Abstract
Background
Food restriction is known to enhance learning and motivation. The neural mechanisms underlying these responses likely involve alterations in gene expression in brain regions mediating the motivation to feed.
Methods
Analysis of gene expression profiles in male C57BL6/J mice using whole-genome microarrays was completed in the medial prefrontal cortex, nucleus accumbens, ventral tegmental area, and the hypothalamus following a five day food restriction. Quantitative PCR was used to validate these findings and determine the time-course of expression changes. Plasma levels of the stress hormone corticosterone (CORT) were measured by ELISA. Expression changes were measured in adrenalectomized animals that underwent food restriction, as well as in animals receiving daily injections of CORT. Progressive ratio responding for food, a measure of motivated behavior, was assessed after CORT treatment in restricted and fed animals.
Results
Brief food restriction results in an upregulation of peripheral stress responsive genes in the mammalian brain. Time-course analysis demonstrated rapid and persistent expression changes in all four brain regions under study. Administration of CORT to non-restricted animals was sufficient to induce a subset of the genes, and alterations in gene expression after food restriction were dependent on intact adrenal glands. CORT can increase the motivation to work for food only in the restricted state.
Conclusions
These data demonstrate a central role for CORT in mediating both molecular and behavioral responses to food restriction. The stress hormone-induced alterations in gene expression described here may be relevant for both adaptive and pathological responses to stress.
Keywords: transcription, neural plasticity, starvation, obesity, motivation, stress hormones
INTRODUCTION
The effect of food restriction in behavioral plasticity has been well established. Food restriction increases motivation to respond for food rewards across a variety of species (1–3). Food restricted (FR) animals are also more sensitive to the effects of drugs of abuse and work harder for drug rewards (4–6). In addition, food restriction produces central adaptations beyond the described effects on motivation (7, 8). Strikingly, the adipocyte-derived hormone leptin can reverse many of the behavioral and physiological effects of food restriction, providing an example of how hormones serve to communicate the metabolic state to the brain (9). Leptin signaling has been primarily studied in the hypothalamus, but recent work from our lab and others has suggested a functional role for leptin in the dopaminergic cells of the ventral tegmental area (VTA) (10, 11). The distributed response to leptin suggests that multiple brain regions may be responsive to metabolic state and underlie leptin’s central effects (12).
Within the hypothalamus, expression of neuropeptides such as melanin concentrating hormone as well as the leptin receptor are regulated following short-term food deprivation (13, 14). Food restriction can have persistent effects; for example, when re-fed mice show increased sensitivity to amphetamine after a previous period of food restriction (15). It is likely that transcriptional changes contribute since studies on learning and drug addiction have identified transcriptional events to be required for establishing neuronal plasticity (16, 17). Transcription factors such as CREB and FosB in mesocorticolimbic regions have been implicated in generating aspects of neuronal plasticity (18). The mesocorticolimbic circuits include the dopaminergic cell bodies within the VTA as well as the nucleus accumbens (Acb) and the medial prefrontal cortex (mPFC) target regions. Since the neuronal circuitry mediating behavioral responses to food and drugs of abuse are partly shared, these same brain circuits are likely candidates for neural plasticity following restriction (19). While studies have suggested altered dopamine signaling in the striatum may underlie effects of food restriction, systematic and unbiased transcriptional analysis within these circuits has not been completed (20, 21). We hypothesized that food restriction and motivation would share this requirement for transcriptional plasticity.
Here we report gene expression changes in the hypothalamus as well as regions of the mesocorticolimbic system following a brief food restriction. A set of genes were validated by quantitative polymerase chain reaction (qPCR) and found to be up-regulated in a rapid and persistent manner in all brain regions examined. The identity of the genes suggested activation of a stress-responsive pathway and led us to explore the role of stress hormones in restricted feeding behavior. Corticosterone (CORT) was sufficient to induce genes in the fed state, while the adrenal glands were necessary for all observed induction upon restriction. CORT induces motivated behavioral responses in a feeding state dependent manner. These results indicate that stress hormones are a key signal to alter gene expression and to produce motivated behavior in response to food restriction.
METHODS AND MATERIALS
Mice
All experiments were performed on 8–9 week old C57/BL6J male mice (Jackson Labs) later sacrificed at 10–12 weeks. Animals had ad libitum water and were on a 12-h light/dark cycle beginning at 7am. Four independent cohorts, fed Research Diets D12450B, were used in the microarray study (n=4 arrays, n=20 animals, pooled as discussed below). Time course cohorts and the 5-day CORT cohort were fed Prolab RMH 3000 chow (n=5 per group). Weighing and feeding were done within one hour of dark onset, as were all sacrifices. Animals were housed five per cage and food intake was measured daily for 7 days. The average intake consumed value was used to generate the amount given during the restriction (75%). This was done for the initial array cohort of 5 day food restriction, as well the three additional cohorts generated for the 1-, 5-, and 10 day restriction and the adrenalectomy cohort. The adrenalectomized mice were ordered from Jackson Labs (surgery performed when 5 weeks old) and then were maintained with two bottles, one with water and another with 1.0% saline, throughout the experiment. All procedures were approved by the animal care committee at Yale University.
Dissections
Animals were rapidly decapitated, trunk blood was collected and the brain was removed and placed in chilled artificial cerebrospinal fluid (NaCl 124 mM, KCl 4 mM, NaHCO3 26 mM, D-glucose 10 mM, CaCl2 1.5 mM, MgSO4 1.5 mM, KH2PO4 1.25 mM, pH 7.5) for one minute. The brain was then sectioned using a 1mm brain block (BrainTree) and each region was microdissected from a single 1 mm slice using a scalpel, except for the Acb where a 15 gauge circular punch was used to collect the core and shell. For the mPFC, we took infralimbic, prelimbic and a part of anterior cingulate. Each piece of tissue was rapidly frozen on dry ice and placed at −80°C until used for molecular analysis. The trunk blood was immediately added to K2 EDTA (Sigma) at a final concentration of 2mg/ml and then spun for 20 minutes at 1000g at room temperature; plasma was collected as the supernatant.
