Abstract
Effective control of the Ca2+ homeostasis in any living cell is paramount to coordinate some of the most essential physiological processes, including cell division, morphological differentiation, and intercellular communication. Therefore, effective homeostatic mechanisms have evolved to maintain the intracellular Ca2+ concentration at physiologically adequate levels, as well as to regulate the spatial and temporal dynamics of Ca2+ signalling at subcellular resolution. Members of the superfamily of EF-hand Ca2+-binding proteins are effective to either attenuate intracellular Ca2+ transients as stochiometric buffers or function as Ca2+ sensors whose conformational change upon Ca2+ binding triggers protein-protein interactions, leading to cell state-specific intracellular signalling events. In the central nervous system, some EF-hand Ca2+-binding proteins are restricted to specific subtypes of neurons or glia, with their expression under developmental and/or metabolic control. Therefore, Ca2+-binding proteins are widely used as molecular markers of cell identity whilst also predicting excitability and neurotransmitter release profiles in response to electrical stimuli. Secretagogin is a novel member of the group of EF-hand Ca2+-binding proteins whose expression precedes that of many other Ca2+-binding proteins in postmitotic, migratory neurons in the embryonic nervous system. Secretagogin expression persists during neurogenesis in the adult brain, yet becomes confined to regionalized subsets of differentiated neurons in the adult central and peripheral nervous and neuroendocrine systems. Secretagogin may be implicated in the control of neuronal turnover and differentiation, particularly since it is re-expressed in neoplastic brain and endocrine tumours and modulate cell proliferation in vitro. Alternatively, and since secretagogin can bind to SNARE proteins, it might function as a Ca2+ sensor/coincidence detector modulating vesicular exocytosis of neurotransmitters, neuropeptides or hormones. Thus, secretagogin emerges as a functionally multifaceted Ca2+-binding protein, whose molecular characterization can unravel a new and fundamental dimension of Ca2+ signalling under physiological and disease conditions in the nervous system and beyond.
Keywords: Alzheimer's disease, Ca2+ buffer, Ca2+ sensor, brain development, neurodegeneration, pancreas
1. Introduction: brief overview of Ca2+-binding proteins with emphasis on the central nervous system
Calcium (Ca2+) is an essential bivalent ion in every organ system of the body. Ca2+ bioavailability determines, e.g., the rate of skeletal and heart muscle contractions, the rigidity of bones, the function of various enzymes as co-factor, and the membrane properties and excitability of cells such as neurons. Ca2+ acts as a second messenger in signal transduction pathways in many fundamental biological processes governing embryonic development and adult physiology [1]. Therefore, the loss of Ca2+ homeostasis invariably manifests in disease. Since the momentarily available intracellular Ca2+ concentration ([Ca2+]i) critically determines the ability of neurons to release neurotransmitters [2] from nerve terminals through soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-dependent exocytosis of neurotransmitter-laden vesicles [3], pathological changes in Ca2+ signalling can precipitate in neuropsychiatric and neurological diseases [4,5] including migraine, ataxia [6], Alzheimer's disease (AD) [7], stroke [8,9] or epilepsy [6,10].
Several mechanisms have evolved to provide efficient homeostatic control of the Ca2+ homeostasome, as well as to prevent irreparable cellular damage, including Ca2+ extrusion by plasmalemmal Ca2+-ATPases [11], dissipating Ca2+ oscillations via Ca2+ uptake by intracellular stores [12], and chelation of free, cytosolic Ca2+ by Ca2+-binding proteins (CBPs) [13]1. CBPs share a unique tandem repeat of a Ca2+ -binding loop flanked by two α helices known as the “EF-hand” Ca2+ -binding site [14,15]. The discovery and localization of Ca2+-binding proteins in the nervous system [13,16-21] together with detailed analysis of the structure and ion binding characteristics of their “EF-hand” domains [14,15] provided an essential impetus to expand and refine our knowledge of the molecular regulation of Ca2+-dependent processes in synaptic neurotransmission and the regulation of neuron-glia interactions.
CBPs can be divided into two major groups [22]: (i) Ca2+ sensors, which undergo significant conformational rearrangements when binding Ca2+, allowing them to interact with specific downstream targets to initiate signal transduction cascades; (ii) cytosolic Ca2+ buffers that bind Ca2+ without conformational changes to dissipate local Ca2+. Synaptotagmin, calmodulin and S100 families are classical representatives of Ca2+ sensors to affect kinases (e.g., Ca2+/calmodulin-dependent kinases), phosphatases (calcineurin) [21], during, e.g., exocytotic processes [3,23], neurite outgrowth [24,25], cell division, gene expression and cell survival [26,27]. Distinction between Ca2+ sensors or buffers is not necessarily clear (Table 1) since the function of individual proteins can depend on their focal concentration, the availability of interacting partners in signaling networks, and the cellular context.
Table 1.
| Parvalbumin | Calbindin | Calretinin | Secretagogin | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Isoform | α | β2 | CB-D28k | CB-9k | Calretinin | CR-22k | Secretagogin | R22 | Setagin |
| Molecular weight (kDa) | 10-12 | 11-12 | 28 | 9 | 29-31 | 22 | 32 | 32 | 7 |
| Ca2+-binding (EF) domains | 3 | 3 | 6 | 2 | 6 | 4 | 6 | 6 | 0 |
| Functional EF domains | 2 | 2 | 4 | 2 | 5 | 4 | 4 | 4 | 0 |
| Mixed Ca2+/Mg2+ binding | + | + | -3 | - | - | - | + | + | - |
| Kd (nM) | 4-9 | 42-45 | (h) 180-240 (m) 410-510 |
(S1) 200-500 (S2) 60-300 |
1,5004 | 1,200 | 25,000 | 25,0005 | not applicable |
| Ca2+-sensor-like properties | - | - | +6 | -7 | +8 | + | + | + | not applicable |
| Co-localization in rodent brain (major isoform) | CB-D28k, calretinin | n/a9 | parvalbumin, calretinin, secretagogin | - | parvalbumin, secretagogin | - | CB-28k, calretinin | -10 | - |
| Expressed in prenatal CNS | -11 | - | + | - | + | - | + | not tested | not tested |
| Neuroendocrine systems | - | - | + | + | - | not tested | + | + | - |
| Tumor | + | + | + | + | + | + | + | not tested | not tested |
β-parvalbumin refers to oncomodulin.
Low-affinity Mg2+ binding (Kd,Mg ≈ 700 μM) can significantly decrease Ca2+-binding affinity [118].
Ca2+ binding to high activity sites (indicated) occurs with moderate co-operativity. Low affinity site Kd = 0.5 mM (calretinin), 1 mM (CR-22k) [119].
Secretagogin contains a signle high-affinity and three low affinity Ca2+-binding sites. Secretagogin-R22's Ca2+-binding capacity has been qualitatively assessed [55,68].
Although calbindin-D28k is considered as a fast Ca2+ buffer, known binding partners include: Ran-binding protein [42], caspase-3 [41], 3′,5′-cyclic nucleotide phosphodiesterase [40], ATPase [39], L-type Ca2+ channel α subunit [37], myo-inositol monophosphatase [120], TRPV5 [38].
Although undergoes conformational changes upon Ca2+ binding, no identified binding partners are known to date [22].
Can qualify as “Ca2+-sensor” [119].
Oncomodulin has only been detected in the nervous system of mice lacking α-parvalbumin expression [50].
Co-localization studies have not been performed although the protein is expressed in the nervous system [68].
Prenatal expression has been reported in human fetal brain [84].
In neurobiology, the parvalbumin (PV) and calbindin subfamilies, the latter including the vitamin D-dependent 28 kDa isoform of calbindin (CB) and calretinin (CR), gained significant attention since they exhibit phylogenetically preserved tissue-specific expression patterns in vertebrates [13,28,29]. These CBPs are restricted to morphologically distinct subpopulations of GABAergic interneurons and local projection cells in rodent, primate and human cerebrum with the exception of CB that is also expressed by cortical pyramidal and dentate granule cells [17]. Besides revealing morphologically distinct types of neurons, PV, CB and CR also contribute to shaping electrophysiologcal characteristics [30,31]: amongst interneurons, PV(CB)-containing cells are generally termed “fast-spiking”, whereas CB or CR-containing interneurons commonly belong to regular or irregular-firing subclasses, respectively [29,32-34].
Varying numbers of Ca2+-binding sites, of which some are non-functional or exhibit different affinity to Mg2+, are found in these CBPs (Table 1). PV, widely considered as the “prototypic” Ca2+ buffer with high-affinity Ca2+ binding, is able to bind both Ca2+ and Mg2+. CB exhibits low affinity Mg2+ binding, whereas CR is unable to bind this bivalent cation [35]. Notwithstanding the broadly held view of being a cytosolic “buffer” protein [36], CB has a significant number of identified target proteins (Table 1) in neurons [37-42] suggesting additional Ca2+ sensor-like functions. CR may also possess Ca2+ sensor properties since it undergoes significant conformation changes upon Ca2+ binding [43].
If cytosolic CBPs are pivotal to coordinate neuronal excitability then their genetic deletion must impact the organization and function of the nervous system. Deletion of PV or CB results in impaired motor coordination [44], and neuronal sensitization towards injury and metabolic hypofunction in epilepsy and aging [45,46]. Similarly, the lack of CR leads to motor coordination defects in mice [47]. Although most neurons express a typifying cytosolic Ca2+ buffer protein in the brain, CBPs can occasionally co-localize [32,48,49]. Nevertheless, neurons appear to commit to certain Ca2+ buffers since genetic removal of a CBP is usually not compensated by up-regulation of expression of another EF-hand CBP superfamily member (see however, [50]).
