Abstract
OBJECTIVE
Vitamin A and D, and their receptors, are important regulators of the immune system, including vaccine immune response. We assessed the association between polymorphisms in the vitamin A (RARA, RARB and RARG) and vitamin D receptor (VDR)/RXRA genes and inter-individual variations in immune responses after two doses of measles vaccine in 745 subjects.
METHODS
Using a tagSNP approach, we genotyped 745 healthy children for the 391 polymorphisms in vitamin A and D receptor genes.
RESULTS
The RARB haplotype (rs6800566/rs6550976/rs9834818) was significantly associated with variations in both measles antibody (global p=0.013) and cytokine secretion levels, such as IL-10 (global p=0.006), IFN-α (global p=0.008), and TNF-α (global p=0.039) in the Caucasian subgroup. Specifically, the RARB haplotype AAC was associated with higher (t-statistic 3.27, p=0.001) measles antibody levels. At the other end of the spectrum, haplotype GG for rs6550978/rs6777544 was associated with lower antibody levels (t-statistic −2.32, p=0.020) in the Caucasian subgroup. In a sensitivity analysis, the RARB haplotype CTGGGCAA remained marginally significant (p<0.02) when the single SNP rs12630816 was included in the model for IL-10 secretion levels. A significant association was found between lower measles-specific IFN-γ Elispot responses and haplotypes rs11102986/rs11103473/rs11103482/rs10776909/rs12004589/rs35780541/rs2266677/rs875444 (global p=0.004) and rs6537944/rs3118571 (global p<0.001) in the RXRA gene for Caucasians. We also found associations between multiple RARB, VDR and RXRA SNPs/haplotypes and measles-specific IL-2, IL-6, IL-10, IFN-α, IFN-γ, IFNλ-1, and TNF-α cytokine secretion.
CONCLUSION
Our results suggest that specific allelic variations and haplotypes in the vitamin A and D receptor genes may influence adaptive immune responses to measles vaccine.
Keywords: Single Nucleotide Polymorphisms, Measles Vaccine Immunity, Vitamin A Receptor, Vitamin D Receptor, Genes, Immunogenetics
INTRODUCTION
Vitamin A (retinol) and vitamin D and their receptors are essential regulators of immune function [1]. Their metabolites, such as retinoic acid and 1,25-dihydroxyvitamin D3, respectively, also exhibit immunoregulatory hormone-like characteristics. Vitamin D receptor (VDR) generally forms a hetero-dimer with the nuclear retinoid X receptor alpha (RXRA) that mediates the retinoic acid receptor signaling pathway. Accordingly, three retinoic acid receptors belonging to the family of nuclear hormone receptors and encoded by the RARA (retinoic acid receptor alpha), RARB (retinoic acid receptor beta) and RARG (retinoic acid receptor gamma) genes are known. Vitamin D receptor gene polymorphisms have been associated with susceptibility to infections, such as HIV-1, hepatitis B virus (HBV), human T-cell lymphotropic virus type-1 (HTLV-1) and tuberculosis [2–5]. Recently, we demonstrated that variations in adaptive immune responses following two doses of rubella vaccine are associated with single nucleotide polymorphisms (SNPs) in the vitamin A and D receptor genes in healthy Caucasian children and young adults, suggesting the importance of these genes more generally in viral vaccine-induced immunity [6;7].
Though an immune response is likely to have multigenic influences, several genes (loci) could contribute more dominant effects to the immune response phenotype. While associations have been found between immune response genes, such as human leukocyte antigen (HLA) [8], cytokine/cytokine receptors [9], Toll-like receptors (TLR) [10], CD46 and signaling lymphocyte activation molecule (SLAM) measles virus (MV) binding receptors [11] and measles vaccine-specific immune responses, limited information exists regarding the role of the vitamin A and VDR/RXRA receptor gene polymorphisms in MV immunity. In this study, we examined associations between SNPs/haplotypes in the vitamin A receptor (RARA, RARB and RARG) and VDR/RXRA genes and measures of measles vaccine-induced humoral and cell-mediated immunity (CMI) in healthy children and young adults following two doses of measles vaccine. We hypothesized that specific SNPs/haplotypes within the vitamin A and D receptor genes are associated with variations in immune responses to measles vaccine.
METHODS
Study participants
Study participants and the laboratory methods described herein are similar to those reported in our previous genetic association manuscripts [7;10;12]. Our study cohort comprised a combined sample of 816 subjects from two independent age-stratified random cohorts of healthy schoolchildren and young adults from all socioeconomic strata in Rochester, Minnesota published elsewhere [10]. In 2006–2007, we enrolled 440 children (study cohort 1) [7;12]. Three hundred ninety-six parents agreed to allow their children to take part in the current measles vaccine study, and from these children we obtained a blood sample on 388 of them. In 2008–2009, we enrolled an additional 376 children (study cohort 2). All 764 subjects (age 11–22 years) had documentation of having received two doses of measles-mumps-rubella (MMR, Merck) vaccine. Of these, 19 subjects lacked sufficient genotyping data, resulting in 745 subjects for analysis. The Institutional Review Board of Mayo Clinic approved the study, and we obtained permission through written informed consent from the parents of all children who participated in the study, as well as assent from age-appropriate children.
Plaque reduction microneutralization assay (PRMN)
Measles-specific neutralizing antibody levels were quantified using a fluorescence-based PRMN [10;13]. Heat-inactivated sera were diluted four-fold from 1:4 to 1:4,096 in Opti-MEM (Gibco, Invitrogen), mixed with an equal volume of low passage challenge virus MVeGFP and Vero cells for 43 h at 37°C. The brightly fluorescent green plaques (syncytia) were scanned and counted on an automated Olympus IX71 Fluorescent microscope using the Image-Pro Plus Software version 6.3 (MediaCybernetics). The 50% end point titer (neutralizing doze, ND50) was calculated using Karber’s formula. The use of the 3rd WHO international anti-measles standard (3,000 mIU/ml, NIBSC code no. 97/648) enabled quantitative ND50 values to be transformed into mIU/ml [13].
Elispot assay
IFN-γ Elispot response was assessed using kits from R&D Systems (Minneapolis, MN) [14]. Briefly, seven wells were plated with 2×105 peripheral blood mononuclear cells (PBMCs) per subject: 3 wells were supplemented with MV at an multiplicity of infection (MOI) of 0.5, 3 wells were supplemented with culture medium to serve as a negative control, and 1 well was supplemented with 5 ug/ml of PHA. Plates were read with an ImmunoSpot S4 Pro Analyzer (Cleveland, OH). The intraclass correlation coefficient (ICCs) comparing the multiple observations per subject was 0.94 for the stimulated values, and 0.85 for the unstimulated values, indicating reasonably high levels of measurement reliability.
Secreted cytokines
ELISA assays were performed to measure the level of seven cytokines [IL-2 (n=739), IL-6 (n=737), IL-10 (n=740), IFN-α (n=734)., IFN-γ (n=737)., IFNλ-1 (n=738), and TNF-α (n=732)] secreted by PBMCs following in vitro stimulation with MV using pre-optimized conditions for culture time and virus MOI as described previously [15;16]. We developed and applied a response surface methodology approach to predict optimal combinations of length in culture and virus MOIs for maximum virus-specific cytokine secretion for each specific cytokine of interest, as shown for other viruses [16;17]. The MOI and incubation time for each cytokine were as follows: IL-2, MOI = 0.5, 48 hours; IL-6, MOI=1.0, 72 hours; IL-10, MOI=0.5, 48 hours; IFN-α, MOI=1.0, 24 hours; IFN-γ, MOI=1.0, 72 hours; IFNλ-1, MOI=1.0, 72 hours; TNF-α, MOI=1.0, 24 hours. IL-2, IL-6, IL-10, IFN-γ, and TNF-α were measured in supernatants using kits from BD Biosciences (San Jose, CA), TNF-α was measured using a kit from Mabtech (Cincinnati, OH), and IFNλ-1 was measured using a kit from R&D Systems. Cytokine-specific ICCs ranged from 0.65 (IL-2, unstimulated values) to 0.94 (IFN-α and IL-6, stimulated values).