Plasma CORT Measurements
CORT ELISAs were performed according to manufacturer’s protocol (Assay Designs). Briefly, 1 μl of plasma, diluted 1:50, was compared to known concentrations. An OD reading at 405nm with correction at 570nm was taken and a standard curve was generated using BioTek GEN5 software.
RNA Preparation
Microarray RNA samples were prepared using the mirVana miRNA Isolation Kit (Ambion) following the total RNA isolation procedure. Time course, adrenalectomized and CORT i.p. RNA samples were purified using Trizol reagent (Invitrogen); NanoDrop ND-1000 (Thermo Scientific) was used to determine final RNA concentration.
Microarrays
Microarrays were conducted in collaboration with the NIH Neuroscience Microarray Consortium using Operon mouse whole genome arrays (4.0) with a reference sample. Detailed protocols are available on the Duke Microarray Facility site (http://microarray.genome.duke.edu/). These data were then subjected to analysis with the GenePix Pro software (Molecular Devices). Resulting files were analyzed using Genespring software (Agilent). Background spot intensity was calculated by taking the median of the medians for each color channel (~60) and each spot had either the Cy5 or Cy3 channel above the background threshold. Data were normalized using Lowess and were then filtered for a fold change of greater than plus or minus 1.2 and a p value of p<0.05 using the volcano plot function. None of these data passed the Benjamini Hochberg False Discovery Rate. The geometrically averaged FR/REF number was divided by the averaged REF/CTL for probes found on at least 2 of the 4 arrays for each treatment to achieve corrected fold changes. A cutoff of greater than plus or minus 1.5 fold was used to generate gene lists.
Quantitative PCR
Microarray and time course cDNA samples were generated using 500 ng total RNA and the Superscript III RNase H-Reverse Transcriptase kit (Invitrogen). qPCR primers were designed to cross an intron-exon boundary (intron of >400 bp) using Primer3 software (http://frodo.wi.mit.edu/primer3-0.4.0/input.htm). Primers were ordered from Integrated DNA Technologies and diluted to 6–12 μM. All primer sets were found to be 90–110% efficient and resulted in one PCR product as confirmed by melt curve and gel electrophoresis analysis. Sequences for primer pairs can be found in Table S3 in the Supplement. qPCR was performed according to manufacturers recommendations using a 7500 Fast Real Time PCR System and the Power SYBR Green master mix (Applied Biosystems). The ΔΔCT method of calculating fold change was used and TATA box binding protein (Tbp) was the control gene (22).
Fluorescent In Situ Hybridization
Protocols used were as described previously from our lab (10). Briefly, riboprobes were generated using an in vitro transcription kit (Roche) with digoxigenin-labeled UTP for Cdkn1a and fluorescein-labeled UTP for Gad1 and Gad2, and hybridized to 14-μm brain sections. Following antibody incubation (1:200 dilution of horseradish peroxidase (HRP)-conjugated mouse-anti digoxigenin antibody (Jackson ImmunoResearch) or 1:2000 dilution of HRP-conjugated rabbit anti-fluorescein antibody (Molecular Probes)), sections were incubated with TSA-direct coupled to either Cy3 or fluorescein (Perkin-Elmer), and then visualized.
CORT administration
Intact mice were injected with 2.4mg/kg i.p. CORT (LKT Laboratories) dissolved in 1% ethanol in saline. The injections occurred at 5pm while kept on an ad libitum diet for 5 days and were sacrificed on the final day two hours after the last CORT injection.
Operant Conditioning
Experiments were performed as previously described (23). Briefly, mice were food restricted to ~93% original body weight and then trained to nose poke for food reinforcement (20 mg; F0071, Bioserv) using standard Med-Associates operant conditioning chambers. Mice were initially trained to nose poke on a single nose poke aperture using a one-hour session and a variable ratio 2 schedule of reinforcement. Upon stable responding, mice were shifted to a linear progressive ratio schedule of reinforcement in which the response requirement increased by 4 responses after each reinforcement was earned. Test sessions ended when animals executed no active responses for 5 consecutive min. After 7 sessions, mice were then injected with saline or acute CORT (2.4 mg/kg, i.p.) in a counter-balanced fashion 30 minutes prior to test. Mice were injected with the opposite compound the next day. This experiment was replicated in mice that were returned to free-feeding at the onset of progressive ratio testing to evaluate whether the effects of CORT were selective to FR, as opposed to the fed state. Break point ratios were analyzed by 2-factor ANOVA with repeated measures and with feeding state and CORT exposure as factors.
RESULTS
Food restriction paradigm
To investigate the gene expression changes found in selected brain regions associated with food restriction, nine week old C57BL6/J male mice were housed 5 per cage and food intake was monitored for 7 days. Average food intake values were calculated and used to provide 75% of that amount each day to the FR group. All food manipulations occurred near the beginning of the dark cycle to minimize disruption of normal activity patterns. One cage was randomly assigned to the FR group while the other cage was the control group and received ad libitum food (Ad Lib).
After 5 days, the expected changes in weight were observed (Ad Lib: start weight 22.9+/−0.3g, FR: start weight 23.1 +/−0.3g, n.s.; Ad Lib: final weight 23.7 +/−0.4g, FR: final weight 21.0 +/− 0.3g, p<0.01; n=20). The mice were rapidly sacrificed at the beginning of the dark cycle and the mPFC, core/shell of the Acb, the hypothalamus (Hyp) and the VTA were quickly microdissected (Fig. S1 in the Supplement). RNA from each brain region was pooled from the five mice per cage to generate sufficient amounts for the array hybridizations. RNA was prepared from these samples and divided in two halves. One portion was used for the microarray while the other portion was saved for validation using qPCR. This procedure was done four independent times to generate a n=4 for this study.
