Abstract
Calmodulin (CaM) is a highly conserved, ubiquitous, calcium-binding protein; it binds to and regulates many different protein targets, thereby functioning as a calcium sensor and signal transducer. CaM contains 9 methionine (Met), 1 histidine (His), 17 aspartic acid (Asp), and 23 glutamine acid (Glu) residues, all of which can potentially react with platinum compounds; thus, one third of the CaM sequence is a possible binding target of platinum anticancer drugs, which represents a major challenge for identification of specific platinum modification sites. Here, top-down electron capture dissociation (ECD) was used to elucidate the transition metal-platinum(II) modification sites. By using a combination of top-down and bottom-up mass spectrometric (MS) approaches, ten specific binding sites for mononuclear complexes, cisplatin and [Pt(dien)Cl]Cl, dinuclear complex [{cis-PtCl2(NH3)}2(μ-NH2(CH2)4NH2)] on CaM were identified. High resolution MS of cisplatin-modified CaM revealed that cisplatin mainly targets Met residues in solution at low molar ratios of cisplatin-CaM (2:1), by cross-linking Met residues. At a high molar ratio of cisplatin:CaM (8:1), up to 10 platinum(II) bind to Met, Asp, and Glu residues. [{cis-PtCl2(NH3)}2(μ-NH2(CH2)4NH2)] forms mononuclear adducts with CaM. The alkanediamine linker between the two platinum centres dissociates due to a trans-labilization effect. [Pt(dien)Cl]Cl forms {Pt(dien)}2+ adducts with CaM, and the preferential binding sites were identified as Met51, Met71, Met72, His107, Met109, Met124, Met144, Met145, Glu45 or Glu47, and Asp122 or Glu123. The binding of these complexes to CaM, particularly when binding involves loss of all four original ligands, is largely irreversible which could result in their failure to reach the target DNA or be responsible for unwanted side-effects during chemotherapy. Additionally, the cross-linking of cisplatin to CaM might lead to the loss of the biological function of CaM or CaM-Ca2+ due to limiting the flexibility of the CaM or CaM-Ca2+ complex to recognize target proteins or blocking the binding region of target proteins to CaM.
INTRODUCTION
Sulfur-containing biomolecules play significant roles in platinum anticancer chemotherapy because of their high affinity for platinum compounds. Studies of cisplatin drug resistance have demonstrated that a failure of a sufficient quantity of platinum to reach the target DNA can lead to resistance.1–4 Further evidence showed that increased levels of cytoplasmic sulfur-rich species, such as glutathione and metallothioneins, causes cisplatin drug resistance and leads to detoxification because platinum binds irreversibly to thiolate sulfur.5–9 Calmodulin (CaM) is a ubiquitous, calcium (Ca2+)-binding protein that senses changes in intramolecular calcium level to coordinate the activity of over thirty different target proteins in eukaryotic cells.10 CaM is expressed in many cell types and can have different subcellular locations, including the cytoplasm, within organelles, or associated with the plasma or organelle membranes. CaM is a methionine-rich protein, with 9 methionine residues out of 148 amino acid residues. Upon calcium activation, methionine-rich binding pockets are exposed in each of the opposing globular domains of CaM.11 These hydrophobic binding sites facilitate CaM association and promote activation of a diversity of conformations depending on the target.12 In fact Met residues contribute as much as 46% of the exposed surface area of the hydrophobic patches on the CaM surface,13 which makes them likely targets for platinum drugs. Oxidation of methionine residues of CaM has been shown to decrease the ability of CaM to activate target enzymes.14–18 Recent studies have suggested that cisplatin can bind strongly to CaM by forming Pt-S bonds with Met residues, which can cause inhibition of calmodulin’s capacity to activate target proteins. For example, inhibition of Ca2+-CaM due to direct interactions with cisplatin could play a major role in stomach distention.19 In addition, CaM is also rich in aspartic acid (Asp) and glutamine acid (Glu) amino acids, nearly half of which are involved in calcium binding. Therefore, the binding of platinum(II) to Met or Asp, or Glu residues, or the single His residue is also likely lead to the malfunction of CaM. However, very little is known about the binding of platinum anti-cancer drugs to CaM. In view of the current widespread chemical use of platinum anticancer drugs and needs to elucidate their mechanism of activity and their side-effects, it is of interest to investigate the interactions between CaM and cisplatin analogs more closely.