Candidate genes
SNPs from genes encoding RARA, RARB, RARG, VDR, and RXRA molecules were selected. SNPs within candidate genes and 5 kb upstream and downstream for each gene were chosen based on the linkage disequilibrium (LD) tagSNP selection algorithm [18] from Hapmap Phase II (http://www.hapmap.org), Seattle SNPs (http://pga.mbt.washington.edu/) and NIEHS SNPs (http://egp.gs.washington.edu/). For each gene, we selected tagSNPs with a minor allele frequency (MAF) ≥0.05 and successful Illumina predictive genotyping scores based on a pairwise LD threshold of r2 ≥0.90 in both the Caucasian and African public source samples using ld Select [18].
Genotyping methods
Using the same cohort of subjects, we have designed several other candidate gene studies, such as cytokine/cytokine receptor, CD46/SLAM receptor, TLR and host antiviral genes that reflect separate specific aims, designed in advance for our ongoing population genetics study on measles vaccine response [10;19;20]. Three hundred ninety-one SNPs from five candidate genes were included in the two custom Illumina GoldenGate SNP panels (Illumina, San Diego, CA) for 1,536 and 768 SNPs. Samples included 764 potentially eligible subjects with adequate DNA samples, a Coriel trio of CEPH controls (mother: NA10859, father: NA10858, daughter: NA11875) run in duplicate for each of the nine plates, and 13 wells without DNA. All SNPs had an Illumina design scores >0.4. Illumina 10% GenCall scores >0.4 and SNP call rates >95% were used as laboratory quality control thresholds. Of the 391 SNPs selected, 28 failed our laboratory quality assurance because of failure to amplify, poor clustering, or SNP or subject call rates less than 95%. An additional 43 SNPs were excluded due to low minor allele frequencies (MAF <0.01), yielding a total of 320 SNPs available for analysis. Subject exclusions were made based on low call rates (n=19), leaving 745 subjects for final analysis.
Statistical analysis
The statistical methods described herein are identical to those carried out for previous genetic association manuscripts we published elsewhere [7;10;12]. Seven measures of MV-specific cytokine secretion (IL-2, IL-6, IL-10, IFN-α, IFN-γ, IFNλ-1, and TNF-α, each reported in pg/ml) were examined; as was a measure of CMI via IFN-γ PBMC responses (evaluated as a count variable using Elispot); and also levels of MV-specific neutralizing antibodies (measured in mIU/ml). Assessments of cytokine secretion resulted in five recorded values per outcome prior to stimulation with MV and five values post-stimulation. CMI resulted in three recorded values prior to stimulation and three post-stimulation. For descriptive purposes, a single response measurement per individual was obtained for each outcome by subtracting the median of the multiple unstimulated values from the median of the multiple stimulated values. Assessments of antibody levels resulted in only one recorded value per individual. Data based on these summary measures were descriptively summarized across individuals using medians and interquartile ranges.
Observed genotypes were used to estimate allele frequencies for each SNP and departures from Hardy-Weinberg equilibrium (HWE) were assessed using either a Pearson goodness-of-fit test or, for SNPs with a MAF of less than 5%, a Fisher exact test [21]. Estimates of pair-wise LD based on the r-squared statistic, and identification of intra-genic haplotype blocks based on the Gabriel method, were obtained using Haploview software, version 3.32 [22].
SNP associations with immune response outcomes were individually evaluated using linear regression models, first in all enrolled subjects and then subset to Caucasians (our largest sub-sample). Simple linear regression was used for MV antibody levels, which had only one measured value per individual. Repeated measures approaches, in the form of linear mixed models, were implemented for the cytokine secretion and CMI variables, simultaneously modeling the multiple observed measurements. In these models, we allowed for within-subject correlations without imposing any constraints on their structure within a person using an unstructured within-subject covariance matrix. The genotype-specific effect was assessed by including the genotype variable in the regression model, together with a variable representing stimulation status. The resulting covariate reflecting the genotype-by-stimulation status interaction was then tested for statistical significance. Primary tests of association assumed an additive SNP effect.
To further explore genomic regions containing statistically significant single-SNP effects for one or more outcomes of interest, we performed post-hoc haplotype analyses on intra-genic haplotype blocks based on the methods outlined by Schaid et al [23]. Haplotype design variables were created that reflected the number of each of the haplotypes that were expected to be carried by each subject. Analyses were performed on these variables using simple least squares regression for antibody levels and the repeated measures approach for the cytokine secretion described above. Because of the imprecision involved in estimating the effects of low-frequency haplotypes, we considered only those occurring with an estimated frequency of greater than 1%. Differences in immune response among all common haplotypes were first assessed simultaneously via a global test to collectively determine whether at least one haplotype in the gene region was associated with a modified immune response. Following these global tests, we examined individual haplotype effects in the spirit of Fisher’s protected least significant difference test; individual associations were not considered statistically significant in the absence of global significance. Each haplotype was included in a separate regression analysis, thus comparing immune response levels for the haplotype of interest against all others combined. To further assess the effect of the haplotype, a sensitivity analysis was performed by fitting a model with the significant haplotype and any significant SNPs within that haplotype. The haplotype is interpreted to add predictive information over and above the SNP if it remains significant in the model. Due to phase ambiguity, haplotype-specific medians and inter-quartile ranges could not be calculated. Thus, descriptive summaries were represented using the back-transformed estimate and confidence interval corresponding to the haplotype main effect term for antibody levels and the t-statistic for the haplotype-by-stimulation status interaction term for cytokine secretion. As with the single-SNP results, analyses were run for all subjects and subset to Caucasians only.
All of the association analyses described above adjusted for age at enrollment, race, gender, age at first measles vaccination, age at second measles vaccination, and cohort status (cohort 1 vs. cohort 2) to account for their potential impact on the measured immune responses. Data transformations were used to correct for data skewness in all linear regression models. An inverse normal transformation was used for all cytokine secretion and CMI outcome variables, and a log transformation was used for the antibody response measure. Due to the large number of statistical tests, only p-values less than 0.01 were considered statistically significant. In addition, we computed q-values for each SNP to estimate the corresponding false discovery rates (FDR) as per Storey and Tibshirani [24;25]. All statistical tests were two-sided and, unless otherwise indicated, all analyses were carried out using the SAS software system (SAS Institute, Inc., Cary, NC).
RESULTS
Associations between SNPs/haplotypes in the RARB gene and antibody responses
Summaries of demographic characteristics and immunological variables of the 745 study subjects are shown in Table 1. SNP data were analyzed for 598 Caucasian subjects. A secondary analysis of SNP data was also carried out for the combined group of 745 subjects and separately for 89 African-American subjects (Supplemental Tables 1–6). Results are reported only for the Caucasians, because the African-American subgroup sample size provided insufficient power. We identified significant associations between two intronic SNPs (rs6800566 and rs13070407) located in the RARB gene and MV-specific antibodies (Table 2). Specifically, in both our combined cohort (Supplemental Table 1) and the Caucasian subjects we found associations for these two intronic SNPs in the RARB gene, and the minor alleles were associated with an allele-dose-related increase (rs6800566, p≤0.002) and an allele-dose-related decrease (rs13070407, p≤0.008), respectively, in MV antibody levels.
Table 1.