Identification of genes regulated upon food restriction
Analysis of gene expression was performed using the mouse v4.0 Operon whole genome longmer microarrays in collaboration with the NIH Neuroscience Consortium. A common reference RNA sample was used to allow for normalization and thus enable comparisons across multiple samples generated at different times. Lowess normalization, filtering for p<0.05, fold changes greater or less than 1.5 and minimal spot intensity were used to generate lists of genes differentially regulated in the FR versus Ad Lib groups. No genes passed the false discovery rate, consistent with the low sample number in the study (24). The gene lists for the four brain regions demonstrated more up-regulated genes than down-regulated genes (Fig. S2 and Table S1 in the Supplement). qPCR was performed on a subset of genes to provide confidence in these data. Stringent criteria (see methods) were used to ensure the primers were efficient and did not amplify genomic DNA. While few of the down-regulated genes validated, a number of up-regulated genes showed validation by qPCR (Table S2 in the Supplement). It is notable that the most consistently regulated genes show regulation across all four brain regions when directly tested by qPCR (Table 1).
Table 1.
| Gene Symbol | Gene Name | mPFC | Acb | Hyp | VTA | References |
|---|---|---|---|---|---|---|
| Nutrient Regulated | ||||||
| Angptl4 | angiopoietin-like 4 | 2.78 | 2.30 | 2.90▯ | 3.08 | 32,34 |
| Pdk4 | pyruvate dehydrogenase kinase, isoenzyme 4 | n.s.▯ | 1.96 | 2.21▯ | 2.17 | 30,43 |
| Slc2a4 | solute carrier family 2 (faciliated glucose transporter),member 4 | UN▯ | n.s.▯ | 1.62 | 1.31 | 31 |
| Slc39a4 | solute carrier family 39 (zinc transporter), member 4 | UN▯ | UN▯ | n.s.▯ | 2.27 | 28 |
| Stress Responsive | ||||||
| Ada | adenosine deaminase | 1.71▯ | 2.46 | 1.83▯ | 3.40 | 40 |
| Arl4d | ADP-ribosylation factor-like 4D | n.s.▯ | n.s. | 2.24 | 2.29 | 30 |
| Arrdc2 | arrestin domain containing 2 | 1.63 | 2.04 | 2.46 | 2.62 | 36 |
| Cdkn1a | cyclin-dependent kinase inhibitor 1a | 1.44▯ | n.s.▯ | 2.82 | 3.39 | 30,37 |
| Fkbp5 | FK506 binding protein 5 | 2.09▯ | n.s.▯ | 2.75 | 2.10 | 35 |
| Mertk | c-mer proto-oncogene tyrosine kinase | n.s.▯ | n.s.▯ | 1.47 | 2.01 | 25,29 |
| Nfkbia | nuclear factor kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | n.s.▯ | 1.37 | 1.71 | 1.78 | 26,38 |
| Sgk1 | serum/glucocorticoid regulated kinase 1 | 1.60▯ | 2.45 | 2.08 | 2.97 | 42 |
| Sgk3 | serum/glucocorticoid regulated kinase 3 | n.s.▯ | n.s.▯ | 1.81 | 2.67 | 33,41 |
| Trp53inp1 | transformation related protein 53 inducible nuclear protein 1 | 1.60 | n.s.▯ | 1.31▯ | 1.39▯ | 30,39 |
| Tsc22d3 | TSC22 domain family, member 3 | 1.68 | 1.86 | 1.87 | 2.05▯ | 27 |
mPFC, medial prefrontal cortex; Acb, nucleus accumbens; Hyp, hypothalamus; VTA, ventral tegmental area. n.s, not significant; UN, untested.
not on array gene lists. References indicate previous findings in other species and tissues to support the inclusion as nutrient regulated or stress responsive.
Rapid and persistent gene expression changes seen in multiple brain regions
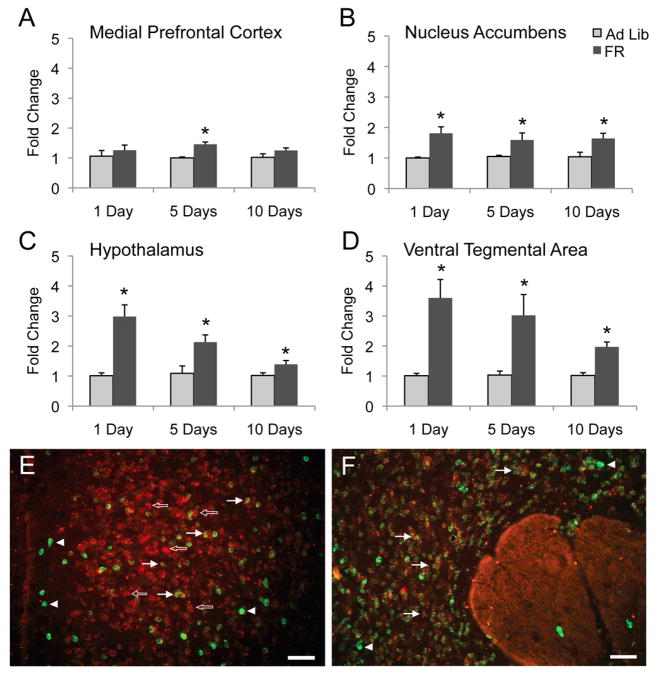
An independent cohort of 5-day FR mice was generated to assess the reproducibility of the findings using qPCR (Ad Lib: start weight 24.5+/−0.8g, FR: start weight 23.5 +/−0.7g, n.s.; Ad Lib: final weight 25.7 +/−1.0g, FR: final weight 22.1 +/− 0.5g, p<0.01). To characterize the time course of gene regulation, FR groups of one day and ten days were generated (Ad Lib-1: start weight 24.0+/−0.2g, FR-1: start weight 23.0 +/−0.5g, n.s.; Ad Lib-1: final weight 24.2 +/−0.2g, FR-1: final weight 22.0 +/− 0.6g, p<0.01) (Ad Lib-10: start weight 23.2+/−1.0g, FR-10: start weight 22.8 +/−0.8g, n.s.; Ad Lib-10: final weight 25.3 +/−0.8g, FR-10: final weight 21.6 +/− 0.7g, p<0.01). One of the validated genes, Cdkn1a, was found to be up-regulated in 3 of 4 brain regions after 10 days of food restriction. This indicates that Cdkn1a remains up-regulated during more prolonged FR conditions (Fig. 1A–D). Surprisingly, Cdkn1a also showed up-regulation upon a single day of food restriction in the same manner. Similar results were found with Arrdc2 and Angptl4 (Fig. S3 in the Supplement). Thus there appears to be a rapid and persistent expression change upon food restriction.
