial vaginosis. Am J Obstet Gynecol. 2011;204:305, e301–e307. doi: 10.1016/j.ajog.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Mobarry BK, Proudnikov D, Stahl DA, Rittmann BE, Mirzabekov AD. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl Environ Microbiol. 1997;63:2397–2402. doi: 10.1128/aem.63.6.2397-2402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. Isme J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashsham SA, Wick LM, Rouillard JM, Gulari E, Tiedje JM. Potential of DNA microarrays for developing parallel detection tools (PDTs) for microorganisms relevant to biodefense and related research needs. Biosens Bioelectron. 2004;20:668–683. doi: 10.1016/j.bios.2004.06.032. [DOI] [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- He Z, Gentry TJ, Schadt CW, et al. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. Isme J. 2007;1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- Holmes S, Alekseyenko A, Timme A, Nelson T, Pasricha PJ, Spormann A. Visualization and statistical comparisons of microbial communities using R packages on phylochip data. Pac Symp Biocomput. 2011:142–153. doi: 10.1142/9789814335058_0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. Omics. 2010;14:9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman J, Ritari J, Romantschuk M, Paulin L, Auvinen P. Universal ligation-detection-reaction microarray applied for compost microbes. BMC Microbiol. 2008;8:237. doi: 10.1186/1471-2180-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J, Salonen A, Nikkila J, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One. 2011;6:e23035. doi: 10.1371/journal.pone.0023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander K, Myllyluoma E, Rajilic-Stojanovic M, et al. Clinical trial: multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment Pharmacol Ther. 2008;27:48–57. doi: 10.1111/j.1365-2036.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, McSweeney CS. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- Kent AD, Triplett EW. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu Rev Microbiol. 2002;56:211–236. doi: 10.1146/annurev.micro.56.012302.161120. [DOI] [PubMed] [Google Scholar]
- Klaassens ES, de Vos WM, Vaughan EE. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl Environ Microbiol. 2007;73:1388–1392. doi: 10.1128/AEM.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B, Monif GR. Understanding the bacterial flora of the female genital tract. Clin Infect Dis. 2001;32:e69–e77. doi: 10.1086/318710. [DOI] [PubMed] [Google Scholar]
- Lee ZM, Bussema C, 3rd, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1 doi: 10.1128/mBio.00129-10. e00129-00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letowski J, Brousseau R, Masson L. Designing better probes: effect of probe size, mismatch position and number on hybridization in DNA oligonucleotide microarrays. J Microbiol Methods. 2004;57:269–278. doi: 10.1016/j.mimet.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Bodrossy L. Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin Chim Acta. 2006;363:106–119. doi: 10.1016/j.cccn.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Manges AR, Labbe A, Loo VG, et al. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridum difficile-associated disease. J Infect Dis. 2010;202:1877–1884. doi: 10.1086/657319. [DOI] [PubMed] [Google Scholar]
- Marchesi JR, Holmes E, Khan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- Martin FP, Sprenger N, Montoliu I, Rezzi S, Kochhar S, Nicholson JK. Dietary modulation of gut functional ecology studied by fecal metabonomics. J Proteome Res. 2010;9:5284–5295. doi: 10.1021/pr100554m. [DOI] [PubMed] [Google Scholar]
- McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol. 2009;12:378–392. doi: 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Mitterer G, Huber M, Leidinger E, Kirisits C, Lubitz W, Mueller MW, Schmidt WM. Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J Clin Microbiol. 2004;42:1048–1057. doi: 10.1128/JCM.42.3.1048-1057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JB, Kellogg CA. Microbial ecology of corals, sponges, and algae in mesophotic coral environments. FEMS Microbiol Ecol. 2010;73:17–30. doi: 10.1111/j.1574-6941.2010.00862.x. [DOI] [PubMed] [Google Scholar]