Demographic characteristics and immunological variables of the study population
| Variable | Outcome |
|---|---|
| Median age at enrollment, years | 15 (IQRa 13; 17) |
| Median age at first measles immunization, months | 15 (IQR 15; 16) |
| Median age at second measles immunization, years | 5 (IQR 4; 11) |
| Median time from second measles immunization to enrollment, years | 7.5 (IQR 5.6; 9.2) |
| Male, no. | 417 (56.0%) |
| Female, no. | 328 (44.0%) |
| Non-white, no. | 147 (19.7%) |
| White, no. | 598 (80.3%) |
| Cohort 1 | 372 (49.9%) |
| Cohort 2 | 373 (50.1%) |
| Median antibody, mIU/ml | 846 (IQR 418; 1,772) |
| Median IFN-γ Elispot response, SFC per 2 × 105 cellsb | 36 (IQR 12; 69) |
| Median cytokine responsec, pg/ml | |
| IL-2 | 37 (IQR 20; 64) |
| IL-6 | 354 (IQR 248; 461) |
| IL-10 | 18 (IQR 11; 28) |
| IFN-α | 551 (IQR 273; 1,025) |
| IFN-γ | 67 (IQR 35; 120) |
| IFNλ-1 | 34 (IQR 14; 73) |
| TNF-α | 14 (IQR 9; 19) |
SFC-spot-forming cells
All results are in median (IQR = inter-quartile range within 1st quartile – 3rd quartile) unless indicated otherwise.
Response is defined as the subject-specific median measles-stimulated response (measured in triplicate) minus the median unstimulated response (also measured in triplicate).
Response is defined as the subject-specific median measles-stimulated response (measured in five replicates) minus the median unstimulated response (also measured in five replicates).
Table 2.
Associations between SNPs in the vitamin A and D receptor genes and humoral and IFN-γ Elispot immune responses to measles vaccine in Caucasians
| Gene | SNP ID | Location | Genotypea | N | Median (IQR)b | p valuec | q valued |
|---|---|---|---|---|---|---|---|
| Antibody Titer (mIU/ml)
| |||||||
| RARB | rs6800566 | intron | GG | 277 | 702 (386, 1,650) | 0.0011 | 0.300 |
| GA | 256 | 996 (458, 1,801) | |||||
| AA | 65 | 1,265 (515, 2,154) | |||||
|
| |||||||
| RARB | rs13070407 | intron | AA | 305 | 998 (469, 1,885) | 0.0082 | 0.676 |
| AG | 245 | 815 (387, 1,699) | |||||
| GG | 46 | 608 (357, 939) | |||||
|
| |||||||
| IFN-γ Elispot (SFC per 2 × 105 PBMCs)
| |||||||
| RXRA | rs2266677 | intron | AA | 312 | 40 (15, 78) | 0.0006 | 0.300 |
| AG | 219 | 33 (14, 61) | |||||
| GG | 32 | 19 (5, 40) | |||||
|
| |||||||
| RXRA | rs6537944 | intron | AA | 473 | 35 (14, 68) | 0.0010 | 0.300 |
| AG | 88 | 41 (17, 78) | |||||
| GG | 2 | 119 (75, 164) | |||||
|
| |||||||
| RARB | rs11916491 | intron | GG | 405 | 35 (13, 66) | 0.0012 | 0.300 |
| GA | 137 | 38 (17, 75) | |||||
| AA | 18 | 79 (18, 106) | |||||
|
| |||||||
| RARB | rs2116703 | intron | GG | 402 | 35 (13, 67) | 0.0041 | 0.676 |
| GA | 137 | 38 (17, 77) | |||||
| AA | 22 | 54 (15, 103) | |||||
|
| |||||||
| VDR | rs11168287 | intron | AA | 147 | 33 (13, 64) | 0.0091 | 0.679 |
| AG | 273 | 36 (14, 73) | |||||
| GG | 142 | 37 (18, 70) | |||||
A-Adenine, C-Cytosine, G-Guanine, T-Thymine, SFC-spot-forming cells, -- no subjects for that genotype.
A total of 391 SNPs were examined; only those found to be statistically significant (p ≤0.01) were included in the table.
Values are presented as homozygous major allele/heterozygous/homozygous minor allele.
IQR, interquartile range, values are median levels in mIU/ml as measured by plaque reduction microneutralization assay, and median levels in SFC per 2×105 PBMCs as measured by IFN-γ Elispot.
Test for trend p value from the repeated measures linear regression analysis. The p values are adjusted for age, gender, age at 1st and 2nd MMR and cohort status (cohort 1 vs. cohort 2) using linear regression analyses.
Corresponding q-values, adjusting for FDR.
Further, we found three RARB haplotypes in the Caucasian sample (global p≤0.035) (Table 3). In particular, the RARB haplotype AAC, resolved by three SNPs (rs6800566/rs6550976/rs9834818) was associated with higher (t-statistic 3.27, p=0.001) measles-specific antibodies in the Caucasian subgroup (global p=0.01).
Table 3.
RARB gene haplotype associations with humoral immune response to measles vaccine in Caucasians
| RARB Haplotypea | Frequency | Back-transformed mean (mIU/ml) | Back-transformed 95% CI (mIU/ml) | Test Statistic | Allele p valueb | Global p value |
|---|---|---|---|---|---|---|
| rs6800566, rs6550976, rs9834818 | ||||||
| AAC | 0.323 | 900.9 | (827.1, 981.2) | 3.27 | 0.001 | 0.013 |
| rs6550978, rs6777544 | ||||||
| AC | 0.484 | 847.2 | (785.3, 913.9) | 2.45 | 0.014 | 0.035 |
| GG | 0.391 | 819.5 | (757.0, 887.0) | −2.32 | 0.020 | |
The string of SNP nucleotides represented in the RARB haplotypes from left to right. Haplotype effects are estimated using the haplotype t-statistic, which reflects the direction and relative magnitude of the estimated haplotypic effect on the antibody measure. Allele p-values compare individual haplotypes to all other haplotypes combined.
One degree-of-freedom log-additive p-value from the linear regression analysis adjusting for age, gender, race (for combined analyses only), age at 1st and 2nd MMR vaccine and cohort status (cohort 1 vs. cohort 2).
Associations between SNPs in the VDR/RXRA and RARB genes and IFN-γ Elispot responses
Three significant associations with intronic polymorphisms in the RXRA (rs6537944, p=0.0010), RARB (rs1626875, p=0.005) and VDR (rs2239181, p=0.008) genes and allele-dose-related MV-specific IFN-γ Elispot responses were found in the combined cohort of subjects (Supplemental Table 1). One out of these three polymorphisms identified in the combined group (RXRA rs6537944, 35 vs. 119 SFC per 2×105 PBMCs, p=0.0010) also demonstrated an association with IFN-γ Elispot response in Caucasians (Table 2). Additionally, pairwise LD analysis found rs11916491 (p=0.001) to be linked to rs2116703 (p=0.004) located in the RARB gene (r2 =0.96), which was associated with an allele-dose-related increase in IFN-γ Elispot responses in the Caucasians. An increased representation of minor allele G for an intronic SNP in the VDR gene (rs11168287, p=0.009) was associated with variation in MV-specific IFN-γ Elispot responses in the Caucasian subgroup.
Associations between SNPs in the RARA, RARB, VDR and RXRA genes and cytokine responses
Specific SNPs in the RARA, RARB, VDR and RXRA genes were associated with MV-induced IL-2, IL-6, IL-10, IFN-α, IFNλ-1, and TNF-α secretion levels (Table 4). Seven SNPs in the RARB and VDR genes were significantly associated (p<0.01) with variations in IL-2 secretion. In both the Caucasian (p=0.008) (Table 3) subjects and the combined cohort (p=0.002) (Supplemental Table 3) we found similar associations for an intronic SNP (rs11574027, LD with rs12581281) in the VDR gene and IL-2 secretion.
Table 4.