Figure 1.
Cdkn1a is widely up-regulated during food restriction and is neuronally expressed. Cdkn1a gene expression was analyzed in independent cohorts of 1-, 5-, and 10-day FR mice. Significant up-regulation occurred only during 5-day food restriction in the mPFC (A), and was induced by 1, 5, and 10 days of food restriction in the Acb (B) hypothalamus (C) and VTA (D). *=P < 0.05, n=5 mice per group. Photomicrographs illustrate the expression of Cdkn1a (red) and Gad1/2 (green, marker for GABAergic neurons) mRNAs within the mPFC (E) and Acb (F). Cdkn1a mRNA was observed within most cortical neurons (open arrows), including many Gad1/2-containing neurons (solid arrows), although not all (arrowheads). Within the Acb, most neurons also expressing Gad1/2 expressed Cdkn1a mRNA, although not all. Scale bars=50mm.
To visualize gene expression within neurons, the expression pattern of Cdkn1a within the mPFC and Acb was determined via in situ hybridization. Cdkn1a mRNA was detected in both mPFC and Acb (Fig. 1E, F). Within the mPFC, Cdkn1a mRNA was observed in many cortical cells including some Gad1/2-containing neurons that act as markers for GABAergic neurons. Within the Acb, Cdkn1a mRNA was again broadly expressed and found in many Gad1/2-positive neurons, which label most of the neurons in this region. Together, these findings suggest that Cdkn1a is generally expressed across cortical and sub-cortical neurons.
The role of CORT as a signal of restriction
Assessment of the qPCR validated genes revealed two potential classes of regulated genes. The first are genes previously shown to be regulated by nutritional state. The second class includes genes previously shown to be stress responsive (Table 1 and references (25–43)). These include genes responsive to systemic stress signals as well as those responsive to cellular stress. Stress and stress hormones may influence feeding behavior, providing a candidate signal for the initiation of gene expression changes found in this study (44, 45).
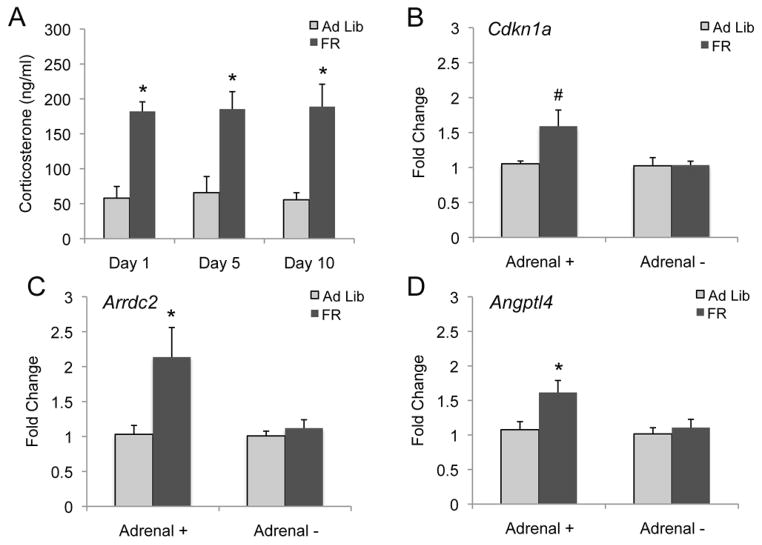
To confirm changes in this candidate signal, plasma CORT levels at the time of feeding were assessed on days one, five and ten to match the gene expression time points. Plasma CORT levels were increased at all three time points (Fig. 2A), raising the possibility that the observed increase in gene expression may be in part mediated by increases in glucocorticoid levels. Adrenal glands were removed from a cohort of animals to ablate the production of CORT, confirmed by analysis of blood plasma levels (Fig. S4 in the Supplement). This cohort of animals underwent the same 5-day FR paradigm used in the original microarray study. Due to its known role in motivation, these experiments focused on changes within the Acb. Cdkn1a, Arrdc2 and Angptl4 required the intact adrenal glands for gene induction upon food restriction (Fig. 2B–D). Strikingly, this was found for all validated genes tested in this study (Fig. S5 in the Supplement). Therefore the adrenal glands, and potentially CORT, are necessary for all the changes in gene expression observed upon food restriction.
Figure 2.
CORT is increased in FR animals and is necessary for gene up-regulation in the food restricted state. Blood plasma CORT levels were found to be significantly increased in 1-, 5-, and 10-day FR animals (A). Cdkn1a was significantly up-regulated in the Acb of an independent cohort of 5-day FR animals, however was not up-regulated in the Acb of 5-day FR adrenalectomized mice (B). Similar results were found with Arrdc2 (C) and Angptl4 (D). #=P=0.051, *= P < 0.05, n=5 mice per group.