Are there yet uncharacterized neuron-specific CBPs in the nervous system? The answer is clearly “yes” since secretagogin has recently been demonstrated to identify developmentally-related neuronal subpopulations in the mammalian brain [51,52], including humans [53,54]. Secretagogin is a hexa EF-hand CBP capable to simultaneously bind four Ca2+ ions [55,56]. Its tertiary structure changes upon Ca2+ -binding [55]. This property, together with an in vitro-identified cellular interaction network [57], suggests that secretagogin may act as a Ca2+ sensor rather than buffer in neurons. This review summarizes experimental and histopathological data hitherto obtained and concerned with secretagogin's structure, expression pattern, physiological and pathological significance and discusses critical experiments to conclusively define secretagogin functions in the central nervous system (CNS).
2. Secretagogin's cloning and transcriptional control
Secretagogin was discovered by using the “expression dictates function” principle: quantal insulin release from pancreatic β cells relies on the Ca2+-dependent exocytosis of insulin-filled large dense core granules [58]. Once insulin-containing secretory vesicles are docked, a Ca2+ “sensor” of the synaptotagmin family [23] is required for vesicle fusion into the plasma membrane to proceed. The obligatory Ca2+ dependence of this release mechanism, alike neurotransmitter release from nerve endings [3], illustrates how the intracellular Ca2+ homeostasis contributes to defining intercellular communication. This notion was the critical stepping stone for Wagner et al. [59] to search for novel “sensors” in pancreatic β cells, and led to the cloning of secretagogin upon immunoscreening a cDNA library expressed in λgt11 phage with a monoclonal antibody (D24) raised against unknown insulinoma-specific antigens [60].
Sequence alignment of the secretagogin cDNA identified a single genomic locus mapping onto chromosome 6 in humans (6p22.1-22.3) [59]. The open reading frame (ORF) encoding secretagogin in human consists of 828-base pairs (bp) [59], flanked by 5′- (156 bp) and 3′-untranslated (450 bp) nucleotide sequences. Although the precise exon map encoding secretagogin's 42 C-terminal amino acids (AAs) remains elusive, the secretagogin mRNA appears to be the splice product of at least 10 exons spanning >33 kbps [59]. The polyadenylation signal is localized to bases 1412–1417, yet a poly(A) tail was not found [59].
Skovhus et al. [61] reported basal constitutive promoter activity when subcloning a 1498 bp fragment (-1427 to +71 bp) upstream to secretagogin's transcription start site into a firefly luciferase construct and expressing this in a rat insulinoma cell line in vitro. This finding and the moderately (∼40%) enhanced activity of secretagogin's putative promoter at near-physiological (5.5 mM) glucose concentrations [61] suggest metabolically regulated secretagogin transcription in pancreatic β cells. Future identification of the minimal essential promoter, and a detailed map of transcription factor binding sites will be necessary to appreciate secretagogin's transcriptional variations under disease conditions. In this context it is worth noting that 11 single nucleotide polymorphisms (SNPs), eight of which modify transcription factor binding sites - such as Oct-1, Pit-1 a, GATA1, Sp1 - map to the 1700 bp 5′ untranslated region (over the putative promoter) upstream from the secretagogin gene [61]. These SNPs are unlikely to contribute to familial type 1 diabetes though [61]. Yet they may be significant in the molecular pathogenesis of neuropsychiatric conditions: secretagogin has recently been implicated in cellular neuroprotection under degenerative conditions [53,54]. SNPs could alter secretagogin's promoter activity and impact Oct-1, Pta-1a, GATA1 and/or Sp1 transcription factor-dependent cellular differentiation [62], transformation [63] and survival programs in glia and neurons, reliant on coordinated Ca2+ signalling, in AD [64,65], Down's syndrome with in utero onset [66] and hypoxia [67].
2.1. Splice variants
The size of secretagogin's major mRNA transcript is ∼1,500 bp in all organs harbouring secretagogin expression. Yet a 2600-bp mRNA variant was identified in the pancreas [59], which may either represent the primary mRNA or be specific to pancreatic β cells.
Alternative RNA editing in both normal neuroendocrine tissues and neoplasias can give rise to secretagogin-R22, which contains a single AA exchange (Q/R) at codon 22 (exon 1) [68]. Since AA22 is positioned outside the closest EF-hand motif, the ensuing secretagogin-R22 protein exhibits unchanged Ca2+-binding activity. In contrast, a 49 AA-long peptide, setagin (∼7 kDa), is generated when exons 2 and 7 (83-153 and 472-527 bp of the ORF, respectively) are lost in pancreatic β cells [68]. Only setagin's 27 N-terminal AAs are identical to those of secretagogin due to a frame shift (also introducing a terminal codon). Thus, setagin lacks EF-hand domains resulting in the complete loss of Ca2+ binding [68]. Since setagin is expressed in the human pancreas, it is surprising that neither setagin's organ system-specific distribution nor the molecular identity of setagin-containing cells has so far not been studied. Functional characterization of this protein may be of interest, too, since setagin can coexist with secretagogin [68] and thus could compete with secretagogin when binding to interacting partners [57] under (patho-)physiological conditions.
3. Protein structure and physicochemical properties
Once translated, human secretagogin is a 276 AA acidic protein with a calculated molecular weight of 32 kDa [51,59]. The bulk (>90%) of secretagogin is localized in the cytosol, with scant quantities likely present in the nucleus [51,52,54,59,69-71].
Secretagogin contains six tandem repeats of the EF-hand Ca2+-binding domain, with predicted loops at all six Ca2+-binding sites [56,59]. Five of the Ca2+-binding sites are 12 AAs in length with a glutamate residue at position 12. Conspicuously, and unlike other CBP families, secretagogin's fourth EF-hand site only consists of 8 AAs [56]. This change may suggest loss-of-function variation with both the functionality and avidity of the fourth EF-hand sequence's for Ca2+ being reduced in comparison to the longer sequences. When crystallized in its Ca2+-free conformation [56], secretagogin appears as a modular protein (radius of gyration: 20.6 Å) consisting of three globular domains (12 Å each). Each of these domains contains a pair of EF-hand motifs, with an overall configuration resulting in a bulky V-shaped molecule. Two flexible linkers (AAs 85-98 and 176-185) are predicted to join secretagogin's Ca2+-binding domains, providing a distinct grove at the interface of secretagogin's globular domains. Within individual globular domains, Ca2+ binding loops of the EF-hand motifs contain the consensus sequence DXDGXGXGXIXXXEF (“X” indicates variable AA residues) [55] and form a solvent-exposed two-stranded anti-parallel β sheet. A four-helix bundle formed from helices of the two EF-hand motifs in each domain gives rise to a domain core, which is stabilized by extensive hydrophobic interactions. Comparative structure prediction suggests that the metal binding loops of secretagogin's EF1,3,4,6 motifs adopt conformations similar to those of calmodulin, an archetypical CBP [56]. Whilst the EF2 motif is uniquely structured, EF5 is in a “Ca2+-ready” conformation and stabilized by electrostatic interactions even in the absence of a metal ion.
Theoretically, each molecule of secretagogin could bind a maximum of six Ca2+ ions [55,56,59]. However, secretagogin may be fully competent for Ca2+-binding in only some species (e.g., Danio rerio, Xenopus laevis, Gallus gallus) [56]. Secretagogin orthologs from higher eukaryotes contain at least one non-functional EF-hand motif due to mutation(s) or deletions (e.g., the EF2 loop might not bind Ca2+ in Primates and human). In accordance with the predicted ability of highly conserved EF3-6 loops to bind Ca2+, isothermal calorimetry confirmed a 1:4 stochiometry of Ca2+ binding per human recombinant secretagogin molecule [55].
Cytosolic CBPs, as said, broadly fall into two main categories: i) intracellular Ca2+ buffers (Kd < 0.1 μM) [13,72] or ii) Ca2+ “sensors”/signal modulators (Kd > 0.1 μM). Ca2+ binding by buffer proteins (e.g., PV, CB) occurs in the absence of major modifications to the quaternary protein structure. In contrast, Ca2+ “sensors” (e.g., calmodulin) undergo conformational modifications upon binding free Ca2+ ions (chelation) [14,15]. None of the Ca2+ sensors are themselves enzymes [14]. Instead, their conformational changes orchestrate specific interactions with target proteins - 87 proteins with discrete binding motifs co-precipitate with calmodulin alone in mouse brain [73] - to initiate signalling events. Secretagogin has two classes of Ca2+-binding sites: a high affinity site is accompanied by three low-affinity sites with the latter class exhibiting a high degree of co-operativity of Ca2+ binding [55]. Secretagogin's half-maximal [Ca2+] binding affinity is ≈25 μM in quasi-physiological salt buffers (e.g., 150 mM KCl) [55]. This is significantly lower than that of PV (equilibrium dissociation constant (Kd) ≈ 0.05 μM) [55] (Table 1) and suggests that only 0.5 – 14% of secretagogin might be saturated at resting intracellular Ca2+ concentrations.
Secretagogin undergoes tertiary structure rearrangement when binding Ca2+ (1 mM), and its Ca2+ affinity is comparable to that of e.g. synaptotagmin I [55]. These properties suggest that secretagogin may not function as a canonical intracellular Ca2+ buffer, but belong to the Ca2+ “sensor” family instead [55]. However, secretagogin's target spectrum appears to be more restricted than that of many other Ca2+ sensors, with prevailing intermolecular interactions probably regulating vesicular exocytosis in both endocrine cells and neurons [55,57].
3.1. Interaction with molecular motors of vesicle trafficking
A prerequisite of dynamic signalling processes is the fine-tuned control of the intracellular Ca2+ concentration. Ca2+ “sensor” proteins coordinate cellular responses by interacting with specific networks of target proteins. A Ca2+ “sensor” protein can exist in Ca2+-free (apo), Ca2+-bound or Ca2+/target-complexed signal-competent configurations. If secretagogin is a Ca2+ sensor protein then it must dynamically and reversibly interact with specific targets to initiate cell state-specific signalling events.