Associations between SNPs in the vitamin A and D receptor genes and secreted cytokine immune responses to measles vaccine in Caucasians
| Secreted cytokine | Gene | SNP ID | Location | Genotypea | Na | Median, pg/ml (IQR)b | p valuec | q valued |
|---|---|---|---|---|---|---|---|---|
| IL-2 | RARB | rs1626875 | intron | GG | 392 | 39 (21, 65) | 0.0013 | 0.300 |
| GA | 188 | 45 (24, 70) | ||||||
| AA | 13 | 44 (21, 56) | ||||||
|
| ||||||||
| RARB | rs11707637 | intron | AA | 177 | 48 (23, 68) | 0.0023 | 0.478 | |
| AG | 285 | 42 (21, 71) | ||||||
| GG | 131 | 36 (20, 59) | ||||||
|
| ||||||||
| RARB | rs1290443 | intron | AA | 404 | 40 (21, 66) | 0.0073 | 0.676 | |
| AG | 178 | 44 (24, 70) | ||||||
| GG | 12 | 44 (16, 56) | ||||||
|
| ||||||||
| RARB | rs13099641 | intron | AA | 449 | 43 (23, 69) | 0.0078 | 0.676 | |
| AT | 136 | 36 (19, 61) | ||||||
| TT | 9 | 46 (37, 56) | ||||||
|
| ||||||||
| RARB | rs1286761 | intron | CC | 387 | 39 (21, 65) | 0.0080 | 0.676 | |
| CG | 192 | 44 (23, 70) | ||||||
| GG | 15 | 44 (21, 56) | ||||||
|
| ||||||||
| VDR | rs11574027 | intron | CC | 583 | 42 (21, 66) | 0.0084 | 0.676 | |
| CA | 11 | 101 (43, 118) | ||||||
| AA | 0 | -- | ||||||
|
| ||||||||
| VDR | rs12581281 | intron | GG | 583 | 42 (21, 66) | 0.0084 | 0.676 | |
| GA | 11 | 101 (43, 118) | ||||||
| AA | 0 | -- | ||||||
|
| ||||||||
| IL-6 | RARB | rs12636182 | intron | GG | 416 | 345 (245, 449) | 0.0012 | 0.300 |
| GA | 167 | 383 (258, 495) | ||||||
| AA | 8 | 360 (234, 538) | ||||||
|
| ||||||||
| RARB | rs6803265 | intron | AA | 390 | 350 (252, 451) | 0.0058 | 0.676 | |
| AT | 191 | 365 (240, 473) | ||||||
| TT | 11 | 444 (221, 549) | ||||||
|
| ||||||||
| RARB | rs3821629 | intron | AA | 339 | 347 (248, 452) | 0.0062 | 0.676 | |
| AG | 226 | 359 (248, 461) | ||||||
| GG | 27 | 444 (244, 516) | ||||||
|
| ||||||||
| RARB | rs12635379 | intron | GG | 358 | 346 (247, 446) | 0.0064 | 0.676 | |
| GA | 214 | 361 (245, 471) | ||||||
| AA | 19 | 464 (298, 526) | ||||||
|
| ||||||||
| RARB | rs6795340 | intron | GG | 337 | 344 (244, 443) | 0.0068 | 0.676 | |
| GA | 224 | 375 (259, 468) | ||||||
| AA | 31 | 416 (245, 476) | ||||||
|
| ||||||||
| VDR | rs7965281 | flanking _3UTR |
AA | 144 | 340 (221, 448) | 0.0079 | 0.676 | |
| AG | 312 | 362 (239, 465) | ||||||
| GG | 135 | 355 (275, 464) | ||||||
|
| ||||||||
| RARB | rs1058378 | 3UTR | AA | 492 | 349 (245, 456) | 0.0091 | 0.679 | |
| AC | 95 | 392 (258, 486) | ||||||
| CC | 3 | 390 (196, 440) | ||||||
|
| ||||||||
| VDR | rs7968585 | flanking _3UTR |
GG | 139 | 345 (221, 451) | 0.0098 | 0.698 | |
| GA | 313 | 357 (235, 462) | ||||||
| AA | 140 | 360 (283, 465) | ||||||
|
| ||||||||
| IL-10 | RARB | rs6800566 | intron | GG | 275 | 21 (12, 30) | 0.0012 | 0.300 |
| GA | 254 | 17 (12, 28) | ||||||
| AA | 65 | 16 (12, 23) | ||||||
|
| ||||||||
| RARB | rs12630816 | flanking _5UTR |
GG | 537 | 18 (12, 28) | 0.0080 | 0.676 | |
| GA | 55 | 24 (15, 32) | ||||||
| AA | 2 | 66 (25, 107) | ||||||
|
| ||||||||
| VDR | rs886441 | intron | AA | 385 | 18 (11, 28) | 0.0098 | 0.698 | |
| AG | 192 | 19 (13, 29) | ||||||
| GG | 16 | 28 (18, 40) | ||||||
|
| ||||||||
| IFN-α | RARB | rs6800566 | intron | GG | 271 | 722 (356; 1,168) | 0.0006 | 0.300 |
| GA | 252 | 541 (254, 983) | ||||||
| AA | 65 | 416 (205, 776) | ||||||
|
| ||||||||
| RARB | rs17016570 | intron | AA | 517 | 612 (290, 1,077) | 0.0012 | 0.300 | |
| AG | 66 | 540 (238, 906) | ||||||
| GG | 5 | 443 (153, 942) | ||||||
|
| ||||||||
| RARB | rs13099641 | intron | AA | 443 | 610 (295, 1,077) | 0.0062 | 0.676 | |
| AT | 136 | 582 (236, 1,037) | ||||||
| TT | 9 | 256 (210, 942) | ||||||
|
| ||||||||
| RARB | rs6550978 | intron | AA | 224 | 543 (259, 977) | 0.0076 | 0.676 | |
| AG | 265 | 630 (307, 1,077) | ||||||
| GG | 97 | 722 (282, 1,239) | ||||||
|
| ||||||||
| IFNλ-1 | RARB | rs2033447 | intron | AA | 472 | 41 (19, 82) | 0.0043 | 0.676 |
| AG | 112 | 29 (13, 57) | ||||||
| GG | 8 | 33 (13, 48) | ||||||
|
| ||||||||
| RARB | rs12630816 | flanking _5UTR |
GG | 535 | 39 (17, 748) | 0.0046 | 0.676 | |
| GA | 55 | 61 (27, 127) | ||||||
| AA | 2 | 23 (6, 40) | ||||||
|
| ||||||||
| TNF-α | RARB | rs6800566 | intron | GG | 271 | 15 (10, 20) | 0.0048 | 0.676 |
| GA | 251 | 14 (9, 18) | ||||||
| AA | 65 | 11 (8, 16) | ||||||
|
| ||||||||
| RARB | rs1483856 | intron | AA | 467 | 14 (10, 20) | 0.0059 | 0.676 | |
| AC | 112 | 13 (9, 17) | ||||||
| CC | 8 | 13 (10, 21) | ||||||
|
| ||||||||
| RARB | rs4681025 | intron | AA | 465 | 14 (10, 20) | 0.0064 | 0.676 | |
| AG | 110 | 13 (9, 17) | ||||||
| GG | 10 | 12 (9, 16) | ||||||
A-Adenine, C-Cytosine, G-Guanine, T-Thymine, -- no subjects for that genotype.
A total of 391 SNPs were examined; only those found to be statistically significant (p ≤ 0.01) were included in the table.
Values are presented as homozygous major allele/heterozygous/homozygous minor allele.
IQR, interquartile range, values are median levels in pg/ml as measured by ELISA.
Test for trend p value from the repeated measures linear regression analysis. The p-values are adjusted for age, gender, age at 1st and 2nd MMR and cohort status (cohort 1 vs. cohort 2) using repeated measures linear regression analyses.
Corresponding q-values, adjusting for FDR.