Mass spectrometry is an extremely powerful tool to study the interactions of drugs with proteins, due to its advantage of sensitivity and the ability to provide direct sequence-specific information on the position and the form of the drug-protein adducts.20 Reactions between proteins (transferrin, cyctochrome C, ubiquitin, insulin, superoxide dismutase and etc.) and metallodrugs have been previously studied by electrospray ionization mass spectrometry (ESI MS).21–27 Recent progress with combinating mass spectrometry and proteomics technologies have made the identification of binding sites much more feasible. There are basically two classes of methods to achieve this, “bottom-up” (peptide level) and “top-down” (intact protein level) approaches.28 “Bottom up” strategies involve cleaving the protein into peptide fragments using proteolytic enzymes prior to mass spectrometry detection. By applying the bottom-up mass spectrometric approach, Allardyce et al identified that cisplatin binds to threonine 457 of transferrin.24 However, as only some tryptic peptides are normally detected, it is possible that other cisplatin modification sites on transferrin have been missed, particularly because there are multiply potential cisplatin binding sites (44 Asp, 19 His, 8 Met, and 42 Glu residues) on transferrin. “Top-down” methods identify proteins by measuring the mass of the whole protein, then using tandem mass spectrometry (such as collisionally activated dissociation (CAD), electron capture dissociation (ECD), or infrared multiphoton dissociation (IRMPD)) to fragment intact proteins in order to generate sequence information, so that all modifications are normally detected. Recently, Moreno-Gordaliza et al were able to determine the binding sites between cisplatin and insulin by combining the top-down approach with nano-electrospray ionization mass spectrometry using a linear ion trap (nESI-LIT-MS).25 The combination of Fourier transform ion cyclotron resonance mass spectrometry (FTICR MS) with top-down and bottom-up proteomic approaches generate effective binding site information; Hartinger et al identified the binding sites of three different platinum anticancer drugs with ubiquitin by top-down high resolution MS approach.26
Top-down analysis of proteins by ECD has developed rapidly in recent years.29–33 ECD cleaves N-Cα bonds to produce mainly c and z• ions, complementary to b and y ions produced in CAD by cleaving CO-NH bonds.29 The combination of ECD and CAD has greatly improved the efficiency and sequence coverage in top-down protein analyses. However, in previous reports, the application of ECD in protein-platinum or peptide-platinum complexes for characterizing platinum binding sites is rather limited.26,34 Previously, ECD has been successfully applied to cisplatin cross-linked peptides and protein-CaM by Li et al.35 Here, the top-down ECD mass spectrometric approach is extended to map the binding sites for platinum anti-cancer drugs on CaM.
Cisplatin and its analogues are widely used in clinical cancer treatments. The mechanisms for transport of platinum anticancer drugs through cell membranes and possible intermediate formed by binding to proteins remain poorly understood, although they may contribute to many of the drugs’ side-effects.36 The objective of this report is to gain insights into the reactivity of various platinum complexes with CaM and to map the binding sites by using top-down and bottom-up high resolution MS approaches. Here, the application of top-down ECD mass spectrometry for mapping the binding sites of the platinum anticancer complexes cisplatin (Pt_1), [Pt(dien)Cl]Cl (Pt_2) and [{cis-PtCl2(NH3)}2(μ-NH2(CH2)4NH2)] (Pt_3) (Figure 1) to CaM is demonstrated.
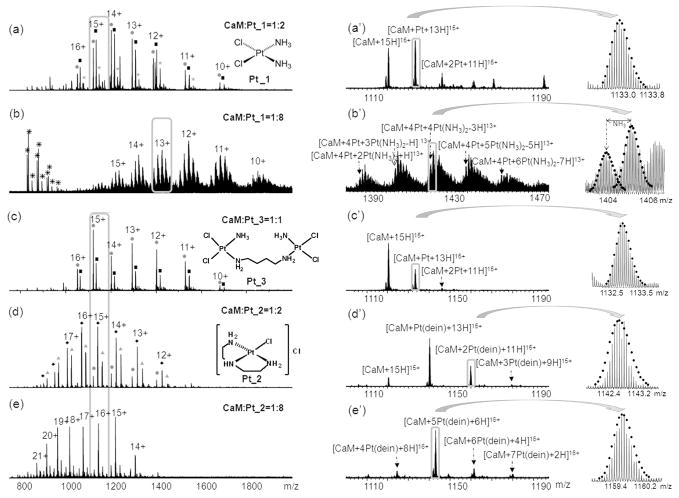
Figure 1.