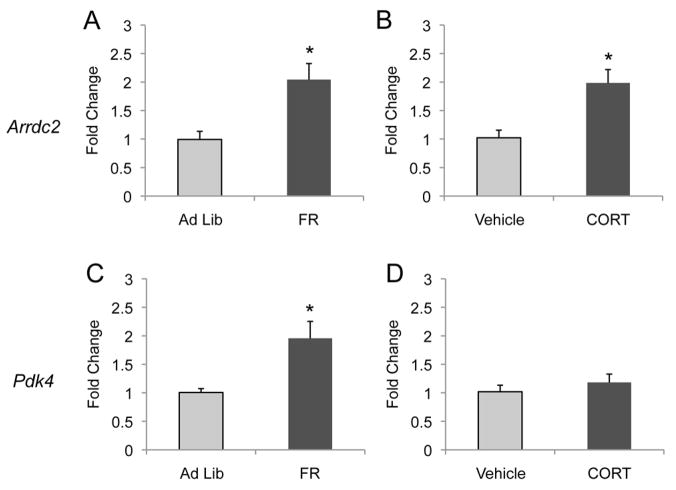
A complementary strategy is to assess whether CORT is sufficient to up-regulate these genes in the fed state. CORT was delivered intraperitoneal (i.p.) for 5 days to model the observed effect seen upon food restriction. Animals were sacrificed 2 hours after the final injection of CORT, and gene expression was assessed by qPCR. Arrdc2 and Pdk4 are validated genes showing up-regulation upon 5 days of food restriction (Table 1, Fig. 3A,C). Arrdc2 mRNA levels were increased after 5 days of CORT exposure, showing that CORT is sufficient for the induction of this gene in the absence of food restriction (Fig. 3B). In contrast, Pdk4 was not induced by this exposure (Fig. 3D). A survey of the validated genes from this study shows that 6 of the 13 genes tested are induced by this exposure (Fig. S6 in the Supplement). Taken with the above results, the adrenal glands are required for the induction of all transcripts in this study and CORT is sufficient to induce a subset of the genes.
Figure 3.
Repeated CORT exposure up-regulated gene expression in a subset of validated genes. Arrdc2 expression was significantly up-regulated within the Acb after 5 days of food restriction (A) and after 5 days of CORT exposure while provided with ad libitum diet (B). In contrast, Pdk4 expression was significantly up-regulated after 5 days of food restriction in the Acb (C) however not after 5 days of CORT exposure while provided with ad libitum food (D). *=P < 0.05, n=5 mice per group.
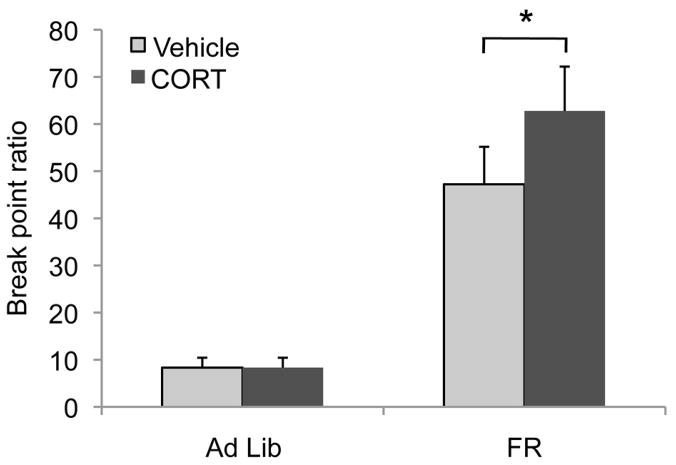
To demonstrate the importance of CORT in feeding behaviors, we used operant responding for food reinforcers on a progressive ratio schedule of reinforcement. This is a well-established index of primary motivation for an appetitive outcome (46). Break point ratios were analyzed by 2-factor (feeding state × CORT) ANOVA with repeated measures. A main effect of feeding state indicated FR mice achieve higher break points than ad libitum fed mice overall [F(1,14)=27.2, p<0.001], as expected (Fig. 4). Interestingly, an interaction between feeding state and CORT was identified [F(1,14)=6.8, p=0.02]. Post-hoc tests indicated that CORT did not affect break points when mice were fed ad libitum (p=0.9), but CORT increased responding in restricted mice (p=0.002) (Fig. 4). Thus CORT administration (at the same dose as in the gene expression studies) before the progressive ratio session further increased responding in restricted animals, with no effects on animals in the fed state. These results show that the stress hormone CORT is able to induce motivated feeding behavior in a feeding state dependent manner. Combined with the above results showing the necessity of adrenal glands and sufficiency of CORT for food restriction-induced gene expression, the progressive ratio results further highlight the role of CORT in potentiating behavioral responding for food specifically in the restricted state.
Figure 4.
Acute CORT increased motivated responding for food only in FR mice. FR mice achieved higher break point ratios on a progressive ratio schedule of reinforcement than did ad libitum fed mice. Acute CORT induced an increase in progressive ratio responding in animals that were food restricted but had no effect on fully fed mice. Break point ratios were analyzed by 2-factor ANOVA with repeated measures and with feeding state and CORT exposure as factors. *= P < 0.05, n=7 ad libitum fed and 9 FR mice.
DISCUSSION
The present results demonstrate that brief food restriction evokes stress hormone induced alterations in gene expression within mesocorticolimbic circuits. A microarray screen was conducted to elucidate the potential molecular mechanisms associated with food restriction-induced behavioral plasticity. A group of genes were identified that are induced after one day of food restriction and were persistently up-regulated after ten days. Many of this novel set of up-regulated genes were previously associated with peripheral stress responses. We show their expression to be critically dependent on food restriction and intact adrenal glands. CORT administration was sufficient to induce a subset of these validated genes in the fed state, confirming a functional role for stress hormone-induction as a physiological signal that produces gene expression changes in response to food restriction. Finally, it was shown that CORT could potentiate motivated responding for food reinforcement in a restricted state, but not in the fed state. The genes identified in this study are argued to be key molecular signals initiated by food restriction that induce behavioral plasticity.