Eight SNPs in the RARB and VDR genes were significantly associated (p<0.01) with variations in IL-6 secretion. Three out of the six polymorphisms identified in the combined group (RARB gene intronic rs12635379, 345 vs. 444 pg/ml, p=0.003 and rs12636182, 347 vs. 276, p=0.010, and VDR gene promoter rs7968585, 360 vs. 351 pg/ml, p=0.006) (Supplemental Table 3) demonstrated associations with IL-6 secretion in the Caucasian subgroup (rs12635379, 346 vs. 464 pg/ml, p=0.006, rs12636182, 345 vs. 360, p=0.001 and rs7968585, 345 vs. 360 pg/ml, p=0.010).
Three SNPs in the RARB and VDR genes were significantly associated (p<0.01) with variations in IL-10 production. Again, in both the Caucasian (p≤0.010) subjects and the combined cohort (p≤0.002) we found similar associations for two SNPs (intronic rs6800566 and promoter rs12630816) in the RARB gene and IL-10 secretion.
Further, four SNPs in the RARB gene were significantly associated (p<0.01) with variations in IFN-α secretion. We found an association between an intronic SNP located in the RARB gene and IFN-α secretion (rs6800566, 722 vs. 416 pg/ml, p=0.0006) in the Caucasian subgroup.
No significant association with MV-specific IFN-γ secretion was found for polymorphisms in the vitamin A and D receptor genes in either the Caucasian subjects or the combined cohort. However, associations were observed for the promoter SNPs in the RARB gene and variations in IFNλ-1 in both the Caucasians (rs2033447, p=0.004 and rs12630816, p=0.005; r2 =0.21) and the combined cohort (rs12630816, p=0.005).
Finally, three intronic SNPs in the RARB gene were significantly associated (p<0.01) with variations in TNF-α secretion in the Caucasians (Table 4). These SNP associations were found with MV-specific TNF-α secretion (RARB rs6800566, p=0.005, rs1483856, p=0.006 and rs4681025, p=0.006; r2 ≥0.87).
Associations between haplotypes in the RARB, VDR and RXRA genes and IFN-γ Elispot and cytokine responses
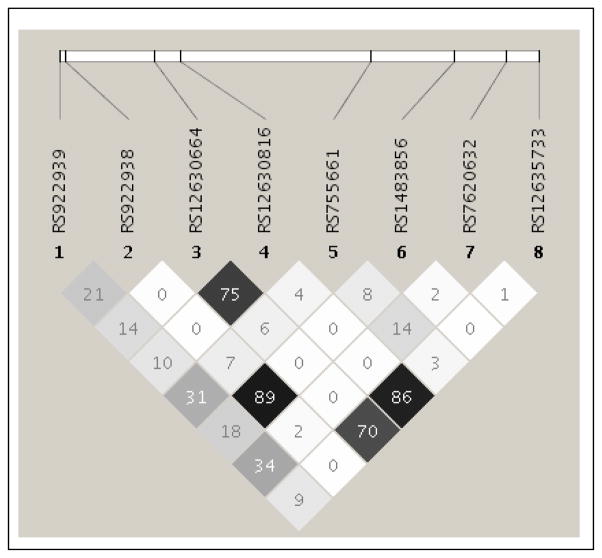
We also studied inferred haplotypes across the vitamin A and vitamin D receptor gene loci in relation to measles-specific cytokine immune responses. A secondary analysis of haplotype data was also carried out for the combined group of 745 subjects and for 89 African-Americans (Supplemental Tables 4–6). Several RARB haplotypes with frequencies ≥1% were identified (Table 5). A haplotype analysis revealed significant associations between IL-2 secretion and two RARB haplotypes (global p-value 0.052 for the Caucasians and global p-value 0.002 for the combined cohort) resolved by two SNPs (rs6550978 and rs6777544). Specifically, the RARB haplotype AG was significantly associated with lower IL-2 secretion, while the AC haplotype was significantly associated with higher IL-2 secretion. Of note, RARB haplotype associations were generally stronger for the Caucasian subgroup than for our combined cohort (Supplemental Table 4). The global tests from the RARB haplotype analyses also demonstrated statistically significant associations between haplotypes and IL-10 (p≤0.020) and IFN-α (p≤0.021) secretion in Caucasians. The common RARB haplotype CTGGGCAA (rs922939/rs922938/rs12630664/rs12630816/rs755661/rs1483856/rs7620632/rs12635733) was associated with lower MV-specific IL-10 secretion levels (t-statistic −2.78, p=0.006), whereas haplotype CAAAGAAG was associated with higher IL-10 secretion levels (t-statistic 2.66, p=0.008) in the Caucasians (Table 5 and Figure 1). In a sensitivity analysis, the haplotype CTGGGCAA remained marginally significant when the single SNP rs12630816 was included in the model for IL-10 secretion levels (p<0.02 for the haplotype and the SNP), and the effects were in the same direction as in the individual models. Further, all other haplotype effects attenuated to non-significance after inclusion of the statistically significant individual SNPs, indicating that most of the effect seen in those haplotypes is sufficiently captured by the individual SNPs.
Table 5.
RARB gene haplotype associations with IFN-γ Elispot and secreted cytokine immune responses to measles vaccine in Caucasians
| Cytokine | RARB Haplotypea | Frequency | Test Statistic | Allele p valueb | Global p value |
|---|---|---|---|---|---|
| rs11706799, rs1299407, rs1286652, rs11916491, rs1286650, rs1286648, rs2568875, rs2056777, rs1406575, rs2116703, rs1153582, rs1286641, rs1153584, rs4260361, rs17016566 | |||||
| IFN-γ Elispot | GATATCAAGAGTAAC | 0.080 | 2.15 | 0.032 | 0.036 |
| GATATCAAGAGTAAG | 0.066 | 1.91 | 0.057 | ||
| rs6550978, rs6777544 | |||||
| IL-2 | AC | 0.484 | 2.13 | 0.034 | 0.052 |
| AG | 0.123 | −1.78 | 0.076 | ||
| rs1730240, rs1286729, rs6795340 | |||||
| IL-6 | CGA | 0.242 | 2.72 | 0.007 | 0.038 |
| rs12635379, rs1286738 | |||||
| IL-6 | AG | 0.212 | 2.74 | 0.006 | 0.022 |
| rs12636182, rs1730220, rs6803265, rs1631354 | |||||
| IL-6 | ACTC | 0.150 | 3.37 | <0.001 | 0.015 |
| rs3821629, rs1730226 | |||||
| IL-6 | AA | 0.560 | −1.75 | 0.080 | 0.023 |
| GT | 0.234 | 2.84 | 0.005 | ||
| rs922939, rs922938, rs12630664, rs12630816, rs755661, rs1483856, rs7620632, rs12635733 | |||||
| IL-10 | CAAAGAAG | 0.048 | 2.66 | 0.008 | 0.012 |
| CTGGGCAA | 0.089 | −2.78 | 0.006 | ||
| rs4416353, rs9284856, rs9284857, rs9871002, rs6550975 | |||||
| IL-10 | AAGTG | 0.076 | 2.02 | 0.044 | 0.020 |
| AGGTC | 0.240 | −2.25 | 0.025 | ||
| GGGTG | 0.197 | 2.09 | 0.037 | ||
| rs6800566, rs6550976, rs9834818 | |||||
| IL-10 | AAC | 0.323 | −3.26 | 0.001 | 0.006 |
| GGG | 0.194 | 2.23 | 0.026 | ||
| rs6800566, rs6550976, rs9834818 | |||||
| IFN-α | AAC | 0.323 | −3.45 | <0.001 | 0.008 |
| rs6550978, rs6777544 | |||||
| IFN-α | AC | 0.484 | −1.80 | 0.073 | 0.021 |
| GG | 0.391 | 2.70 | 0.007 | ||
| rs1560633, rs1286645, rs1997353, rs17016570, rs6767543 | |||||
| IFN-α | AGGGA | 0.058 | −3.47 | <0.001 | 0.009 |
| GGAAA | 0.565 | 1.89 | 0.059 | ||
| rs6550978, rs6777544 | |||||
| IFN-γ | AC | 0.484 | 2.29 | 0.023 | 0.074 |
| GG | 0.391 | −1.89 | 0.059 | ||
| rs922939, rs922938, rs12630664, rs12630816, rs755661, rs1483856, rs7620632, rs12635733 | |||||
| IFNλ-1 | CAAAGAAG | 0.048 | 2.54 | 0.011 | 0.167 |
| rs922939, rs922938, rs12630664, rs12630816, rs755661, rs1483856, rs7620632, rs12635733 | |||||
| TNF-α | CTGGGCAA | 0.089 | −2.58 | 0.010 | 0.063 |
| rs6800566, rs6550976, rs9834818 | |||||
| TNF-α | AAC | 0.323 | −2.83 | 0.005 | 0.037 |
The string of SNP nucleotides represented in the RARB haplotypes from left to right. Haplotype effects are estimated using the haplotype t-statistic, which reflects the direction and relative magnitude of the estimated haplotypic effect on the cytokine measure. Allele p-values compare individual haplotypes to all other haplotypes combined.