High resolution of ESI-FTMS analyses of reaction mixtures of platinum complexes with CaM at different molar ratios. (a) CaM:Pt_1 (1:2); (b) CaM:Pt_1 (1:8); (c) CaM:Pt_3 (1:1); (d) CaM:Pt_2 (1:2); (e) CaM:Pt_2 (1:8). Figures 1a′–1e′ are expansions of the corresponding parts in Figures 1a–1e. In Figure 1a, 1c, and 1d are the structures of platinum(II) complexes, cisplatin (Pt_1), [PtCl(dien)]Cl (Pt_2), and [{cis-PtCl2(NH3)}2(μ-NH2(CH2)4NH2)] (Pt_3). * Represents chemical noise; ● CaM; ■ CaM+Pt; ★CaM+2Pt; ◆ CaM+Pt(dien), and ▲ CaM+2Pt(dien).
EXPERIMENTAL METHODS
Materials
Bovine calmodulin, trypsin (TPCK treated from bovine pancreas), ammonium acetate (CH3COONH4), and ammonium bicarbonate (NH4HCO3) were purchased from Sigma (St. Louis, MO). HPLC grade of methanol, acetic acid (HAc), and acetonitrile (ACN) were obtained from Fisher Scientific (Pittsburgh, PA). Cisplatin (Pt_1), [PtCl(dien))Cl] (Pt_2), and [{cis-PtCl2(NH3)}2(μ-NH2(CH2)4NH2)] (Pt_3) were synthesized and characterized by standard methods.37–39
Reaction of CaM with Pt_1, Pt_2, and Pt_3
Aqueous solutions of CaM (500 μM) and platinum complex (Pt_1, Pt_2, and Pt_3, 500 μM each) were prepared and mixed to give a 200 μL (40 μM) solution of protein:platinum complex at molar ratios of 1:1, 1:2, and 1:8. The samples were incubated at 37 °C for 24 h. To remove free platinum complexes and desalt, amicon filters (MW cut off = 3 kDa, Millipore, Watford, UK) were used at 13000 rpm for 30 min at room temperature, and washed twice with 200 μL water. The sample was diluted to 0.4 μM with 50% MeOH-1% CH3COOH buffer immediately before mass spectrometry analysis.
Protein Digestion
The CaM-platinum adducts in the 1:1, 1:2, and 1:8 molar ratios of CaM:Pt_1 or CaM:Pt_2 mixtures were diluted to 20 μM with 50 mM NH4HCO3 (pH 7.8) and then subjected to trypsin digestion at a protein to enzyme ratio of 40:1 (w/w) at 37 °C for 4 h. As a control, 20 μM CaM without platinum reagents was digested under the same conditions. The sample was diluted to 0.4 μM with 50% MeOH-1% CH3COOH buffer immediately before mass spectrometry analysis.
FTICR Mass Spectrometry
ESI-MS was performed on a Bruker solariX FTICR mass spectrometer with an ESI source and a 12 T actively shielded magnet. Samples were electrosprayed at a flow rate of ~300 μL/hour at a concentration of 0.4 μM in 50:50 MeOH:H2O with 1% acetic acid. For ECD experiments, the parent ions were first isolated in the first quadrupole (Q1) and externally accumulated in the collision cell for 2–20 s. After being transferred to the Infinity cell,40 ions were irradiated with 1.5 eV electrons from a 1.7 A heated hollow cathode dispenser for 10 to 100 ms.41 A one millisecond single frequency shot at m/z 100 was given at the beginning of the ECD event to improve the overlap between electron beam and the trapped ions.42–43 Full spectra were internally calibrated using the unmodified c ion series which were previously defined in the ECD spectra of CaM.