Peripheral stress response to food restriction triggers molecular plasticity in the brain
By using a 5-day food restriction procedure, this molecular study was designed to identify early response genes that potentially mediate the initial transcriptional responses to restriction. Most of the genes identified have not been directly associated with food restriction but have been studied in other contexts. The previous identification of Angptl4 and Pdk4 as regulated by feeding state in peripheral tissues is consistent with a general role for these genes in response to restriction. However their role in mesocorticolimbic circuits has yet to be explored. Angiopoietin-like 4 (Angptl4) was originally identified as fasting induced adipose factor in the mouse liver and was subsequently shown to be a novel member of the angiopoietin family of secreted peptides due to homology (32). Pyruvate dehydrogenase kinase 4 (Pdk4) is an isoform of a mitochondrial kinase that has been shown to be induced by starvation in the rat heart (43). Cyclin-dependent kinase inhibitor p21 (Cdkn1a) is well known for its important roles in negatively regulating the cell cycle through interactions with CDK2 and PCNA, as well as being shown to inhibit apoptosis (47). Cdkn1a has been shown to be induced in the rodent brain after ischemia, although the reason for its induction in postmitotic neurons is unclear (48). There have been reports suggesting that Cdkn1a may protect against cell death in postmitotic neurons, or may have a non-enzymatic function by inhibiting stress-activated kinases (49, 50). Arrestin domain containing 2 (Arrdc2) was found to be up-regulated in the rat PFC, hippocampus and the midbrain by 90 minutes of exposure to lysergic acid diethylamide (LSD) (36). While the specific cellular and molecular function of these genes after food restriction is unclear, their regulation suggests a possible novel role for these genes in neuronal function and plasticity that is initiated by a peripheral stress response. Interestingly, some of these genes have been shown to be responsive to changes in glucose or insulin, suggesting that that the stress-mediated transcriptional response is integrated with metabolic changes during restriction (31, 43, 51).
A strength of the current study is the comprehensive use of qPCR as a validation measure, which allowed for the identification of a set of robust gene changes after a brief food restriction. As in all large-scale screening, microarray analysis is expected to have false negatives. Some genes, such as those previously identified as regulated by food deprivation in the hypothalamus were not seen in the current study (13, 14). While studies of long-term caloric restriction have identified additional genes not seen here, changes in cellular stress-responsive genes were also identified (e.g. (52)). Moreover, recent studies of short-term food restriction have also supported a role for stress responses in the amygdala and lateral hypothalamus (53, 54). The present work supports and extends these findings by testing the specific role of CORT in both molecular and behavioral responses to restriction.
Role of CORT
To test the role of stress hormones in mediating the observed expression changes, CORT production was ablated by removing the adrenal glands and then exposing these animals to the 5-day restriction paradigm. None of the genes identified after food restriction were up-regulated in the absence of adrenal glands, suggesting that factors released from these glands are necessary to initiate these changes in gene expression. To further determine the specific role of CORT, the hormone was administered in a manner that modeled what was seen in the restricted state. CORT was sufficient to induce 6/13 genes in the fully fed state, highlighting the role of this hormone in the observed gene up-regulation. CORT acts via glucocorticoid and mineralocorticoid receptors and expression studies have defined CORT responsive genes in the brain (55). Some of the genes identified here, such as Angptl4, are directly regulated by the glucocorticoid receptor (34). On the other hand, other direct targets, such as Tsc22d3, Pdk4, and Fkbp5 are not regulated following injections of CORT (27, 35, 56). There are at least two explanations for this: 1) some of the genes may be sensitive to the specific dose and timing of the CORT administration, and 2) some genes might require additional factors (e.g. metabolic) to be CORT responsive. Interestingly, the behavioral studies showed a state-dependent effect of CORT. Using a task of motivated feeding, it was demonstrated that CORT can increase motivation in a restricted state yet has no effect in the fully fed state. This finding is consistent with work that indicates similar levels of CORT, produced by different stressors, can lead to distinct behavioral responses (57). While results from our unbiased genomic screen demonstrate that a common molecular response is initiated throughout the brain under conditions of food restriction, the functional effects of these common pathways may be distinct in the different brain regions. Future work will better define these effects and reveal how the CORT signal is interpreted and integrated.
Studies have suggested a role for stress in setting the homeostatic response of the animal (58), and stress pathways are likely critical for short-term behavioral adaptations that are essential for survival. The potentiation of food motivation and consumption after restriction is an example of an adaptive behavioral response that is mediated by stress hormones. The transcriptional changes described here are also likely to be relevant to our understanding and treatment of maladaptive behavioral responses to stress. Stress often plays a role in potentiating responses to drugs of abuse (59–61), and can drive intake of palatable food (45). Molecular responses that are adaptive in the restrictive state likely underlie these stress-sensitive behaviors that are frequently detrimental to human health.
Supplementary Material
Acknowledgments
We thank Drs. M. Sarhan and B. Land for assistance in making figures, Monica Lu for expert brain processing and dissection, as well as the entire DiLeone lab for thoughtful discussions throughout the project. The microarray was done with the NIH Neuroscience Microarray Consortium. We thank Drs. J. Ploski, S. Sathyanesan, and R. Duman for assistance in early stages of microarray analysis.
This work was supported by UL-DE19586 and RL1AA017537 (R.J.D. and J.R.T.) as well by the State of Connecticut, Department of Mental Health and Addiction Services (R.J.D. and J.R.T.).