One degree-of-freedom additive p-value from the repeated measures linear regression analysis adjusting for age, gender, race (for combined analyses only), age at 1st and 2nd MMR vaccine and cohort status (cohort 1 vs. cohort 2).
Fig. 1. The linkage disequilibrium output for RARB SNPs from Haploview.
Haplotype block structure of the RARB gene region in the Caucasian subjects. The haplotype (rs922939, rs922938, rs12630664, rs12630816, rs755661, rs1483856, rs7620632, rs12635733) from the RARB gene was associated with measles virus-specific IL-10 secretion. The r2 color scheme is: white (r2 = 0), shades of gray (0 < r2 < 1), black (r2 = 1).
Similarly, two RARB haplotypes AAC (rs6800566/rs6550976/rs9834818, t-statistic −3.45, p<0.001) and AGGGA (rs1560633/rs1286645/rs1997353/rs17016570/rs6767543, t-statistic −3.47, p< 0.001) were associated with lower IFN-α secretion (Table 5).
Both global tests and individual haplotype analyses revealed significant associations between RXRA gene haplotypes and measles-specific CMI responses in Caucasians. Our haplotype analysis revealed an association between IL-6 secretion and VDR haplotype (resolved by two SNPs: rs7965281/rs7968585, p=0.023) in the combined cohort (Supplemental Table 6).
Lastly, the global tests from the RXRA haplotype analyses suggested possible associations between several haplotypes and IFN-γ Elispot (p≤0.008) responses, and IFNλ-1 (p=0.07) secretion levels in the Caucasian subgroup (Table 6). The specific RXRA haplotype GG resolved by two SNPs (rs6537944/rs3118571) was associated with higher IFN-γ Elispot responses (t-statistic 3.49, p<0.001), as well as with higher IFNλ-1 secretion levels (t-statistic 2.27, p=0.024).
Table 6.
RXRA gene haplotype associations with IFN-γ Elispot and secreted cytokine immune responses to measles vaccine in Caucasians
| Cytokine | RXRA Haplotypea | Frequency | Test Statistic | Allele p valueb | Global p value |
|---|---|---|---|---|---|
| rs11102986, rs11103473, rs11103482, rs10776909, rs12004589, rs35780541, rs2266677, rs875444 | |||||
| IFN-γ Elispot | GAAGAAAG | 0.035 | 1.97 | 0.050 | 0.008 |
| GAAGCGGG | 0.039 | −1.84 | 0.067 | ||
| GTAGCGAG | 0.045 | 1.76 | 0.078 | ||
| rs6537944, rs3118571 | |||||
| IFN-γ Elispot | AG | 0.287 | −2.43 | 0.015 | <0.001 |
| GG | 0.080 | 3.49 | <0.001 | ||
| rs6537944, rs3118571 | |||||
| IFNλ-1 | GG | 0.080 | 2.27 | 0.024 | 0.070 |
The string of SNP nucleotides represented in the RXRA haplotypes from left to right. Haplotype effects are estimated using the haplotype t-statistic, which reflects the direction and relative magnitude of the estimated haplotypic effect on the cytokine measure. Allele p-values compare individual haplotypes to all other haplotypes combined.
One degree-of-freedom additive p-value from the repeated measures linear regression analysis adjusting for age, gender, race (for combined analyses only), age at 1st and 2nd MMR vaccine and cohort status (cohort 1 vs. cohort 2).
DISCUSSION
Vitamin A and D and their receptors belong to the family of nuclear receptors that regulate multiple important immune functions, including lymphocyte proliferation and antibody production, and hence are important candidates for population-based immunogenetic vaccine studies. We previously genotyped 714 Caucasian subjects for vitamin A and D receptor gene polymorphisms and reported associations between intronic and promoter SNPs in RARA, RARB, RARG, and RXRA genes and rubella vaccine-induced immunity (388 of these children participated in the current measles vaccine study) [6;7]. Similarly, variations in the VDR gene have been associated with susceptibility to several infectious diseases, such as HIV-1, tuberculosis, and HBV [2–4;26]. However, the role of genetic variations in these genes in host immune responses to measles vaccine remains unclear.
Studies to date have focused mainly on circulating serum levels of vitamin A and D and the effect of vitamin A and D supplementation on immune responses to measles. The effect of vitamin A supplementation (VAS) on improved antibody response following one and/or two doses of measles vaccine and on antibody response to oral polio vaccine in children has been described [27;28]. Also, it was suggested that VAS may influence susceptibility to measles infection in Guinea-Bissau newborn children in a gender-dependent manner [29]. A rationale for vitamin D supplementation (a ‘seasonal stimulus’) in the prevention of respiratory infections and seasonal influenza has been proposed [30;31]. The results from several studies demonstrated modulation of cytokine responses by vitamin D, such as differential up-regulation of TNF-α, IL-4, IL-10, and TGF-β and down-regulation of IL-12 and IFN-γ [32–36]. Data also suggest genetic susceptibility to tuberculosis infection and a relationship between vitamin D deficiency and disease susceptibility [4;37]. Thus, it is becoming more obvious that vitamins A and D and their metabolites have important effects on immune responses [1].
Vitamin receptors are also crucial in initiating signaling pathways and mediating vitamin effects on immunity. Given the above, the purpose of our study was to understand the role of genetic variation in vitamin A (RARA, RARB and RARG) and vitamin D (VDR/RXRA) receptor genes likely to impact immune responses to measles vaccine. A candidate gene approach was used to study common genetic variations in these genes and their effects on measles vaccine immune response heterogeneity. Haplotype blocks (where possible) were also studied to determine the potential role for specific haplotypes in the development of measles vaccine immunity.
Our data from single SNP and haplotype analyses indicate the significant contribution of polymorphic variants within intronic and promoter regions of the RARB gene in measles-specific immune response variations, including neutralizing antibody levels. Two RARB SNPs of highest interest (Table 2) were associated with MV antibodies in an allele-dose-dependent manner. While these SNPs are located within introns, they may affect alternative or differential splicing of mRNA and potentially modify the binding site of a transcription factor [38]. Further, the RARB three-SNP intronic haplotype AAC was associated with increased antibody levels (p=0.001) in the Caucasian subjects (Table 3). Associations between RARB haplotypes and multiple MV-specific cellular immunity measures (IFN-γ Elispot, IL-2, IL-6, IL-10, IFN-α, IFN-γ, and TNF-α) were also discovered (Table 5). Importantly, we identified a specific RARB haplotype (resulting from SNPs rs6550978 and rs6777544) that was associated with variations in measles antibody levels and measles-specific IL-2 and IFN-γ secretion in Caucasians (Tables 3 and 5), suggesting potential cross-regulation influences that polymorphic variations in the RARB gene may have on vaccine immunity. Similarly, another RARB AAC haplotype was concurrently associated with variations in measles antibody levels, and IL-10, IFN-α, and TNF-α secretion (Tables 3 and 5). Furthermore, one particular RARB haplotype (CTGGGCAA) (Figure 1) associated with measles-specific IL-10 secretion, remained statistically significant in a sensitivity analysis even after including the significant SNP effects in the model, indicating that at least part of the effect seen in this specific haplotype is not sufficiently captured by the individual SNPs in this case. Thus, RARB gene polymorphisms appear to play an important role in regulating measles vaccine-induced immune response pathways.