Soluble N-ethylmaleimide-sensitive factor attachment protein of 25 kDa (SNAP-25) has been identified as the first candidate to bind secretagogin, when tissue homogenates from either brain or pancreatic cell lines were superfused over immobilized recombinant secretagogin bait [55]. This high-affinity interaction proceeds in the presence of Ca2+ (Kd = 1.2 × 10-7 M) yet becomes significantly reduced when apo-secretagogin participates in the absence of Ca2+ (Kd = 1.5 × 10-6 M) [55]. It is likely that secretagogin's domain participating in this molecular interaction maps onto its internal or C-terminal AA sequence since setagin, with only its N-terminal extremity homologous to that of secretagogin, cannot bind SNAP-25 [55]. More recently, high content protein array screening has identified at least nine proteins – SNAP-23, double C2-like domain-containing protein α (DOC2α), ADP-ribosylation factor GTPase-activating protein 2 (ARFGAP2) and its homolog (ARFGAP3), kinesin (KIF5B), rootletin, β-tubulin, N,N-dimethylarginine dimethylaminohydrolase 2 (DDAH-2), ATP-synthase O subunit, and myeloid leukemia factor 2 – as potential interaction partners [57]. The Kd of these targets from secretagogin ranges from 100 pM - 10 nM. Putative members of this interactome network specifically contribute to intracellular trafficking (KIF5B, β-tubulin, ARFGAP2/3), docking (SNAP-23, DOC2α), scaffolding (rootletin; in retina [74], and fusion (SNAP-23) of transport vesicles during exocytosis. Cumulatively, these data suggest that secretagogin may be essential in neuroendocrine (hormone) and synaptic (neurotransmitter) release by fine-tuning short-range Ca2+ signalling.
The finding that SNAP-25, and its homologue SNAP-23 (exhibiting ∼60% sequence identity) can bind secretagogin in vitro fuels the hypothesis that secretagogin might be involved in the focal control of vesicle exocytosis in both neural and endocrine systems [55,57]. Secretagogin's probable molecular inter-actors have been identified by using high-density protein arrays and validated by surface plasmon resonance and pull-down assays of the proteins co- and over-expressed in heterologous cell systems in vitro [57]. While these methods are suggestive of putative molecular interactions, they lack inherent instrumental capacity to reveal whether or not these in vitro interactions are of physiological relevance (that is, whether the proteins are natively co-expressed in any given cell type of the organism). In fact, neither vesicular glutamate nor vesicular GABA transporter appears to co-exist with secretagogin in forebrain neurons of mammals, including primates (AA, THö & THa, unpublished data). Therefore, future functional studies must clarify the precise cellular context(s), in which secretagogin can drive (patho-)physiological processes by modulating cellular responsiveness through the Ca2+-dependent formation of effector complexes.
4. Secretagogin expression: focus on the neuroendocrine axis and central nervous system
Secretagogin expression has been mapped in virtually all organ systems of rodents, primates and human (Fig. 1 and 2). Initial investigations highlighted secretagogin's neuroendocrine attributes (Fig. 1A-B): Northern blotting revealed robust secretagogin mRNA expression in the pancreas, and to a lesser extent in the adrenal medulla and cortex, thyroid, pituitary, and the luminous organs of the gut (stomach, small intestine, colon) surfaced by simple epithelia (Fig. 1B) [59]. In the digestive tract, the localization of secretagogin+ cells typically reflected the histological pattern of enteroendocrine cells [59], and secretagogin's co-localization with specific neuroendocrine markers in the prostate confirmed cell identity [75]. In a case study, secretagogin expression was also reported in an exocrine gland, the submucosal gland of the human nasal mucosa [76]. These studies were significantly extended by showing secretagogin expression in subpopulations of developing or adult neurons [51,52], and implicating secretagogin in cellular defences against neuronal injury in neurodegenerative diseases, particularly AD [53,77].
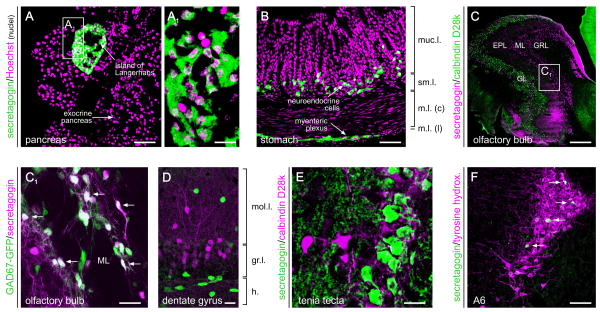
Fig. 1. Organ systems distribution of secretagogin during adulthood.
(A, A1) Secretagogin is expressed at particularly high levels by pancreatic β cells in the islands of Langerhans. Neuroendocrine cells, such as those in the principal part of gastric glands (B), also harbour secretagogin. In the nervous system, secretagogin is expressed by interneurons in the periglomerular and plexiform layers of the olfactory bulb (C, C1), where it can co-exist with glutamic acid decarboxylase (GAD, arrows), the enzyme synthesizing GABA. Secretagogin has also been found in granular cells of the dentate gyrus (D) and tenia tecta (E), as well as the locus coeruleus (F) containing noradrenergic cells (group A6). Arrows in (F) identify secretagogin+ cells intermingled with tyrosine hydroxylase-expressing neurons. Abbreviations: EPL, external plexiform layer; GL, glomerular layer; GRL, granular layer; ML, mitral layer; m.l. (c/l), muscular layer (central/lateral); muc.l., mucosal layer; sm.l., submucosal layer. Scale bar = 300 μm (C), 70 μm (A,B,F), 20 μm (A1,C1,D,E).
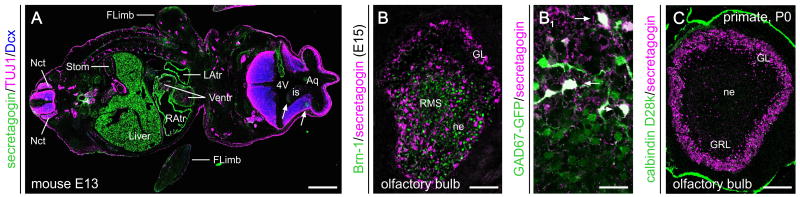
Fig. 2. Secretagogin expression during foetal development.
(A) Secretagogin immunoreactivity in the central nervous system (arrows) and at the periphery (e.g., heart, liver, stomach, gonads) of a mouse embryo at embryonic day (E) 13. (B) Secretagogin+ neurons populate the olfactory bulb by E15, and differentiate into GABAergic neurons as revealed in GAD67-EGFP reporter mice (B1). (C) Secretagogin expression is confined predominantly to periglomerular neurons in the olfactory bulb of grey mouse lemur (Microcebus murinus, Primates) by birth. Abbreviations: 4V, fourth ventricle; Aq, aqueduct; FLimb, forelimb; is, isthmus; GL, glomerular layer; GRL, granular layer of the olfactory bulb; L/RAtr, left/right atrium of the heart; Nct, neural crest; ne, neuroepithel; RMS, rostral migratory stream; Stom, stomach; Ventr, ventricle of the heart. Scale bar = 500 μm (A), 200 μm (C), 70 μm (B), 20 μm (B1).
The first attempt to demonstrate secretagogin expression in the human brain was through histopathological examination of post-mortem tissues [59,78], and led to a secretagogin “proto-map” including immunoreactive structures in the temporal cortex, basal ganglia, hippocampus, hypothalamus and cerebellum. Detailed neurochemical and neuroanatomy studies in mouse [51] showed secretagogin mRNA and protein at varying levels in the granular and periglomerular layers of the main olfactory bulb, non-cholinergic cells in the medial septum, granule cells in the anterior hippocampal continuation (including the dentate gyrus, indusium griseum and tenia tecta) along the corticolimbic axis, locus coeruleus, and cerebellum (Fig. 1). The overall conclusion based on presently available localization data [51] is that i) secretagogin-positive cells are invariably neurons. ii) Secretagogin's expression pattern complements that of PV. Yet secretagogin has been found associated with CR in olfactory periglomerular cells in mouse [51] and human (J.A., A.A., T.H., unpublished data). More infrequent is the co-localization of secretagogin and CB, e.g. in the ventral pallidal territory, which still contains appreciable quantities of dual-labelled neurons [51]. iii) New insights in secretagogin expression might help resolving the origins of particular cell assemblies in the brain, like the anterior hippocampal continuation whose olfactory vs. hippocampal origins [79,80] are still debated. Based on their secretagogin immunoreactivity and electrophysiological characteristics it seems that a substantial proportion of neurons in the indusium griseum might share developmental origins with granule cells of the dentate gyrus [51].
Developmental studies have recently addressed whether secretagogin is expressed in the foetal brain. The long-held view is that CBPs are late markers of postmitotic GABA cells in both the mouse forebrain [81]: CB-positive(+) pioneer neurons populate the cerebral cortex by embryonic day (E) 14 in mouse [82], and are also present in human fetal brain by week 14 of pregnancy [83]. CR-expressing neurons invade the developing cerebrum by mid-gestation in both rodents and human [84,85]. While PV first appears at E13 in the sensory ganglia and ascending pathways in the spinal cord, the onset of PV expression in forebrain GABAergic neurons is restricted to the first postnatal week [20,86], except in human telencephalon where PV-immunoreactive Cajal-Retzius cells were noted by gestational weeks 20-24 [84]. In the mouse forebrain, secretagogin emerges as early as E11 in the anterior wall of the cerebral vesicle, i.e. the prospective olfactory bulb and in the subpial area of the ganglionic eminence [52]. A day later, these cells transit the differentiation zone and were identified as post-mitotic, non-pyramidal neurons at the subpial surface (Fig. 2). Neurons from this pool then migrate either to the olfactory bulb or the subpallium caudally to commit neurons to the extended amygdala, where they can acquire neurochemical characteristics of cholinergic or GABAergic neurons. Since postmitotic neurons express secretagogin, it is not surprising that they retain expression of this CBP postnatally [51,52]. Accordingly, secretagogin+ neurons populate the bed nucleus of the stria terminals, the interstitial nucleus of the posterior limb of the anterior commissure, ventral pallidum, the dorsal substantia innominate and the central and medial amygdaloid nuclei in neonates and adults.