Footnotes
FINANCIAL DISCLOSURES: The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferster CB, Skinner BF. Schedules of Reinforcement. Acton: Copley Publishing Group; 1957. [Google Scholar]
- 2.Clark FC. The effect of deprivation and frequency of reinforcement on variable-interval responding. J Exp Anal Behav. 1958;1:221–228. doi: 10.1901/jeab.1958.1-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forestell CA, Schellinck HM, Boudreau SE, LoLordo VM. Effect of food restriction on acquisition and expression of a conditioned odor discrimination in mice. Physiol Behav. 2001;72:559–566. doi: 10.1016/s0031-9384(00)00439-x. [DOI] [PubMed] [Google Scholar]
- 4.Olds J. Effects of hunger and male sex hormone on self-stimulation of the brain. J Comp Physiol Psychol. 1958;51:320–324. doi: 10.1037/h0040783. [DOI] [PubMed] [Google Scholar]
- 5.Carroll ME, France CP, Meisch RA. Food deprivation increases oral and intravenous drug intake in rats. Science. 1979;205:319–321. doi: 10.1126/science.36665. [DOI] [PubMed] [Google Scholar]
- 6.Cabeza de Vaca S, Carr KD. Food restriction enhances the central rewarding effect of abused drugs. J Neurosci. 1998;18:7502–7510. doi: 10.1523/JNEUROSCI.18-18-07502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz MW, Dallman MF, Woods SC. Hypothalamic response to starvation: implications for the study of wasting disorders. Am J Physiol. 1995;269:R949–957. doi: 10.1152/ajpregu.1995.269.5.R949. [DOI] [PubMed] [Google Scholar]
- 9.Ahima RS, Prabakaran D, Mantzoros C, Qu D, Lowell B, Maratos-Flier E, et al. Role of leptin in the neuroendocrine response to fasting. Nature. 1996;382:250–252. doi: 10.1038/382250a0. [DOI] [PubMed] [Google Scholar]
- 10.Hommel JD, Trinko R, Sears RM, Georgescu D, Liu ZW, Gao XB, et al. Leptin receptor signaling in midbrain dopamine neurons regulates feeding. Neuron. 2006;51:801–810. doi: 10.1016/j.neuron.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 11.Fulton S, Pissios P, Manchon RP, Stiles L, Frank L, Pothos EN, et al. Leptin regulation of the mesoaccumbens dopamine pathway. Neuron. 2006;51:811–822. doi: 10.1016/j.neuron.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Myers MG, Münzberg H, Leinninger GM, Leshan RL. The geometry of leptin action in the brain: more complicated than a simple ARC. Cell Metab. 2009;9:117–123. doi: 10.1016/j.cmet.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu D, Ludwig DS, Gammeltoft S, Piper M, Pelleymounter MA, Cullen MJ, et al. A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature. 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- 14.Baskin DG, Seeley RJ, Kuijper JL, Lok S, Weigle DS, Erickson JC, et al. Increased expression of mRNA for the long form of the leptin receptor in the hypothalamus is associated with leptin hypersensitivity and fasting. Diabetes. 1998;47:538–543. doi: 10.2337/diabetes.47.4.538. [DOI] [PubMed] [Google Scholar]
- 15.Cabib S, Orsini C, Le Moal M, Piazza PV. Abolition and reversal of strain differences in behavioral responses to drugs of abuse after a brief experience. Science. 2000;289:463–465. doi: 10.1126/science.289.5478.463. [DOI] [PubMed] [Google Scholar]
- 16.McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- 17.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 18.Nestler EJ. Molecular neurobiology of addiction. Am J Addict. 2001;10:201–217. doi: 10.1080/105504901750532094. [DOI] [PubMed] [Google Scholar]
- 19.Trinko R, Sears RM, Guarnieri DJ, DiLeone RJ. Neural mechanisms underlying obesity and drug addiction. Physiol Behav. 2007;91:499–505. doi: 10.1016/j.physbeh.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Haberny SL, Carr KD. Comparison of basal and D-1 dopamine receptor agonist-stimulated neuropeptide gene expression in caudate-putamen and nucleus accumbens of ad libitum fed and food-restricted rats. Brain Res Mol Brain Res. 2005;141:121–127. doi: 10.1016/j.molbrainres.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Carr KD. Chronic food restriction: enhancing effects on drug reward and striatal cell signaling. Physiol Behav. 2007;91:459–472. doi: 10.1016/j.physbeh.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Gourley SL, Kiraly DD, Howell JL, Olausson P, Taylor JR. Acute hippocampal brain-derived neurotrophic factor restores motivational and forced swim performance after corticosterone. Biol Psychiatry. 2008;64:884–890. doi: 10.1016/j.biopsych.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 25.Anwar A, Keating AK, Joung D, Sather S, Kim GK, Sawczyn KK, et al. Mer tyrosine kinase (MerTK) promotes macrophage survival following exposure to oxidative stress. J Leukoc Biol. 2009;86:73–79. doi: 10.1189/JLB.0608334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 27.D’Adamio F, Zollo O, Moraca R, Ayroldi E, Bruscoli S, Bartoli A, et al. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7:803–812. doi: 10.1016/s1074-7613(00)80398-2. [DOI] [PubMed] [Google Scholar]
- 28.Dufner-Beattie J, Wang F, Kuo YM, Gitschier J, Eide D, Andrews GK. The acrodermatitis enteropathica gene ZIP4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J Biol Chem. 2003;278:33474–33481. doi: 10.1074/jbc.M305000200. [DOI] [PubMed] [Google Scholar]
- 29.Graham DK, Dawson TL, Mullaney DL, Snodgrass HR, Earp HS. Cloning and mRNA expression analysis of a novel human protooncogene, c-mer. Cell Growth Differ. 1994;5:647–657. [PubMed] [Google Scholar]
- 30.Han ES, Muller FL, PÈrez VI, Qi W, Liang H, Xi L, et al. The in vivo gene expression signature of oxidative stress. Physiol Genomics. 2008;34:112–126. doi: 10.1152/physiolgenomics.00239.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaestner KH, Christy RJ, Lane MDCP. Mouse insulin-responsive glucose transporter gene: characterization of the gene and trans-activation by the CCAAT/enhancer binding protein. Proc Natl Acad Sci U S A. 1990;87:251–255. doi: 10.1073/pnas.87.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kersten S, Mandard S, Tan NS, Escher P, Metzger D, Chambon P, et al. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J Biol Chem. 2000;275:28488–28493. doi: 10.1074/jbc.M004029200. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi T, Deak M, Morrice N, Cohen PCP. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J. 1999;344(Pt 1):189–197. [PMC free article] [PubMed] [Google Scholar]