Our findings are in agreement with previous findings from our laboratory, which demonstrated significant associations between polymorphisms/haplotypes in the vitamin A (RARB) receptor gene and rubella-specific antibody, and IFN-γ, and IL-10 secretion levels [6;7]; thus lending support to the hypothesis of a more generalized mechanism for RARB polymorphic loci in modulating both measles and rubella vaccine-induced immune responses.
In addition, the retinoid X receptor, RXRA, plays a key regulatory effect on pathways linked to vitamin D and VDR function. Hence, we speculated that genetic polymorphism in the RXRA gene may have an effect on the function of VDR and the subsequent immune response after vaccination. Both VDR and RXRA are highly polymorphic genes, of which we studied 59 and 61 SNPs, respectively. We found individual intronic RXRA and VDR SNPs, as well as RXRA specific haplotypes associated with measles cellular immunity in Caucasians, such as IFN-γ Elispot responses and secreted cytokines (Tables 2, 4 and 6). Interesting results include two highly significant intronic RXRA polymorphisms (rs2266677, p=0.0006 and rs6537944, p=0.0010, Table 2) associated with significant variations in IFN-γ Elispot response as a primary measure of cellular immunity to measles vaccine. Importantly, we also found a specific RXRA haplotype GG (resulting from SNPs rs5637944 and rs3118571) that was associated with both higher IFN-γ Elispot and IFNλ-1 responses in Caucasians (Table 6). Our results point out that allelic variation in the VDR gene may affect MV-induced cytokine responses, such as IFN-γ Elispot response, IL-2, IL-6, IL-10, and IFN-α secretion. The possible functional mechanism for these associations is not known yet, other than potential alternative splicing of mRNA and subsequent translation into multiple protein isoforms due to SNPs located within intronic regions [39]. As these SNPs/haplotypes may also be important in SNP-SNP interactions, additional research is needed to verify our findings and elucidate the mechanisms that account for these associations. Future replication, fine-mapping and functional studies are planned to investigate how intronic polymorphisms or nearby regulatory regions in VDR/RXRA may affect the immune response to measles vaccine.
Protective measles vaccine-induced humoral and cellular immune responses were identified in our study subjects. No seronegative subjects (RPMN antibody titer <14 mIU/ml) were found in this study cohort. The overall cytokine secretion pattern was consistent with a Th1-dominant innate/proinflammatory response, characterized by higher levels of MV-specific IFN-α, IFN-γ, IFNλ-1, IL-6, and IL-2. Measles-specific IFN-γ T-cell memory responses were also detected by Elispot in these vaccinated subjects. The interpretation of these immunological data in the context of the presented associations and the complexity of the immune system is difficult; however, it is important to note the observed considerable immune variation associated with some of the identified SNPs. For example, the homozygous minor allele genotype of RARB rs6800566 was associated with an 80% increase in neutralizing antibody levels compared to the homozygous major allele genotype, while the homozygous minor allele genotype of RXRA rs6537944 was associated with a more than 3-fold increase in IFN-γ response compared to the homozygous major allele genotype. In addition, our data demonstrate haplotype associations and cross-regulation patterns for some of the genes/genetic variants (found in separate independent analyses) across multiple inter-related vaccine-induced immune outcome measures (such as antibody levels and IFN-γ responses), which further strengthens our confidence in the observed effects.
This is the first study to show that genetic polymorphisms in vitamin A and D receptors have significant associations with measles vaccine-induced immunity. The strengths of our population-based study include a large sample size with documented MMR vaccine coverage and no identified wild type MV circulating during their lifetimes, and the use of a vigorous SNP tagging approach. To account for population stratification, we restricted our primary analyses of SNP/haplotype associations to self-declared Caucasians (n=598). Assessment of several outcomes of MV-specific humoral and CMI responses in our study cohort permitted us to examine both allele-dose-related responses and cross-regulation patterns for genetic polymorphisms.
There are also limitations in our study. A major concern is that of false-positive associations and multiple tests. Although we originally designed the study based on thresholds for p-values, to address the multiple comparisons issue we have supplemented the results with q-values to account for a possible false discovery rate (FDR). While the q-values for the presented associations are not particularly impressive and indicate that some of the results are likely false positives, we feel this doesn’t diminish the importance of our findings (as this is the first study examining the role of these genes/SNPs in measles vaccine immunity), but rather provides a list of plausible genetic targets/genetic regions of interest for future studies. As a next step we plan to replicate these findings in an independent cohort with an increased sample size. To assess the contribution of SNPs/haplotypes in vitamin A and D receptor genes to vaccine immunity, genetic variants in other loci, for example, TLR1- or TLR2-mediated pathways known to increase the expression of VDR, should also be studied.
Taken together, our results indicate that allelic variation in vitamin A (RARB) receptor and vitamin D receptor/RXRA genes are likely to be associated with inter-individual immune responses to measles vaccine. Vitamin A and D receptor gene polymorphisms appear to be important host genetic factors for vaccine-induced immunity, indicating that specific allelic variations and haplotypes in these genes may influence the outcome of humoral and cellular responses to measles. Clearly, further studies designed to study the mechanistic function of these polymorphisms and their clinical applications are needed. As these SNPs/haplotypes may have potential translational implications in designing new vaccination strategies, additional research is required to replicate and verify our findings. For example, finding a common SNP that codes for a decreased ability to convert 25-hydroxyvitamin D3 into active 1,25-dihydroxyvitamin D3 metabolites needed for proper immune functioning in association with significantly decreased immune responses to live viral vaccines, would suggest a corrective strategy of public health importance. Similarly, a SNP coding for a defective vitamin receptor that precluded developing protective immune responses to live viral vaccines might provide an opportunity for “reverse engineering” new viral vaccines designed to overcome such genetic restrictions. This information will improve our understanding of the genetic determinants of measles vaccine-induced immunity and may lead to novel vaccination strategies and novel candidate prophylactic vaccines.
Supplementary Material
Acknowledgments
We thank the Mayo Clinic Vaccine Research Group staff and subjects who participated in our studies. We thank V. Shane Pankratz and Matthew J. Phan for their help with this manuscript. This work was supported by NIH grants AI 33144, AI 48793 (which recently received a MERIT Award) and 5UL1RR024150-03 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health, and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
CONFLICT OF INTEREST
Dr. Poland is the chair of a safety evaluation committee for novel non-measles vaccines undergoing clinical studies by Merck Research Laboratories. Dr. Jacobson serves on a Safety Review Committee for a post-licensure study of Gardasil for Kaiser-Permanente. Drs. Ovsyannikova, Haralambieva, O’Byrne and Vierkant declare no potential conflict of interest.