Another key site of secretagogin expression is the mammalian retina, where it is seen in well-defined subtypes of cone, but not rod, bipolar cells with prominent labelling of dendrites, perinuclear cytoplasm, axon, and axon terminals [69]. Secretagogin mRNA and protein expression exhibits rapid developmental onset during postnatal days 4-6 in mouse [87], preceding eye opening and the critical period of cortical plasticity during which sensory neuronal circuitries undergo robust activity-dependent refinement. This, together with the temporal expression pattern of the developing mouse retina OFF and ON bipolar cells suggests secretagogin's contribution to synapse maturation and functionality [69].
Secretagogin's distribution in the nervous system may suggest the following inferences: i) If secretagogin effectively contributes to shaping [Ca2+]i then it may affect the excitability of secretagogin+ neurons. This notion forecasts identification of a “secretagogin-containing interneuron” subtype amongst GABA cells. ii) Secretagogin's developmental expression [52], together with its retained presence in new-born neurons during long-range migration in the adult brain [51] suggest a role in defining cellular motility, or repression of Ca2+-regulated mechanisms of morphological differentiation (that is, to keep cells in migratory streams). Therefore, perturbed secretagogin expression may precipitate congenital disorders associated with cell misplacement or aberrant patterning of neuronal network connectivity. iii) Secretagogin-expressing cell groups often align the brain's ventricular system with dendrites positioned along the pial surfaces (e.g., indusium griseum, tenia tecta, hypothalamus) [51,52,78]. If secretagogin is indeed a Ca2+ sensor in vivo then it could trigger the release of neuroactive substances from neurons into the cerebrospinal fluid (CSF). This hypothesis is supported by circulating secretagogin in the CSF [78]. iv) Secretagogin is found at high concentrations in heart muscle, with its blood levels elevated after cardiac ischemia or stroke [88,89]. Therefore, secretagogin's presence in the circulation may be secondary to cell death and lysis or represent the regulated release of a bioactive substance to trigger (neuro-) protective responses.
4.1 Phylogenetic variations
EF-hand CBPs exhibit a high degree of sequence homology: secretagogin's sequence is in 37% and 38% identical to CB and CR, respectively. AA sequences of zebra fish (Danio rerio), rat, mouse, primate or human secretagogin are highly homologous (Danio rerio's secretagogin is 73% identical to human secretagogin) with considerable variations within its N-terminal, resulting in the loss of two EF-hand motifs in rat secretagogin [90]. This evolutionarily conserved homology is also reflected by secretagogin's tissue expression patterns with the pancreas, adrenal glands and neuroendocrine cells consistently immunoreactive for secretagogin [90].
Despite the above predictions, detailed neuroanatomical analyses have uncovered critical differences amongst otherwise seemingly similar subsets of telencephalic neurons in different species: i) in the rostral migratory stream, where adult-born neuroblasts migrate towards the olfactory bulb, secretagogin identifies neuroblasts in primates. In mice, however, secretagogin immunoreactivity is largely to post-migratory granular and periglomerular olfactory bulb neurons [51]. ii) A similar tendency of secretagogin to label non-differentiated neurons was seen in the primate dentate gyrus where even new-born granule cells, positioned in deep cell layers adjacent to the neurogenic subgranular zone [91], were secretagogin+. In contrast, only mature granule cells facing the molecular layer of the dentate gyrus contain secretagogin in mouse (Fig. 1D). In primate or human hippocampus, morphologically-diverse secretagogin+ cells include bistratified-like cells, interneurons, as well as pyramidal cells [51,54]. iii) In spite of gross similarities in the positioning and extent of the anterior hippocampal formation, neurons of the ventral tenia tecta showed secretagogin immunoreactivity only in primates but not in rodents. iv) In stark contrast with the mouse basal forebrain, secretagogin co-exists in the vast majority of choline acetyltransferase immunopositive projection and interneurons in primates [51,52]. Instead, secretagogin+ neurons are GABAergic with spiny dendritic tufts in both the caudatoputamen complex and the ventral pallidal region of the mouse.
Phylogenetic differences across mammalian species are apparent already during embryonic development. In primates, the early cholinergic lineage commitment associated with a selective up-regulation of secretagogin expression is a striking characteristics of higher-order mammals [52]. In addition, and in contrast to rodents, the density of secretagogin+ neurons colonizing amygdaloid nuclei is relatively low in primates.
Evolutionary variations in secretagogin expression are surprising, particularly since the cellular identity of PV, CB and CR immunoreactive neurons, at least in the corticolimbic system, is phylogenetically preserved [13]. The significance of species-specific variations of secretagogin expression in developing and adult neurons is unknown. We hypothesize that secretagogin's distribution relate to the increased complexity of brain structure – underpinning higher cognitive performance – in primates and humans. For example, the appearance of secretagogin expression in forebrain cholinergic projection neurons may be a hallmark of refined cholinergic neurotransmission and more precise integration of cholinergic afferents into cortical circuitries through the increased temporal precision of neurotransmitter release via modulating Ca2+ signalling both at cholinergic presynapses, as well as the somatodendritic axis of cholinergic neurons.
4.2 Secretagogin functions in the nervous system and beyond
Scientific evolution dictates that any new field of scientific endeavour is first concerned with the localization and followed by the functional characterisation of its target. Accordingly, presently available data are inconclusive to define secretagogin's cellular role(s). It remains critical to establish whether secretagogin is a “buffer” or “sensor” in living organisms, and whether it recruits specific target(s) to trigger signalling cascades. In neurons, new understanding will be imperative to characterize whether secretagogin predominantly affects the somatodendritic compartment by stabilizing cytosolic [Ca2+]i or Ca2+-dependent neurotransmitter release. “Is secretagogin in CSF or blood (see below) active or merely a by-product of cellular injury or turnover?” Although secretagogin's presence in the blood has repeatedly been demonstrated, our view is curtailed by the lack of primary mechanistic experimental data. Yet secretagogin's localization in neuroendocrine cells and exocrine organs [59,78] is suggestive of activity-dependent secretagogin release. Elevated blood secretagogin levels after brain or cardiac tissue damage lend further support to this argument [88,89]. Nevertheless, we at present cannot rule out the passive, damage-induced release of the protein. Deciphering secretagogin's (patho-)physiological significance clearly dictates the generation of new experimental models, including – but not restricted to - pharmacological and transgenic in vivo technologies, intravital imaging of tagged secretagogin and large-scale interactome analyses in living cells. It will be pertinent to determine the molecular cascade underpinning secretagogin release, the receptor(s) of secreted CBPs, and their putative sites of action to fully understand the functional role, as well as to appreciate the diagnostic significance of altered secretagogin expression and cellular localization under pathological conditions. Future investment into methodological development might reward investigators by identifying novel modes of intercellular communication, including neuron-neuron, neuron-glia or glia-glia (including microglia oligodendroglia and astroglia) communication, through regulated secretagogin release. These considerations will also be critical to exploit the clinical potential of this protein (see below).
5. Secretagogin: a dynamically released Ca2+-binding protein in cancer?
Disease diagnosis - often through retrospective analysis - includes histopathological examination and blood testing. Considering its tissue expression profile (see above), secretagogin received interest as an emerging biomarker with particular promise for the differential diagnosis of gastrointestinal, gonadal and brain tumours [70,71,75,92,93], as well as acute ischemic stroke [88,89].
Tumorigenesis is associated with significant changes in secretagogin expression at both the mRNA and protein levels [94]. For example, while secretagogin is physiologically expressed in neuroendocrine cells situated in the basal layer of the colorectal mucosa [71], its protein level becomes reduced (2 to 4 fold) in benign hyperplastic-metaplastic polyps, tubular and tubulovillous adenomas of the colon [71]. In colorectal carcinomas, remnant secretagogin expression (>10 fold decrease in protein as well as mRNA) is confined to proliferating cells with malignant morphology [93]. Similarly, secretagogin is expressed in the anterior pituitary gland [78,95]. Yet both secretagogin (2.2-6.9 fold) protein and mRNA (1.8-18.6 fold) were found downregulated in non-functional pituitary adenomas [94] with protein and mRNA levels exhibiting a strong - whether positive or negative - relationship. These findings were confirmed and extended [92] by showing secretagogin expression in a subset of hormone (ACTH, prolactin and growth hormone)-producing pituitary adenomas.
Secretagogin has also been found expressed with high probability in many neuronal tumours (neurocytomas, neuroblastomas, medulloblastomas), while lower abundance of secretagogin+ cells was encountered in oligodendrogliomas, ependymomas, and meningoblastomas [92]. These data suggest that secretagogin levels undergo significant changes in tumour cells relative to their non-neoplastic cell analogues, allowing the use of secretagogin as a marker of cell differentiation and transformation.
The regulated release of a signalling molecule from a given cell implicates the secreted substance in sculpting intercellular communication with discrete temporal and spatial constraints. CBPs can be released from tumour cells, immortalized cell lines or neurons to underpin pathologic or non-physiological modes of intercellular communication. Although not typical, the release of other CBPs is an acknowledged phenomenon. Amongst the “classical” neuron-associated CBPs, secreted α and β paralbumins [96] and calbindins [97] have been found to impact predatory behaviours and venom secretion in lower vertebrates. In particular, α and β paralbumins from frog skin mucus function as unexpectedly efficacious chemo-attractants, and elicit predatory attack responses in snakes [96]. S100 (its name derived from soluble in ammonium sulphate at neutral pH) is another example of secreted cytosolic CBPs: in the brain, S100 can be released from astrocytes to modulate neuronal differentiation and axonal growth responses [24,98], cell survival [25,99] and synaptic plasticity [100]. Both S100α and S100β isoforms can be released, particularly during the rapid proliferation phase of cultured cells [101]. Notably, the plasma concentration of released S100β is considered as a prognostic marker for melanoma cells [102]. Similarly, elevated serum concentrations of secreted CR, and its splice variant (calretinin 22k; Table 1) have been detected in colorectal cancer patients [19].