- 34.Koliwad SK, Kuo T, Shipp LE, Gray NE, Backhed F, So AY, et al. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor) is a direct glucocorticoid receptor target and participates in glucocorticoid-regulated triglyceride metabolism. J Biol Chem. 2009;284:25593–25601. doi: 10.1074/jbc.M109.025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee RS, Tamashiro KL, Yang X, Purcell RH, Harvey A, Willour VL, et al. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology. 2010;151:4332–4343. doi: 10.1210/en.2010-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols CD, Sanders-Bush E. Molecular genetic responses to lysergic acid diethylamide include transcriptional activation of MAP kinase phosphatase-1, C/EBP-beta and ILAD-1, a novel gene with homology to arrestins. J Neurochem. 2004;90:576–584. doi: 10.1111/j.1471-4159.2004.02515.x. [DOI] [PubMed] [Google Scholar]
- 37.Ring RH, Valo Z, Gao C, Barish ME, Singer-Sam J. The Cdkn1a gene (p21Waf1/Cip1) is an inflammatory response gene in the mouse central nervous system. Neurosci Lett. 2003;350:73–76. doi: 10.1016/s0304-3940(03)00883-8. [DOI] [PubMed] [Google Scholar]
- 38.Scheinman RI, Cogswell PC, Lofquist AK, Baldwin AS. Role of transcriptional activation of I kappa B alpha in mediation of immunosuppression by glucocorticoids. Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- 39.Tomasini R, Samir AA, Vaccaro MI, Pebusque MJ, Dagorn JC, Iovanna JL, et al. Molecular and functional characterization of the stress-induced protein (SIP) gene and its two transcripts generated by alternative splicing. SIP induced by stress and promotes cell death. J Biol Chem. 2001;276:44185–44192. doi: 10.1074/jbc.M105647200. [DOI] [PubMed] [Google Scholar]
- 40.Tullo A, Mastropasqua G, Bourdon JC, Centonze P, Gostissa M, Costanzo A, et al. Adenosine deaminase, a key enzyme in DNA precursors control, is a new p73 target. Oncogene. 2003;22:8738–8748. doi: 10.1038/sj.onc.1206967. [DOI] [PubMed] [Google Scholar]
- 41.von Hertzen LS, Giese KP. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J Neurosci. 2005;25:1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster MK, Goya L, Ge Y, Maiyar AC, Firestone GLCP. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu P, Sato J, Zhao Y, Jaskiewicz J, Popov KM, Harris RACP. Starvation and diabetes increase the amount of pyruvate dehydrogenase kinase isoenzyme 4 in rat heart. Biochem J. 1998;329(Pt 1):197–201. doi: 10.1042/bj3290197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adam TC, Epel ES. Stress, eating and the reward system. Physiol Behav. 2007;91:449–458. doi: 10.1016/j.physbeh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Dallman MF, Pecoraro NC, La Fleur SE, Warne JP, Ginsberg AB, Akana SF, et al. Glucocorticoids, chronic stress, and obesity. Prog Brain Res. 2006;153:75–105. doi: 10.1016/S0079-6123(06)53004-3. [DOI] [PubMed] [Google Scholar]
- 46.Hodos W. Progressive ratio as a measure of reward strength. Science. 1961;134:943–944. doi: 10.1126/science.134.3483.943. [DOI] [PubMed] [Google Scholar]
- 47.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Lookeren Campagne M, Gill R. Cell cycle-related gene expression in the adult rat brain: selective induction of cyclin G1 and p21WAF1/CIP1 in neurons following focal cerebral ischemia. Neuroscience. 1998;84:1097–1112. doi: 10.1016/s0306-4522(97)00580-0. [DOI] [PubMed] [Google Scholar]
- 49.Park DS, Morris EJ, Padmanabhan J, Shelanski ML, Geller HM, Greene LA. Cyclin-dependent kinases participate in death of neurons evoked by DNA-damaging agents. J Cell Biol. 1998;143:457–467. doi: 10.1083/jcb.143.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shim J, Lee H, Park J, Kim H, Choi EJ. A non-enzymatic p21 protein inhibitor of stress-activated protein kinases. Nature. 1996;381:804–806. doi: 10.1038/381804a0. [DOI] [PubMed] [Google Scholar]
- 51.Kim HK, Youn BS, Shin MS, Namkoong C, Park KH, Baik JH, et al. Hypothalamic Angptl4/Fiaf is a novel regulator of food intake and body weight. Diabetes. 2010;59:2772–2780. doi: 10.2337/db10-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto Y, Tanahashi T, Kawai T, Chikahisa S, Katsuura S, Nishida K, et al. Changes in behavior and gene expression induced by caloric restriction in C57BL/6 mice. Physiol Genomics. 2009;39:227–235. doi: 10.1152/physiolgenomics.00082.2009. [DOI] [PubMed] [Google Scholar]
- 54.Pankevich DE, Teegarden SL, Hedin AD, Jensen CL, Bale TL. Caloric restriction experience reprograms stress and orexigenic pathways and promotes binge eating. J Neurosci. 2010;30:16399–16407. doi: 10.1523/JNEUROSCI.1955-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datson NA, Morsink MC, Meijer OC, de Kloet ER. Central corticosteroid actions: Search for gene targets. Eur J Pharmacol. 2008;583:272–289. doi: 10.1016/j.ejphar.2007.11.070. [DOI] [PubMed] [Google Scholar]
- 56.Huang B, Wu P, Bowker-Kinley MM, Harris RA. Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor-alpha ligands, glucocorticoids, and insulin. Diabetes. 2002;51:276–283. doi: 10.2337/diabetes.51.2.276. [DOI] [PubMed] [Google Scholar]
- 57.Covington HE, Miczek KA. Intense cocaine self-administration after episodic social defeat stress, but not after aggressive behavior: dissociation from corticosterone activation. Psychopharmacology (Berl) 2005;183:331–340. doi: 10.1007/s00213-005-0190-5. [DOI] [PubMed] [Google Scholar]
- 58.McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- 59.Koob GF. A role for brain stress systems in addiction. Neuron. 2008;59:11–34. doi: 10.1016/j.neuron.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piazza PV, Le Moal M. The role of stress in drug self-administration. Trends Pharmacol Sci. 1998;19:67–74. doi: 10.1016/s0165-6147(97)01115-2. [DOI] [PubMed] [Google Scholar]
- 61.Sinha R. Chronic stress, drug use, and vulnerability to addiction. Ann N Y Acad Sci. 2008;1141:105–130. doi: 10.1196/annals.1441.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.