RESULTS AND DISSCUSSION
FTICR MS Analyses of the interaction of platinum anti-cancer drugs with CaM
Figures 1a–1e show the mass spectra of CaM upon reaction with Pt_1, Pt_2 and the dinuclear platinum compound Pt_3 at different molar ratios. At a lower molar ratio (1:2) of CaM:Pt_1, up to two platinum molecules coordinate to CaM with all cisplatin ligands (NH3 and Cl) displaced in CaM-Pt_1 mixtures (Figure 1a&a′); increasing the molar ratio of platinum complexes in the mixtures leads to more platinum complexes coordinating to CaM (Figure 1b). Taking the 13+ charge states of CaM-Pt_1 complexes (Figure 1b′) for example, five different groups of CaM-Pt_1 complexes were detected, varying from six up to ten cisplatin complexes bound to CaM in a variety of forms. The isotopic patterns of the [CaM+4Pt+nPt(NH3)2+H]13+ (n=2~6) ions were compared with the theoretical isotopic patterns, which fit with mean absolute deviation within 1.5 ppm range (see the inserts of Figure 1b′ and Table S-1). As previously observed, cisplatin preferentially binds to Met sites of CaM with all four cisplatin ligands displaced in low molar ratios of cisplatin-CaM mixtures;35 herein, the maintenance of ligands, such as NH3, indicates that cisplatin might also bind to His, Asp or Glu residues in CaM sequence when increasing the concentration of cisplain in the cisplatin-CaM mixture. In addition, all the modified peaks shift to higher m/z region, namely, lower charge states, which supports the assumption that cisplatin binds to the carboxyl groups of Asp or Glu residues because deprotonation of a carboxyl group is needed before it binds to platinum(II). Therefore, the two positive charges of platinum(II) are neutralized upon platinum coordinating to two carboxyl groups.
Similar phenomena were observed in CaM-Pt_3 mixtures (Figure 1c&c′), where the bridge linking the two platinum molecules is broken and all the ligands are replaced. To address the questions as to whether the loss of all the ligands and the breakage of the bridge of Pt_3 are due to the reaction of platinum complexes to CaM or to over aggressive spray condition, the reactions were monitored and recorded at every 30 minutes. As shown in Figure S-1(a and a′), after 60 minutes’ reaction, both Pt_1 and Pt_3 bind to CaM in the same form, namely, CaM+Pt, CaM+Pt(NH3), CaM+Pt(NH3)Cl, and CaM+Pt(NH3)2Cl; and the intensities of the CaM+Pt species keeps increasing with time, which suggest that the losses of all the ligands of Pt_1 and Pt_3 are because of the binding to CaM. Similar results have also been observed in other Met-rich peptides or proteins upon reactions with cisplatin due to the trans-labilization effect.44–46 In other words, once the displacement of chlorine ligand by sulfur of the Met or Cys residue has occurred, the Pt-NH3 bond trans to the sulfur is significantly labilized and thus the amine group is readily substituted. The observation of CaM+Pt(NH3)Cl species is in agreement with the trans-labilization effect; in addition, this observation also indicates that following the initial replacement of the trans chlorine ligand by sulfur of the Met residue, the loss of amine ligand is a reactively fast process. Therefore, the trans-labilization effect contributes to the release of {Pt(NH3)Cl}+ species from the dinuclear platinum compound Pt_3 after the displacement of chlorine by sulfur of the Met residue. A similar result was reported by Farrell and co-workers in the reaction of polynuclear platinum antitumor compounds with reduced glutathione, the final product was observed in the form of a dinuclear species [{trans-Pt(SG)(NH3)2}2-μ-SG] with the linkage chain replaced.47
In contrast, as shown in Figure 1d&d′, Pt_2 maintains its {Pt(dien)}2+ fragment and with one other coordination site binding to CaM. At a molar ratio 1:2 of CaM:Pt_2, up to three {Pt(dien)}2+ species were found binding to CaM. Increasing the molar ratio to 1:8, mainly five {Pt(dien)}2+ species bind to CaM, but the binding of up to seven {Pt(dien)}2+ fragments to CaM was observed (Figure 1e&e′). As shown in Figure 1e′, all the modified peaks in the sample of CaM:[PtCl(dien)]Cl (1:8) shift to lower m/z region (higher charge states). Although the shifting to higher charge states usually indicates that the conformation of the protein has been altered, the positive charge of the {Pt(dien)}2+ fragments also contributes to the shift in charge state in the case of [PtCl(dien)]Cl binding to CaM because each {Pt(dien)}2+ fragment has two positive charges and only one binding site available on Pt. Therefore, {Pt(dien)}2+ contributes at least one charge upon each binding.