Multiple levels of pre-clinical and clinic-pathological evidence suggest that secretagogin may be released from various tissues upon cytotoxic injury [60,88,92,103]. Constitutively released secretagogin was initially recovered from a variety of stably transfected tumour-derived cell lines under resting conditions [59]. In addition, immunogenic stimuli can facilitate secretagogin release from, e.g., Jurkat cells [59]. Secretagogin can also be detected in the plasma of patients (8-1,800 pg/ml by means of sandwich ELISA) with metastasising carcinoid tumours [71]. Notably, Gartner et al. [104] have recently reported the presence of elevated secretagogin levels (1,424 – 2,436 pg/ml) in the plasma of two patients one year prior to their diagnosis with aggressive malignant glioma. This finding could reinforce secretagogin's potential as a prospective cancer biomarker. However, these patients were diagnosed with either glioblastoma multiforme or anaplastic astrocytoma. In contrast, large-scale histopathological profiling excluded the association of secretagogin expression in astrocytoma, while only revealed sporadic changes in glioblastomas [92]. This paradox may be resolved by either assuming that tissue-infiltrating gliomas might impact the structural integrity of secretagogin-expressing territories in the human brain or tumour-related disruption of the blood-brain barrier can facilitate secretagogin extravasation into the systematic circulation.
Secretagogin expression may stochastically change in cancerous non-neural systems, compromising its prognostic value. Thus, although secretagogin has been identified in human prostate, no changes could be traced in blood samples or immunohistochemically in adenocarcinomas [75]. In the gastrointestinal system, in contrast to a marked down-regulation of secretagogin expression in most colorectal cancerous tissues, some colorectal cancers were characterized by secretagogin-expressing cells [93]. These cells showed malignant morphology which queries a “protective role” for secretagogin, seen in other diseases [54]. In the urinary system in turn, secretagogin levels are not down- but up-regulated in renal cell carcinomas as shown by peptide mass fingerprinting [105]. This finding was clinic-pathologically rationalized by Ilhan et al. [106] who showed a correlation between the secretagogin level and incidence of distant metastases in clear-cell renal carcinomas. Similarly, a fivefold increase in secretagogin protein level was shown in an animal model with cancer bone invasion that could be reversed by radiation [107]. Cumulatively, these data suggest that secretagogin localization in both specific organ systems and serum may be of prognostic value, particularly since it correlates closely with the stage, grade, progression and metastases of cancer. Nevertheless, opposite and potentially contradictory changes in secretagogin levels in different tumour histories make further advances indispensable to clarify general vs. organ- or tumour-specific mechanisms.
5.1. Secretagogin release: at best a surrogate marker in stroke?
Stroke is a life-threatening condition when blood supply to the brain, in whole or in part, is acutely disrupted by obstruction of its vasculature, precluding minimally sufficient blood supply [108]. Acute haemorrhagic stroke is associated with plasma extravasation or bleeding due to blood-tissue barrier damage or the physical rupture of blood vessels. Clinically, acute stroke manifests as a sudden focal neurological deficit of presumed vascular origin [88]. A major challenge in stroke therapy is unequivocal and rapid differential diagnosis since disease prognosis and recovery expectations progressively worsen within the first 24 hours. Therefore, it is of clinical benefit to characterize reliable predictive biomarkers [109].
First, Gartner et al. [78] have suggested secretagogin as a candidate (yet delayed) biomarker of neuronal damage since this CBP displayed elevated plasma levels in patients days after hypoxic brain injury or stroke. Subsequently, secretagogin, as a putative “secreted” protein, has been assayed in two consecutive studies with progressively increasing patient cohorts [88,89]. Secretagogin has been confirmed as a protein detectable in plasma shortly after stroke. However, its levels were initially reported as being constant – irrespective of the type of stroke (e.g., cardioembolic or atherothrombotic aetiology) in a cohort of 707 subjects – unless diabetes was assigned as a predisposing variable [89]. Yet significant association of six biomarkers, including secretagogin, was demonstrated upon increasing the population size (to 915 cases), and by testing stroke cases against stroke-mimicking conditions [88]. Although statistically significant, the magnitude of difference between plasma secretagogin concentrations of stroke (median: 150 ng/ml; range: 70-320 ng/ml) vs. stroke-mimicking patient groups (median: 190 ng/ml; range: 100-490 ng/ml) was limited, relative to e.g., caspase-3. Notably, plasma secretagogin levels exhibited pronounced association with age, smoking, dislipidemia and myocardiopathy. Hypothetically, since secretagogin is expressed in the heart [52], this latter parameter may be reflective of either an active, or a damage-induced passive secretagogin release from the myocardium under chronically hypoxic conditions. It is noteworthy though that data on a relationship between secretagogin and atrial fibrillation-associated silent white matter lesions, without presenting acute neurological deficits, failed to associate secretagogin in plasma (∼106 pg/ml (disease) vs. ∼76 pg/ml (control)) with either atrial deficits or lesion score over a sample size of 222 patients [103]. Overall, these data suggest that secretagogin is a poor stroke biomarker - relative to D-dimer, caspase-3, S100β, matrix metalloproteases, brain natriuretic peptide -, unless specific conditions with massive infarcts probably resulting in secretagogin “leaking” from damaged neurons are considered. It is therefore conspicuous whether secretagogin may be classified as a putative marker of “neuronal death”, particularly since secretagogin distribution in the human brain is confined to neurogenic and secretory niches and cell lineages [52-54,77] (L. Wagner, personal communication), precluding the use of this CBP as a ubiquitous neuronal marker.
“Does secretagogin in plasma or interstitial extracellular fluid exert any specific function?” This remains one of the key questions for future studies. Based on the data discussed above, one may predict that secretagogin release is a secondary, passive consequence of cellular destruction and damage in the nervous system. In contrast, recent advances in the study of secretagogin in cancer might suggest otherwise. Highly (more than 10-fold) increased serum secretagogin levels may reflect homeostatic responses to control cell proliferation, and differentiation. This might be in line with the similar intracellular effect of secretagogin, i.e. an impact upon growth control and differentiation [59,71]. As such, a molecular mechanism reminiscent of S100β's action profile may be a tempting hypothesis.
6. Aging and neurodegeneration: Ca2+ sensor meets Ca2+ buffer?
Impaired Ca2+ homeostasis has been broadly implicated in the aetiology of neurodegenerative diseases [7,27,110-113]. Therefore, CBPs attracted substantial attention, particularly in AD, the most frequent form of dementia [7,27,110-113]. PV and CR were indicated to be neuroprotective in AD although their mechanism of action remains elusive [114,115].
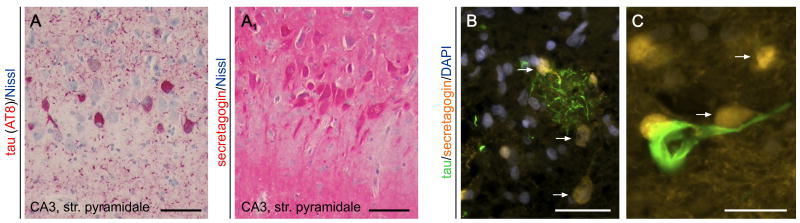
Recently, a series of studies have addressed secretagogin's cellular localization in the hippocampal formation of AD subjects [53,54], as well as experimental models [77]. Secretagogin immunoreactivity in aged humans is present in the hippocampal formation including the cornu ammonis (CA) subfields and subiculum [54]. Neuronal expression was restricted to pyramidal cell-like neurons, as suggested by their characteristic cytoarchitecture. In contrast, somatic secretagogin immunoreactivity was not found in either the entorhinal cortex or dentate gyrus. Instead, secretagogin+ neuropil was seen, reminiscent of a dense meshwork of fine calibre processes. Quantitative analyses using age, gender or dementia stage as co-variables showed retained secretagogin expression in all CA subfields in AD [54]. Subsequent morphometric analysis demonstrated that secretagogin-expressing neurons were spared of hyperphosphorylated tau pathology (e.g. neurofibrillary tangles, a hallmark of AD pathology [116]), as the percentage of secretagogin or tau immunoreactive somata showed no correlation, and co-localization was sporadic in any CA subfield [53] (Fig. 3). An appealing interpretation of these findings is that secretagogin+ expressing neurons are largely resistant to neurodegeneration in AD.
Fig. 3. Secretagogin distribution in Alzheimer's disease.
(A) Hyperphosphorylated tau immunoreactivity (AT8 staining, in red) in the CA3 subfield of the human hippocampus in Alzheimer's. (A1) Secretagogin immunoreactivity in a consecutive (adjacent) section decorates neurons whose distribution is complementary to AT8+ cells in (A). Nissl stain was used as counterstain (in blue). (B) Secretagogin+/AT8-neurons (arrows) in the vicinity of AT8+ dystrophic neurites accumulating in a putative neuritic plaque. Note the mutually exclusive patterns of immunoreactivity. (C) High-power image of a hyperphosphorylated tau-laden neuron surrounded by secretagogin+ cells (arrows). Scale bar = 100 μm (AA1). 50 μm (B), 20 μm (C).