- Olson JC, Cuff CF, Lukomski S, et al. Use of 16S ribosomal RNA gene analyses to characterize the bacterial signature associated with poor oral health in West Virginia. BMC Oral Health. 2011;11:7. doi: 10.1186/1472-6831-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliy O, Foy B. Mathematical modeling of 16S ribosomal DNA amplification reveals optimal conditions for the interrogation of complex microbial communities with phylogenetic microarrays. Bioinformatics. 2011;27:2134–2140. doi: 10.1093/bioinformatics/btr326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliy O, Kenche H, Abernathy F, Michail S. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol. 2009;75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplies J, Glockner FO, Amann R. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl Environ Microbiol. 2003;69:1397–1407. doi: 10.1128/AEM.69.3.1397-1407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preza D, Olsen I, Willumsen T, Grinde B, Paster BJ. Diversity and site-specificity of the oral microflora in the elderly. Eur J Clin Microbiol Infect Dis. 2009;28:1033–1040. doi: 10.1007/s10096-009-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preza D, Olsen I, Willumsen T, Boches SK, Cotton SL, Grinde B, Paster BJ. Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis. 2009;28:509–517. doi: 10.1007/s10096-008-0662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H, de Vos WM. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigsbee L, Agans R, Foy BD, Paliy O. Optimizing the analysis of human intestinal microbiota with phylogenetic microarray. FEMS Microbiol Ecol. 2011;75:332–342. doi: 10.1111/j.1574-6941.2010.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Nikkila J, Jalanka-Tuovinen J, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Schatz MC, Phillippy AM, Gajer P, DeSantis TZ, Andersen GL, Ravel J. Integrated microbial survey analysis of prokaryotic communities for the PhyloChip microarray. Appl Environ Microbiol. 2010;76:5636–5638. doi: 10.1128/AEM.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Suau A. Molecular tools to investigate intestinal bacterial communities. J Pediatr Gastroenterol Nutr. 2003;37:222–224. doi: 10.1097/00005176-200309000-00003. [DOI] [PubMed] [Google Scholar]
- Van den Abbeele P, Grootaert C, Marzorati M, et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Applied and Environmental Microbiology. 2010;76:5237–5246. doi: 10.1128/AEM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogert B, de Vos WM, Zoetendal EG, Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77:2071–2080. doi: 10.1128/AEM.02477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, Beggs ML, Erickson BD, Cerniglia CE. DNA microarray analysis of predominant human intestinal bacteria in fecal samples. Mol Cell Probes. 2004;18:223–234. doi: 10.1016/j.mcp.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Wu CH, Sercu B, Van de Werfhorst LC, et al. Characterization of coastal urban watershed bacterial communities leads to alternative community-based indicators. PLoS One. 2010;5:e11285. doi: 10.1371/journal.pone.0011285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Thompson DK, Li G, Hurt RA, Tiedje JM, Zhou J. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl Environ Microbiol. 2001;67:5780–5790. doi: 10.1128/AEM.67.12.5780-5790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Thompson DK, Liu X, Fields MW, Bagwell CE, Tiedje JM, Zhou J. Development and evaluation of microarray-based whole-genome hybridization for detection of microorganisms within the context of environmental applications. Environ Sci Technol. 2004;38:6775–6782. doi: 10.1021/es049508i. [DOI] [PubMed] [Google Scholar]
- Xu W, Seok J, Mindrinos MN, et al. Human transcriptome array for high-throughput clinical studies. Proc Natl Acad Sci U S A. 2011;108:3707–3712. doi: 10.1073/pnas.1019753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SM, Lee SY, Chang KH, et al. High-throughput identification of clinically important bacterial pathogens using DNA microarray. Mol Cell Probes. 2009;23:171–177. doi: 10.1016/j.mcp.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Zabarovsky ER, Petrenko L, Protopopov A, et al. Restriction site tagged (RST) microarrays: a novel technique to study the species composition of complex microbial systems. Nucleic Acids Res. 2003;31:e95. doi: 10.1093/nar/gng096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. Microarrays for bacterial detection and microbial community analysis. Curr Opin Microbiol. 2003;6:288–294. doi: 10.1016/s1369-5274(03)00052-3. [DOI] [PubMed] [Google Scholar]
- Zhou J, He Z, Van Nostrand J, Wu L, Deng Y. Applying GeoChip Analysis to Disparate Microbial Communities. Microbe. 2010;5:60–65. [Google Scholar]
- Zhou JZ, He ZL, Van Nostrand JD, Deng Y. Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Frontiers of Environmental Science & Engineering in China. 2011;5:1–20. [Google Scholar]
- Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]