Mapping the binding sites of cisplatin (Pt_1) to CaM by top-down and bottom-up MS approaches
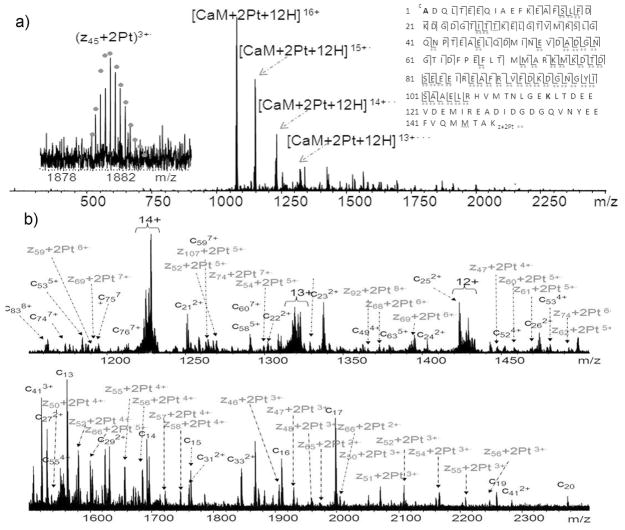
For CaM:Pt_1 (1:2) sample, the CaM+2Pt species was chosen for mapping the Pt-modification sites by performing top-down ECD experiment. Figure 2 shows the ECD spectra of CaM+2Pt species. The peaks are assigned to fragments expected from the sequence of CaM with platinum modifications. In all, 86 cleavages of the total 147 available N-Cα backbone bonds were generated and assigned, representing overall backbone cleavage efficiency (fraction of inter-residue bonds cleavage) of 58%. These cleavages allow localization of the two platinum modification sites to the region of CaM(106-148). MS3 is a reasonable option to find out the exact Pt-binding points. However, the intensities of the Pt-modified c/z· ions (as shown in Figure 2b) are often very low, which makes getting quality data out of MS3 very difficult. To further localize the binding sites of platinum to CaM, CaM-cisplatin adducts at different molar ratios (1:1, 1:2, and 1:8) were trypsin-digested and analyzed by MS. In the 1:1 molar ratio sample of trypsin-digested CaM-cisplatin, in addition to the previously reported cross-linked species,35 another platinum cross-linked species CaM(107-126)+Pt+CaM(142-148) as well as platinum modified CaM(38-74), CaM(107-126), and CaM(127-148) were also observed. The same Pt-modified peaks were also observed in the CaM-cisplatin (1:2) sample; however, when the molar concentration of cisplatin is eight times higher than CaM, the intensities of all Pt-modified peaks dropped significantly, and most of them could be barely detected (Figure S-2). This result is in agreement with the MS data for 1:8 CaM-cisplatin (Figure 1b′), that is, at higher molar ratio of cisplatin to CaM, multiple sites of CaM react with cisplatin giving a complicated mixtures of products and significantly decreasing the intensities of each product overall. Thus, as shown in Figure 1b and Figure S-2, the intensities of many of the product peaks are low or even undetectable.
Figure 2.
MS/MS spectra of the 1:2 CaM:Pt_1 sample. (a) ECD spectra of [CaM+2Pt+12H]16+ ions at m/z 1074, the inset is the fragmentation map from ECD spectra of CaM+2Pt; (b) the expanded ECD spectra [CaM+2Pt+12H]16+ ions. Double dots represent doubly Pt-modified fragments. Full peak list is available in supplementary information, Table S-2.
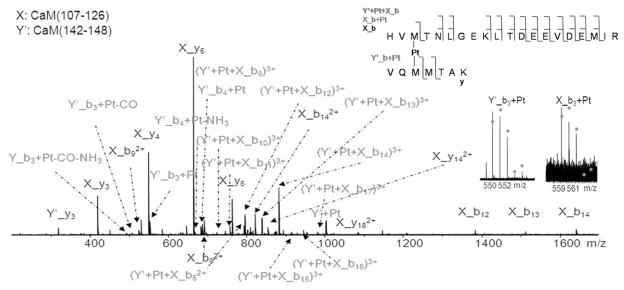
Figure 3 shows the CAD spectrum of the cross-linked product at m/z 681; by matching the fragments from the precursor with digested CaM, the two platinum-cross-linking species were identified as CaM(107-126) and CaM(142-CaM148). The CaM(142-148) species, (F)VQMMTAK was unexpected in the trypsin digested samples because trypsin preferentially cleaves peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, but not the carboxyl side of phenyalanine(F141). Although unusual cleavages can happen in trypsin digestion,48 it is also possible that a small amount of chymotrypsin is active in the trypsin used.49 In addition to the cleavage at the carboxyl side of Phe141, cleavages between Leu69 and Thr70, Met71 and Met72 were also observed (Figure S-2). To simplify the labeling of the spectrum, CaM(107-126) is represented by X, and CaM(142-148) was represented by Y′. The observation of X_b3+Pt and Y_b3+Pt ions indicates that platinum cross-links CaM at Met109 and Met144, although it does not necessarily rule out the possibility that platinum can also bind to Met145.