However, these histopathological findings in human brain tissue may be challenged by data from transgenic models of tau pathology (P301L mice) [77], displaying widespread tau accumulation in the hippocampus and neocortex. Although regions devoid of tau expression, i.e. bed nucleus of stria terminalis, showed unaltered secretagogin immunoreactivity, greatly diminished neuronal secretagogin labelling was reported in the hippocampus affected by tau over-expression. Yet, and similar to findings in human brain, cellular co-localization of secretagogin and tau was not observed in the P301L hippocampus [77]. Data from the above lines of research might be reconciled by suggesting that tau overexpression exceeding a critical threshold will ultimately impair neuronal integrity such that secretagogin expression becomes down-regulated. The eventual interdependency of secretagogin and tau has been further strengthened by data from insulin-secreting cells [117]: association of secretagogin and the tau's 4R isoform were demonstrated by pull-down assays and immunohistochemistry, fuelling the hypothesis that secretagogin might participate in the Ca2+-dependent fine-tuning of microtubule dynamics [117]. Nevertheless, and without conclusive evidence on secretagogin's cellular function(s), the role of this CBP in neurodegeneration ultimately remains elusive.
7. Concluding remarks
Since its identification ∼10 years ago, secretagogin received attention from research groups working on tumour monitoring, medical endocrinology, pathology and neurobiology. These studies have clearly achieved milestone results by interrogating from different aspects of fundamental and clinical (applied) biology. Yet secretagogin has thus far attracted surprisingly low interest from the scientific community, failed to enter the textbooks and become part of mainstream research. This is probably because secretagogin-related studies appear fragmented and critical proof-of-concept experiments have evaded contemporary investigations. To change the landscape of appreciation, two fundamental points need urgent clarification: In which cellular or intercellular compartment secretagogin functions as a “pure” Ca2+ buffer, sensor or dual-action protein? What is its precise mode of action? To achieve tangible clinical benefit, secretagogin's role in tumour pathology should be clarified to prevent further contradictions, as well as to assert a causal relationship between tumour growth and secretagogin's expressional regulation. Similarly, future milestones in neurobiology will be to conclusively establish secretagogin's cellular function, providing much-needed information for investigations in its role in processes of neuronal specification, synaptic communication and survival under disease conditions including AD, epilepsy or pain. If secretagogin truly is “the fourth musketeer of neuronal CBPs” (M.R. Celio, http://ejnblog.wordpress.com/2010/07/03/secretagogin-the-fourth-'musketeer/) then this protein has tremendous discoveries on offer, most of which just have started to emerge.
Highlights.
Secretagogin can serve as Ca2+ buffer as well as sensor in neurons.
Secretagogin expression is developmentally and evolutionary regulated.
Secretagogin can regulate vesicular exocytosis through protein interaction networks.
Secretagogin-expressing neurons are unharmed in Alzheimer's disease.
Secretagogin emerges as a diagnostic marker in tumour biology.
Acknowledgments
The authors thank M. Uhlén and L. Wagner for making polyclonal anti-secretagogin antibodies available to the morphological studies cited and Y. Yanagawa for glutamic acid decarboxylase 67 kDa isoform (GAD67)-EGFP mice. This work was supported by the Scottish Universities Life Science Alliance (SULSA; A.A., T.Ha.), European Commission (HEALTH-F2-2007-201159; T.Ha.), National Institutes of Health (DA023214, T.Ha.), and the Swedish Research Council (T.Hö., T.Ha.).
List of non-standard abbreviations
- AA
amino acid
- AD
Alzheimer's disease
- ARFGAP2/3
ADP-ribosylation factor GTPase-activating protein 2/3
- bp
base pair
- CA
cornu ammonis subfields of the hippocampus
- [Ca2+]i
intracellular Ca2+ concentration
- CB
calbindin D28k
- CBP
Ca2+-binding protein
- CNS
central nervous system
- CR
calretinin
- CSF
cerebrospinal fluid
- DDAH-2
N,N-dimethylarginine dimethylaminohydrolase 2
- DOC2α
double C2-like domain-containing protein α
- E
embryonic day
- GAD
glutamic acid decarboxylase
- Kd
equilibrium dissociation constant
- ORF
open reading frame
- PV
parvalbumin
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- SNAP-25
soluble N-ethylmaleimide-sensitive factor attachment protein of 25 kDa
- SNAP-23
soluble N-ethylmaleimide-sensitive factor attachment protein of 23 kDa
- SNP
single nucleotide polymorphism
Footnotes
Conflict of interest statement: The Authors declare no conflict of interest.
Author contributions: All Authors have participated in preparing this manuscript for publication, and approved its submission to Cellular Signalling.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clapham DE. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Burnashev N, Rozov A. Cell Calcium. 2005;37:489–495. doi: 10.1016/j.ceca.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Pang ZP, Sudhof TC. Curr Opin Cell Biol. 2010;22:496–505. doi: 10.1016/j.ceb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipton SA, Nicotera P. Cell Calcium. 1998;23:165–171. doi: 10.1016/s0143-4160(98)90115-4. [DOI] [PubMed] [Google Scholar]
- 5.Lipton SA, Rosenberg PA. N Engl J Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- 6.Lory P, Mezghrani A. IDrugs. 2010;13:467–471. [PubMed] [Google Scholar]
- 7.Mattson MP, Cheng B, Davis D, Bryant K, Lieberburg I, Rydel RE. J Neurosci. 1992;12:376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toescu EC. Cell Calcium. 2004;36:187–199. doi: 10.1016/j.ceca.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Ohta K, Graf R, Rosner G, Heiss WD. Stroke. 2001;32:535–543. doi: 10.1161/01.str.32.2.535. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Ari Y. Epilepsia. 2001;42 3:5–7. doi: 10.1046/j.1528-1157.2001.042suppl.3005.x. [DOI] [PubMed] [Google Scholar]
- 11.Strehler EE, Filoteo AG, Penniston JT, Caride AJ. Biochem Soc Trans. 2007;35:919–922. doi: 10.1042/BST0350919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholls DG. Biochim Biophys Acta. 2009;1787:1416–1424. doi: 10.1016/j.bbabio.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andressen C, Blumcke I, Celio MR. Cell Tissue Res. 1993;271:181–208. doi: 10.1007/BF00318606. [DOI] [PubMed] [Google Scholar]
- 14.Kretsinger RH. Ann N Y Acad Sci. 1980;356:14–19. doi: 10.1111/j.1749-6632.1980.tb29594.x. [DOI] [PubMed] [Google Scholar]
- 15.Kretsinger RH. CRC Crit Rev Biochem. 1980;8:119–174. doi: 10.3109/10409238009105467. [DOI] [PubMed] [Google Scholar]
- 16.Baimbridge KG, Miller JJ. Brain Research. 1982;245:223–229. doi: 10.1016/0006-8993(82)90804-6. [DOI] [PubMed] [Google Scholar]
- 17.Celio MR. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- 18.Blumcke I, Hof PR, Morrison JH, Celio MR. J Comp Neurol. 1990;301:417–432. doi: 10.1002/cne.903010307. [DOI] [PubMed] [Google Scholar]
- 19.Schwaller B, Meyer-Monard S, Gander JC, Pugin P, Celio MR, Ludwig C. Anticancer Res. 1998;18:3661–3667. [PubMed] [Google Scholar]
- 20.Solbach S, Celio MR. Anat Embryol (Berl) 1991;184:103–124. doi: 10.1007/BF00942742. [DOI] [PubMed] [Google Scholar]
- 21.Heizmann CW. J Cardiovasc Pharmacol. 1986;8 8:S7–12. [PubMed] [Google Scholar]
- 22.Skelton NJ, Kordel J, Akke M, Forsen S, Chazin WJ. Nat Struct Biol. 1994;1:239–245. doi: 10.1038/nsb0494-239. [DOI] [PubMed] [Google Scholar]
- 23.Gauthier BR, Wollheim CB. Am J Physiol Endocrinol Metab. 2008;295:E1279–E1286. doi: 10.1152/ajpendo.90568.2008. [DOI] [PubMed] [Google Scholar]
- 24.Whitaker-Azmitia PM, Wingate M, Borella A, Gerlai R, Roder J, Azmitia EC. Brain Res. 1997;776:51–60. doi: 10.1016/s0006-8993(97)01002-0. [DOI] [PubMed] [Google Scholar]