References
- 1.Mora JR, Iwata M, Von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol. 2008;8:685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Torre MS, Torres C, Nieto G, Vergara S, Carrero AJ, Macias J, et al. Vitamin D receptor gene haplotypes and susceptibility to HIV-1 infection in injection drug users. J Infect Dis. 2008;197:405–410. doi: 10.1086/525043. [DOI] [PubMed] [Google Scholar]
- 3.Motsinger-Reif AA, Antas PR, Oki NO, Levy S, Holland SM, Sterling TR. Polymorphisms in IL-1beta, vitamin D receptor Fok1, and Toll-like receptor 2 are associated with extrapulmonary tuberculosis. BMC Med Genet. 2010;11:37. doi: 10.1186/1471-2350-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellamy R, Ruwende C, Corrah T, McAdam KP, Thursz M, Whittle HC, et al. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis. 1999;179:721–724. doi: 10.1086/314614. [DOI] [PubMed] [Google Scholar]
- 5.Saito M, Eiraku N, Usuku K, Nobuhara Y, Matsumoto W, Kodama D, et al. ApaI polymorphism of vitamin D receptor gene is associated with susceptibility to HTLV-1-associated myelopathy/tropical spastic paraparesis in HTLV-1 infected individuals. J Neurol Sci. 2005;232:29–35. doi: 10.1016/j.jns.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Ovsyannikova IG, Haralambieva IH, Dhiman N, O’Byrne MM, Pankratz VS, Jacobson RM, et al. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. J Infect Dis. 2010;201:207–213. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ovsyannikova IG, Dhiman N, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Human Genet. 2010;127:207–221. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ovsyannikova IG, Pankratz SV, Vierkant R, Jacobson RM, Poland GA. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J Infect Dis. 2006;193:655–663. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- 9.Dhiman N, Ovsyannikova IG, Cunningham JM, Vierkant RA, Kennedy RB, Pankratz VS, et al. Associations between measles vaccine immunity and single nucleotide polymorphisms in cytokine and cytokine receptor genes. J Infect Dis. 2007;195:21–29. doi: 10.1086/510596. [DOI] [PubMed] [Google Scholar]
- 10.Ovsyannikova IG, Haralambieva IH, Vierkant RA, Pankratz VS, Poland GA. The role of polymorphisms in Toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Hum Genet. 2011 doi: 10.1007/s00439-011-0977-x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhiman N, Cunningham JM, Jacobson RM, Vierkant RA, Wu Y, Ovsyannikova IG, et al. Variations in measles vaccine-specific humoral immunity by polymorphisms in SLAM and CD46 measles virus receptors. J Allergy Clin Immunol. 2007;120:666–672. doi: 10.1016/j.jaci.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 12.Haralambieva IH, Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, et al. Oligoadenylate synthetase single-nucleotide polymorphisms and haplotypes are associated with variations in immune responses to rubella vaccine. Human Immunol. 2010;71:383–391. doi: 10.1016/j.humimm.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haralambieva IH, Ovsyannikova IG, Vierkant RA, Poland GA. Development of a Novel Efficient Fluorescence-based Plaque Reduction Microneutralization Assay for Measles Immunity. Clin Vaccine Immunol. 2008;15:1054–1059. doi: 10.1128/CVI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan JE, Ovsyannikova IG, Poland GA. Detection of measles virus-specific IFN-gamma-secreting T-cells by ELISPOT. In: Kalyuzhyny AE, editor. Handbook of ELISPOT: Methods and Protocols. Totowa, NJ: Humana Press Inc; 2005. pp. 207–217. [DOI] [PubMed] [Google Scholar]
- 15.Ovsyannikova IG, Dhiman N, Jacobson RM, Vierkant RA, Pankratz VS, Poland GA. HLA homozygosity does not adversely effect measles vaccine-induced cytokine responses. Virology. 2007;364:87–94. doi: 10.1016/j.virol.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Ryan JE, Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Poland GA. Response surface methodology to determine optimal cytokine responses in human peripheral blood mononuclear cells after smallpox vaccination. J Immunol Methods. 2009;341:97–105. doi: 10.1016/j.jim.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhiman N, Haralambieva IH, Vierkant RA, Pankratz VS, Ryan JE, Jacobson RM, et al. Predominant inflammatory cytokine secretion pattern in response to two doses of live rubella vaccine in health vaccinees. Cytokine. 2010;50:24–29. doi: 10.1016/j.cyto.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. The Association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses--A replication study and examination of novel polymorphisms. Hum Hered. 2011 doi: 10.1159/000331585. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haralambieva IH, Ovsyannikova IG, Kennedy RB, Vierkant RA, Pankratz VS, Jacobson RM, et al. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011 doi: 10.1016/j.vaccine.2011.08.083. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir BS. Genetic data Analysis II: methods for discrete population genetic data. Sinauer Associates, Inc; 1996. pp. 98–99. [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. American J Human Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storey JD. A direct approach to false discovery rates. J R Statist Soc B. 2002;64:479–498. [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suneetha PV, Sarin SK, Goyal A, Kumar GT, Shukla DK, Hissar S. Association between vitamin D receptor, CCR5, TNF-alpha and TNF-beta gene polymorphisms and HBV infection and severity of liver disease. J Hepatol. 2006;44:856–863. doi: 10.1016/j.jhep.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 27.Benn CS, Aaby P, Bale C, Olsen J, Michaelsen KF, George E, et al. Randomised trial of effect of vitamin A supplementation on antibody response to measles vaccine in Guinea-Bissau, west Africa. Lancet. 1997;350:101–105. doi: 10.1016/S0140-6736(96)12019-5. [DOI] [PubMed] [Google Scholar]
- 28.Bahl R, Bhandari N, Kant S, Molbak K, Ostergaard E, Bhan MK. Effect of vitamin A administered at Expanded Program on Immunization contacts on antibody response to oral polio vaccine. Eur J Clin Nutr. 2002;56:321–325. doi: 10.1038/sj.ejcn.1601325. [DOI] [PubMed] [Google Scholar]
- 29.Diness BR, Martins CL, Bale C, Garly ML, Ravn H, Rodrigues A, et al. The effect of high-dose vitamin A supplementation at birth on measles incidence during the first 12 months of life in boys and girls: an unplanned study within a randomised trial. Br J Nutr. 2011:1–4. doi: 10.1017/S0007114510005532. [DOI] [PubMed] [Google Scholar]
- 30.Hope-Simpson RE. The role of season in the epidemiology of influenza. J Hyg (London) 1981;86:35–47. doi: 10.1017/s0022172400068728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannell JJ, Vieth R, Umhau JC, Holick MF, Grant WB, Madronich S, et al. Epidemic influenza and vitamin D. Epidemiol Infect. 2006;134:1129–1140. doi: 10.1017/S0950268806007175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cantorna MT, Mahon BD. D-hormone and the immune system. J Rheumatol Suppl. 2005;76:11–20. [PubMed] [Google Scholar]
- 33.Abu-Amer Y, Bar-Shavit Z. Regulation of TNF-alpha release from bone marrow-derived macrophages by vitamin D. J Cell Biochem. 1994;55:435–444. doi: 10.1002/jcb.240550404. [DOI] [PubMed] [Google Scholar]
- 34.Staeva-Vieira TP, Freedman LP. 1,25-dihydroxyvitamin D3 inhibits IFN-gamma and IL-4 levels during in vitro polarization of primary murine CD4+ T cells. J Immunol. 2002;168:1181–1189. doi: 10.4049/jimmunol.168.3.1181. [DOI] [PubMed] [Google Scholar]
- 35.Penna G, Adorini L. 1 Alpha,25-dihydroxyvitamin D3 inhibits differentiation, maturation, activation, and survival of dendritic cells leading to impaired alloreactive T cell activation. J Immunol. 2000;164:2405–2411. doi: 10.4049/jimmunol.164.5.2405. [DOI] [PubMed] [Google Scholar]
- 36.Farquharson C, Law AS, Seawright E, Burt DW, Whitehead CC. The expression of transforming growth factor-beta by cultured chick growth plate chondrocytes: differential regulation by 1,25-dihydroxyvitamin D3. J Endocrinol. 1996;149:277–285. doi: 10.1677/joe.0.1490277. [DOI] [PubMed] [Google Scholar]
- 37.Bellamy RJ, Hill AVS. Host genetic susceptibility to human tuberculosis. In: Chadwick DJ, Cardew G, editors. Genetics and tuberculosis. Chichester, UK: Wiley; 1998. pp. 3–23. [DOI] [PubMed] [Google Scholar]
- 38.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Research. 2006;34:635–641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.