Figure 3.
CAD spectrum of the cross-linked product of [CaM(107-126)+Pt+CaM(142-CaM148)+3H]5+ ion at m/z 681. To simplify the labeling of the spectrum, CaM(107-126) is represented by X, and CaM(142-148) is represented by Y′. The insets are the CAD fragmentation map of [CaM(107-126)+Pt+CaM(142-CaM148)+3H]5+ ions, and characteristic fragment ions, Y′_b3+Pt and X_b3+Pt, which show that Pt(II) cross-links Met109 and Met144 residues.
To further refine other platinum binding sites on CaM, CAD and ECD experiments were performed on the platinum modified species, [CaM(38-74)+Pt+2H]4+ ion at m/z 1067, [CaM(107-126)+Pt+H]3+ ion at m/z 865, and [CaM(127-148)+Pt+H]3+ ion at m/z 895. A common feature for these three cisplatin-modified species is that all the original NH3 and Cl ligands are displaced from cisplatin, which suggests they are also platinum cross-linked species, and with at least two Met residues binding to one platinum atom in each case. In general, for the top-down analyses of the whole proteins, ECD yields more useful fragment information; for most tryptic peptides, the results were varied with CAD and ECD in some peptides, showing complementarities in that CAD worked better than ECD and vice verse. However, for the intra-chain Pt cross-linked peptides, clearly CAD worked better than ECD (see supplementary information Figure S-3). Therefore, by combination of top-down and bottom-up MS approaches, cisplatin modification sites in CaM were identified as Met51, Met71 or/and Met72, Met 109, Met124, Glu127 or Asp129, Met144, and Met145 residues (Figure S-3); more likely, platinum cross-links Met51 and Met71/Met72 residues, Met109 and Met124 residues, Glu127/Asp129, Met144, and Met145 residues.
Figure 2a (CaM+2Pt) presents a high quality ECD Top-down spectrum; however, no fragments were observed in the region of CaM(106-148). Further ECD experiments on CaM without platinum were performed (Figure S-4), up to 91% backbone cleavages were assigned in the full CaM sequence and nearly 30% of the total cleavages were generated in CaM(106-148) region. Therefore, by comparison, it is reasonable to conclude that the cross-linking of Pt between CaM(109) and CaM(144) contributes to repressed detections of cleavages in the region of CaM(106-148) in the top-down analyses possibly because the intramolecular cross-linking limits the flexibility of modified CaM molecules in the gas phase. It is also likely due to the same reason that only charge reduced species were observed in the ECD spectra of trypsin digested Pt-modified species (Figure S-3). Therefore, the combinations of top-down and bottom-up MS approaches, and also CAD and ECD are necessary for the identification of multiple cisplatin cross-linking CaM sites.
Top-down MS with ECD and CAD for mapping the binding sites of [Pt(dien)Cl]Cl on CaM
The ECD spectrum of [CaM+2Pt(dien)+15H]19+ ions at m/z 916 in the CaM:Pt_2 (1:2) sample is shown in Figure S-5. In this spectrum, the observation of z11+2Pt(dien)3+• indicates that there are two binding sites in the region of CaM(138-148). In addition, the detection of c/z· complementary ion pairs, such as c82+Pt(dien)8+/z66+Pt(dien)8+• (see the inserts of supplementary Figure S-4), suggests that there is one Pt(dien) binding site in the region of CaM(1-81) and the other in the region of CaM(131-148). Therefore, there are at least two isoforms for the CaM+2Pt(dien) species in the CaM:Pt_2 (1:2) sample.