- 25.Winningham-Major F, Staecker JL, Barger SW, Coats S, Van Eldik LJ. J Cell Biol. 1989;109:3063–3071. doi: 10.1083/jcb.109.6.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCue HV, Haynes LP, Burgoyne RD. Cold Spring Harb Perspect Biol. 2010;2:a004085. doi: 10.1101/cshperspect.a004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braunewell KH. Trends Pharmacol Sci. 2005;26:345–351. doi: 10.1016/j.tips.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Klausberger T, Somogyi P. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freund TF, Buzsaki G. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 30.Caillard O, Moreno H, Schwaller B, Llano I, Celio MR, Marty A. Proc Natl Acad Sci U S A. 2000;97:13372–13377. doi: 10.1073/pnas.230362997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collin T, Chat M, Lucas MG, Moreno H, Racay P, Schwaller B, Marty A, Llano I. J Neurosci. 2005;25:96–107. doi: 10.1523/JNEUROSCI.3748-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cauli B, Audinat E, Lambolez B, Angulo MC, Ropert N, Tsuzuki K, Hestrin S, Rossier J. J Neurosci. 1997;17:3894–3906. doi: 10.1523/JNEUROSCI.17-10-03894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaguchi Y, Kubota Y. Neuroscience. 1998;85:677–701. doi: 10.1016/s0306-4522(97)00685-4. [DOI] [PubMed] [Google Scholar]
- 34.Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, Fairen A, Feldmeyer D, Fishell G, Fregnac Y, Freund TF, Gardner D, Gardner EP, Goldberg JH, Helmstaedter M, Hestrin S, Karube F, Kisvarday ZF, Lambolez B, Lewis DA, Marin O, Markram H, Munoz A, Packer A, Petersen CC, Rockland KS, Rossier J, Rudy B, Somogyi P, Staiger JF, Tamas G, Thomson AM, Toledo-Rodriguez M, Wang Y, West DC, Yuste R. Nat Rev Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwaller B. Cold Spring Harb Perspect Biol. 2010;2:a004051. doi: 10.1101/cshperspect.a004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwaller B. Cell Mol Life Sci. 2009;66:275–300. doi: 10.1007/s00018-008-8564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christakos S, Dhawan P, Peng X, Obukhov AG, Nowycky MC, Benn BS, Zhong Y, Liu Y, Shen Q. J Steroid Biochem Mol Biol. 2007;103:405–410. doi: 10.1016/j.jsbmb.2006.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambers TT, Mahieu F, Oancea E, Hoofd L, de LF, Mensenkamp AR, Voets T, Nilius B, Clapham DE, Hoenderop JG, Bindels RJ. EMBO J. 2006;25:2978–2988. doi: 10.1038/sj.emboj.7601186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan DW, Welton AF, Heick AE, Christakos S. Biochem Biophys Res Commun. 1986;138:547–553. doi: 10.1016/s0006-291x(86)80531-9. [DOI] [PubMed] [Google Scholar]
- 40.Reisner PD, Christakos S, Vanaman TC. FEBS Lett. 1992;297:127–131. doi: 10.1016/0014-5793(92)80342-e. [DOI] [PubMed] [Google Scholar]
- 41.Bellido T, Huening M, Raval-Pandya M, Manolagas SC, Christakos S. J Biol Chem. 2000;275:26328–26332. doi: 10.1074/jbc.M003600200. [DOI] [PubMed] [Google Scholar]
- 42.Lutz W, Frank EM, Craig TA, Thompson R, Venters RA, Kojetin D, Cavanagh J, Kumar R. Biochem Biophys Res Commun. 2003;303:1186–1192. doi: 10.1016/s0006-291x(03)00499-6. [DOI] [PubMed] [Google Scholar]
- 43.Billing-Marczak K, Kuznicki J. Pol J Pharmacol. 1999;51:173–178. [PubMed] [Google Scholar]
- 44.Farre-Castany MA, Schwaller B, Gregory P, Barski J, Mariethoz C, Eriksson JL, Tetko IV, Wolfer D, Celio MR, Schmutz I, Albrecht U, Villa AE. Behav Brain Res. 2007;178:250–261. doi: 10.1016/j.bbr.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Schwaller B, Tetko IV, Tandon P, Silveira DC, Vreugdenhil M, Henzi T, Potier MC, Celio MR, Villa AE. Mol Cell Neurosci. 2004;25:650–663. doi: 10.1016/j.mcn.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Moreno H, Burghardt NS, Vela-Duarte D, Masciotti J, Hua F, Fenton AA, Schwaller B, Small SA. Hippocampus. 2011 doi: 10.1002/hipo.20957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bearzatto B, Servais L, Roussel C, Gall D, Baba-Aissa F, Schurmans S, de Kerchove dA, Cheron G, Schiffmann SN. FASEB J. 2006;20:380–382. doi: 10.1096/fj.05-3785fje. [DOI] [PubMed] [Google Scholar]
- 48.del Rio MR, Defelipe J. J Chem Neuroanat. 1997;12:165–173. doi: 10.1016/s0891-0618(96)00191-3. [DOI] [PubMed] [Google Scholar]
- 49.Wouterlood FG, Grosche J, Hartig W. Brain Res. 2001;922:310–314. doi: 10.1016/s0006-8993(01)03220-6. [DOI] [PubMed] [Google Scholar]
- 50.Csillik B, Schwaller B, Mihaly A, Henzi T, Losonczi E, Knyihar-Csillik E. Neuroscience. 2010;165:749–757. doi: 10.1016/j.neuroscience.2009.10.048. [DOI] [PubMed] [Google Scholar]
- 51.Mulder J, Zilberter M, Spence L, Tortoriello G, Uhlen M, Yanagawa Y, Aujard F, Hokfelt T, Harkany T. Proc Natl Acad Sci U S A. 2009;106:22492–22497. doi: 10.1073/pnas.0912484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulder J, Spence L, Tortoriello G, Dinieri JA, Uhlen M, Shui B, Kotlikoff MI, Yanagawa Y, Aujard F, Hokfelt T, Hurd YL, Harkany T. Eur J Neurosci. 2010;31:2166–2177. doi: 10.1111/j.1460-9568.2010.07275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Attems J, Preusser M, Grosinger-Quass M, Wagner L, Lintner F, Jellinger K. Neuropathol Appl Neurobiol. 2008;34:23–32. doi: 10.1111/j.1365-2990.2007.00854.x. [DOI] [PubMed] [Google Scholar]
- 54.Attems J, Quass M, Gartner W, Nabokikh A, Wagner L, Steurer S, Arbes S, Lintner F, Jellinger K. Exp Gerontol. 2007;42:215–222. doi: 10.1016/j.exger.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 55.Rogstam A, Linse S, Lindqvist A, James P, Wagner L, Berggard T. Biochem J. 2007;401:353–363. doi: 10.1042/BJ20060918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bitto E, Bingman CA, Bittova L, Frederick RO, Fox BG, Phillips GN., Jr Proteins. 2009;76:477–483. doi: 10.1002/prot.22362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bauer MC, O'Connell DJ, Maj M, Wagner L, Cahill DJ, Linse S. Mol Biosyst. 2011;7:2196–2204. doi: 10.1039/c0mb00349b. [DOI] [PubMed] [Google Scholar]
- 58.Prentki M, Matschinsky FM. Physiol Rev. 1987;67:1185–1248. doi: 10.1152/physrev.1987.67.4.1185. [DOI] [PubMed] [Google Scholar]
- 59.Wagner L, Oliyarnyk O, Gartner W, Nowotny P, Groeger M, Kaserer K, Waldhausl W, Pasternack MS. J Biol Chem. 2000;275:24740–24751. doi: 10.1074/jbc.M001974200. [DOI] [PubMed] [Google Scholar]
- 60.Wagner L, Templ E, Reining G, Base W, Weissel M, Nowotny P, Kaserer K, Waldhausl W. J Endocrinol. 1998;156:469–476. doi: 10.1677/joe.0.1560469. [DOI] [PubMed] [Google Scholar]
- 61.Skovhus KV, Bergholdt R, Erichsen C, Sparre T, Nerup J, Karlsen AE, Pociot F. Scand J Immunol. 2006;64:639–645. doi: 10.1111/j.1365-3083.2006.01854.x. [DOI] [PubMed] [Google Scholar]
- 62.Kiyota T, Kato A, Altmann CR, Kato Y. Dev Biol. 2008;315:579–592. doi: 10.1016/j.ydbio.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 63.Liu L, Li Y, Van Eldik LJ, Griffin WS, Barger SW. J Neurochem. 2005;92:546–553. doi: 10.1111/j.1471-4159.2004.02909.x. [DOI] [PubMed] [Google Scholar]
- 64.Citron BA, Dennis JS, Zeitlin RS, Echeverria V. J Neurosci Res. 2008;86:2499–2504. doi: 10.1002/jnr.21695. [DOI] [PubMed] [Google Scholar]
- 65.Adwan LI, Basha R, Abdelrahim M, Subaiea GM, Zawia NH. Curr Alzheimer Res. 2011 doi: 10.2174/156720511795745285. [DOI] [PubMed] [Google Scholar]
- 66.Heald B, Hilden JM, Zbuk K, Norton A, Vyas P, Theil KS, Eng C. Nat Clin Pract Oncol. 2007;4:433–438. doi: 10.1038/ncponc0876. [DOI] [PubMed] [Google Scholar]
- 67.Hota SK, Hota KB, Prasad D, Ilavazhagan G, Singh SB. Free Radic Biol Med. 2010;49:178–191. doi: 10.1016/j.freeradbiomed.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 68.Zierhut B, Daneva T, Gartner W, Brunnmaier B, Mineva I, Berggard T, Wagner L. Biochem Biophys Res Commun. 2005;329:1193–1199. doi: 10.1016/j.bbrc.2005.02.093. [DOI] [PubMed] [Google Scholar]
- 69.Puthussery T, Gayet-Primo J, Taylor WR. J Comp Neurol. 2010;518:513–525. doi: 10.1002/cne.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai M, Lu B, Xing X, Xu E, Ren G, Huang Q. Virchows Arch. 2006;449:402–409. doi: 10.1007/s00428-006-0263-9. [DOI] [PubMed] [Google Scholar]
- 71.Birkenkamp-Demtroder K, Wagner L, Brandt SF, Bording AL, Gartner W, Scherubl H, Heine B, Christiansen P, Orntoft TF. Neuroendocrinology. 2005;82:121–138. doi: 10.1159/000091207. [DOI] [PubMed] [Google Scholar]
- 72.Pauls TL, Cox JA, Berchtold MW. Biochim Biophys Acta. 1996;1306:39–54. doi: 10.1016/0167-4781(95)00221-9. [DOI] [PubMed] [Google Scholar]
- 73.Berggard T, Arrigoni G, Olsson O, Fex M, Linse S, James P. J Proteome Res. 2006;5:669–687. doi: 10.1021/pr050421l. [DOI] [PubMed] [Google Scholar]
- 74.Yang J, Li T. Exp Cell Res. 2005;309:379–389. doi: 10.1016/j.yexcr.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 75.Adolf K, Wagner L, Bergh A, Stattin P, Ottosen P, Borre M, Birkenkamp-Demtroder K, Orntoft TF, Torring N. Prostate. 2007;67:472–484. doi: 10.1002/pros.20523. [DOI] [PubMed] [Google Scholar]
- 76.Lee JY, Byun JY, Lee SH. Clin Biochem. 2009;42:692–700. doi: 10.1016/j.clinbiochem.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 77.Attems J, Ittner A, Jellinger K, Nitsch RM, Maj M, Wagner L, Gotz J, Heikenwalder M. J Neural Transm. 2011;118:737–745. doi: 10.1007/s00702-011-0626-5. [DOI] [PubMed] [Google Scholar]
- 78.Gartner W, Lang W, Leutmetzer F, Domanovits H, Waldhausl W, Wagner L. Cereb Cortex. 2001;11:1161–1169. doi: 10.1093/cercor/11.12.1161. [DOI] [PubMed] [Google Scholar]
- 79.Adamek GD, Shipley MT, Sanders MS. Brain Res Bull. 1984;12:657–668. doi: 10.1016/0361-9230(84)90147-3. [DOI] [PubMed] [Google Scholar]
- 80.Wyss JM, Sripanidkulchai K. J Comp Neurol. 1983;219:251–272. doi: 10.1002/cne.902190302. [DOI] [PubMed] [Google Scholar]
- 81.Flames N, Marin O. Neuron. 2005;46:377–381. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 82.Sanchez MP, Frassoni C, varez-Bolado G, Spreafico R, Fairen A. J Neurocytol. 1992;21:717–736. doi: 10.1007/BF01181587. [DOI] [PubMed] [Google Scholar]
- 83.Brun P, Dupret JM, Perret C, Thomasset M, Mathieu H. Pediatr Res. 1987;21:362–367. doi: 10.1203/00006450-198704000-00008. [DOI] [PubMed] [Google Scholar]
- 84.Verney C, Derer P. J Comp Neurol. 1995;359:144–153. doi: 10.1002/cne.903590110. [DOI] [PubMed] [Google Scholar]
- 85.Meyer G, Soria JM, Martinez-Galan JR, Martin-Clemente B, Fairen A. J Comp Neurol. 1998;397:493–518. [PubMed] [Google Scholar]
- 86.Meyer AH, Katona I, Blatow M, Rozov A, Monyer H. J Neurosci. 2002;22:7055–7064. doi: 10.1523/JNEUROSCI.22-16-07055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim DS, Ross SE, Trimarchi JM, Aach J, Greenberg ME, Cepko CL. J Comp Neurol. 2008;507:1795–1810. doi: 10.1002/cne.21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Montaner J, Mendioroz M, Ribo M, Delgado P, Quintana M, Penalba A, Chacon P, Molina C, Fernandez-Cadenas I, Rosell A, varez-Sabin J. J Intern Med. 2010 doi: 10.1111/j.1365-2796.2010.02329.x. [DOI] [PubMed] [Google Scholar]
- 89.Montaner J, Perea-Gainza M, Delgado P, Ribo M, Chacon P, Rosell A, Quintana M, Palacios ME, Molina CA, varez-Sabin J. Stroke. 2008;39:2280–2287. doi: 10.1161/STROKEAHA.107.505354. [DOI] [PubMed] [Google Scholar]
- 90.Gartner W, Vila G, Daneva T, Nabokikh A, Koc-Saral F, Ilhan A, Majdic O, Luger A, Wagner L. Am J Physiol Endocrinol Metab. 2007;293:E347–E354. doi: 10.1152/ajpendo.00055.2007. [DOI] [PubMed] [Google Scholar]
- 91.Gage FH. Journal of Neuroscience. 2002;22:612–613. doi: 10.1523/JNEUROSCI.22-03-00612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pipp I, Wagner L, Rossler K, Budka H, Preusser M. APMIS. 2007;115:319–326. doi: 10.1111/j.1600-0463.2007.apm_590.x. [DOI] [PubMed] [Google Scholar]
- 93.Xing X, Lai M, Gartner W, Xu E, Huang Q, Li H, Chen G. Proteomics. 2006;6:2916–2923. doi: 10.1002/pmic.200401355. [DOI] [PubMed] [Google Scholar]
- 94.Zhan X, Evans CO, Oyesiku NM, Desiderio DM. Pituitary. 2003;6:189–202. doi: 10.1023/b:pitu.0000023426.99808.40. [DOI] [PubMed] [Google Scholar]
- 95.Desiderio DM, Zhan X. Cell Mol Biol (Noisy -le-grand) 2003;49:689–712. [PubMed] [Google Scholar]
- 96.Leroy B, Toubeau G, Falmagne P, Wattiez R. Mol Cell Proteomics. 2006;5:2114–2123. doi: 10.1074/mcp.M600205-MCP200. [DOI] [PubMed] [Google Scholar]
- 97.Goncalves LR, Yamanouye N, Nunez-Burgos GB, Furtado MF, Britto LR, Nicolau J. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1997;118:207–211. doi: 10.1016/s0742-8413(97)00130-8. [DOI] [PubMed] [Google Scholar]
- 98.Marshak DR, Umekawa H, Watterson DM, Hidaka H. Arch Biochem Biophys. 1985;240:777–780. doi: 10.1016/0003-9861(85)90086-4. [DOI] [PubMed] [Google Scholar]
- 99.Barger SW, Van Eldik LJ, Mattson MP. Brain Res. 1995;677:167–170. doi: 10.1016/0006-8993(95)00160-r. [DOI] [PubMed] [Google Scholar]
- 100.Nishiyama H, Knopfel T, Endo S, Itohara S. Proc Natl Acad Sci U S A. 2002;99:4037–4042. doi: 10.1073/pnas.052020999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van Eldik LJ, Zimmer DB. Brain Res. 1987;436:367–370. doi: 10.1016/0006-8993(87)91681-7. [DOI] [PubMed] [Google Scholar]
- 102.Banfalvi T, Boldizsar M, Gergye M, Gilde K, Kremmer T, Otto S. Pathol Oncol Res. 2002;8:183–187. doi: 10.1007/BF03032392. [DOI] [PubMed] [Google Scholar]
- 103.Gartner W, Zierhut B, Mineva I, Sodeck G, Leutmezer F, Domanovits H, Prayer D, Wolf F, Base W, Weissel M, Wagner L. Clin Biochem. 2008;41:1434–1439. doi: 10.1016/j.clinbiochem.2008.09.096. [DOI] [PubMed] [Google Scholar]
- 104.Gartner W, Ilhan A, Neziri D, Base W, Weissel M, Wohrer A, Heinzl H, Waldhor T, Wagner L, Preusser M. Neuro Oncol. 2010;12:1004–1008. doi: 10.1093/neuonc/noq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim DS, Choi YP, Kang S, Gao MQ, Kim B, Park HR, Choi YD, Lim JB, Na HJ, Kim HK, Nam YP, Moon MH, Yun HR, Lee DH, Park WM, Cho NH. J Proteome Res. 2010;9:3710–3719. doi: 10.1021/pr100236r. [DOI] [PubMed] [Google Scholar]
- 106.Ilhan A, Neziri D, Maj M, Mazal PR, Susani M, Base W, Gartner W, Wagner L. Hum Pathol. 2011;42:641–648. doi: 10.1016/j.humpath.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 107.Park HC, Seong J, An JH, Kim J, Kim UJ, Lee BW. Int J Radiat Oncol Biol Phys. 2005;61:1523–1534. doi: 10.1016/j.ijrobp.2004.12.070. [DOI] [PubMed] [Google Scholar]
- 108.Luiten PG, Buwalda B, Traber J, Nyakas C. Brain Res Dev Brain Res. 1994;79:10–18. doi: 10.1016/0165-3806(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 109.Saenger AK, Christenson RH. Clin Chem. 2010;56:21–33. doi: 10.1373/clinchem.2009.133801. [DOI] [PubMed] [Google Scholar]
- 110.Mattson MP, Chan SL. Cell Calcium. 2003;34:385–397. doi: 10.1016/s0143-4160(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 111.Busche MA, Eichhoff G, Adelsberger H, Abramowski D, Wiederhold KH, Haass C, Staufenbiel M, Konnerth A, Garaschuk O. Science. 2008;321:1686–1689. doi: 10.1126/science.1162844. [DOI] [PubMed] [Google Scholar]
- 112.Harkany T, Abraham I, Konya C, Nyakas C, Zarandi M, Penke B, Luiten PGM. Reviews in the Neurosciences. 2000;11:329–382. doi: 10.1515/revneuro.2000.11.4.329. [DOI] [PubMed] [Google Scholar]
- 113.Nimmrich V, Ebert U. Rev Neurosci. 2009;20:1–12. doi: 10.1515/revneuro.2009.20.1.1. [DOI] [PubMed] [Google Scholar]
- 114.Hof PR, Cox K, Young WG, Celio MR, Rogers J, Morrison JH. Neuropathol J Exp Neurol. 1991;50:451–462. doi: 10.1097/00005072-199107000-00006. [DOI] [PubMed] [Google Scholar]
- 115.Hof PR, Nimchinsky EA, Celio MR, Bouras C, Morrison JH. Neurosci Lett. 1993;152:145–148. doi: 10.1016/0304-3940(93)90504-e. [DOI] [PubMed] [Google Scholar]
- 116.Braak E, Braak H, Mandelkow EM. Acta Neuropathol. 1994;87:554–567. doi: 10.1007/BF00293315. [DOI] [PubMed] [Google Scholar]
- 117.Maj M, Gartner W, Ilhan A, Neziri D, Attems J, Wagner L. J Endocrinol. 2010;205:25–36. doi: 10.1677/JOE-09-0341. [DOI] [PubMed] [Google Scholar]
- 118.Berggard T, Miron S, Onnerfjord P, Thulin E, Akerfeldt KS, Enghild JJ, Akke M, Linse S. J Biol Chem. 2002;277:16662–16672. doi: 10.1074/jbc.M200415200. [DOI] [PubMed] [Google Scholar]
- 119.Faas GC, Schwaller B, Vergara JL, Mody I. PLoS Biol. 2007;5:e311. doi: 10.1371/journal.pbio.0050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Berggard T, Szczepankiewicz O, Thulin E, Linse S. J Biol Chem. 2002;277:41954–41959. doi: 10.1074/jbc.M203492200. [DOI] [PubMed] [Google Scholar]