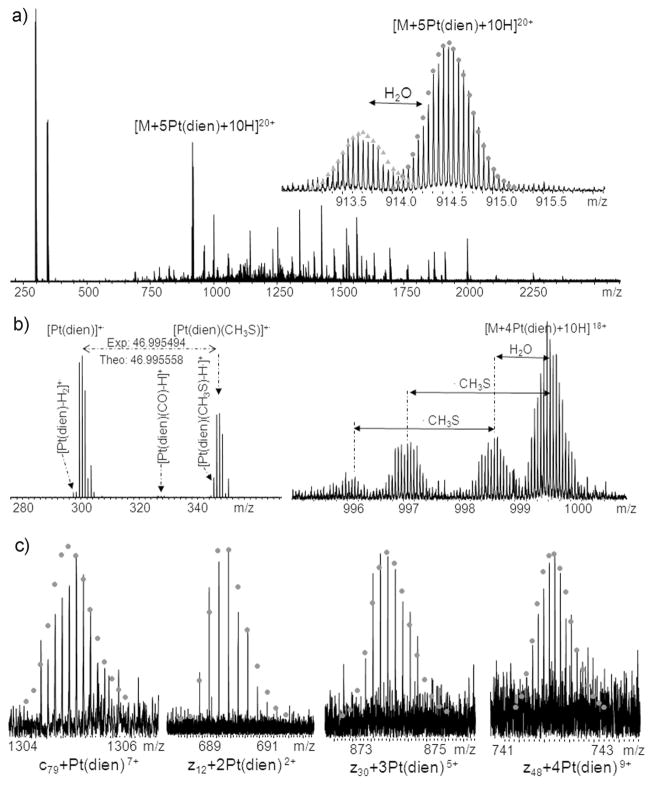
Figure 4 shows the ECD spectrum of [CaM+5Pt(dien)+10H]20+ ions at m/z 914 in the CaM:Pt_2(1:8) sample. In the low m/z region (Figure 4b), fragment ions corresponding to Pt(dien)+•, [Pt(dien)-2H]+•, [Pt(dien)(CO)-H•]+, and Pt(dien)(CH3S)+• ions were observed. Similar results have been previously observed by O’Hair and coworkers;34 however, the CAD product ion of [Pt(II)(dien)(CH3SH)-H]+ and ECD product ion Pt(I)(dien)(CH3S)+• were assigned as Pt(dien)(CH3S)+ in both cases. Although the masses for Pt(II)(dien)(CH3S)+ and Pt(I)(dien)(CH3S)+·· are nearly the same, their origins are different. The [Pt(II)(dien)(CH3S)]+ ions detected in CAD are created by even electron process rather than odd electron dissociation. The masses for Pt(II)(dien)(CH3S)+ and Pt(I)(dien)(CH3S)+·· differ by one electron, 0.55 mDa in mass, which can only be differentiated by FTICR MS due to its superior mass accuracy. As shown in the left side of Figure 4b, the experimental result gave a mass difference of 46.995494 Da between Pt(I)(dien)+• (298.075730) and Pt(I)(dien)(CH3S)+• (345.071224) peaks, which is 0.064 mDa higher than the theoretical mass of CH3S (46.995558 Da). In the case of Pt(II)(dien)(CH3S)+, the mass difference between experimental and theoretical masses of CH3S· (46.995008 Da) is 0.486 mDa. For this spectrum, the internally calibrated mass accuracy (see Table S-3) shows a standard deviation of 0.88 ppm, so that a mass error of 0.486 mDa at ~ 345 Da is 1.6δ. Thus, with a confidence of 89%, we can say that the peak is most likely not Pt(II)(dien)(CH3S)+, and a more consistent assignment is that this peak is Pt(I)(dien)(CH3S)+•. Furthermore, this interpretation is consistent with the radical chemistry related to ECD. In addition, complementary ion pairs for Pt(I)(dien)+• and Pt(I)(dien)(CH3S)+• ions, namely, [CaM+4Pt(dien)+10H]18+ and [CaM+4Pt(dien)+10H-CH3S]18+ ions were also detected, as shown in the right side of Figure 4b, which also support the assignment as Pt(I)(dien)(CH3S)+•. The Pt(dien)(CH3S) +• ion indicates that Pt(dien) coordinates to the sulfur atom of Met residues; similarly, [Pt(dien)(CO)-H•]+ suggests that Pt(dien) also binds to the carboxyl group(s) of Asp or Glu residues.
Figure 4.
(a) ECD spectrum of [CaM+5Pt(dien)+10H]20+ ions at m/z 914; the insert shows the isotopic distribution of precursor ions; (b) Complementary ion pairs, [Pt(dien)]+• and [CaM+4Pt(dien)+10H]18+ ions, [Pt(dien)(CH3S)]+• and [CaM+4Pt(dien)+10H-CH3S]18+ ions; (c) Isotopic distributions of Pt(dien)-modified c and z• ions. For full peak list, see Table S-3.
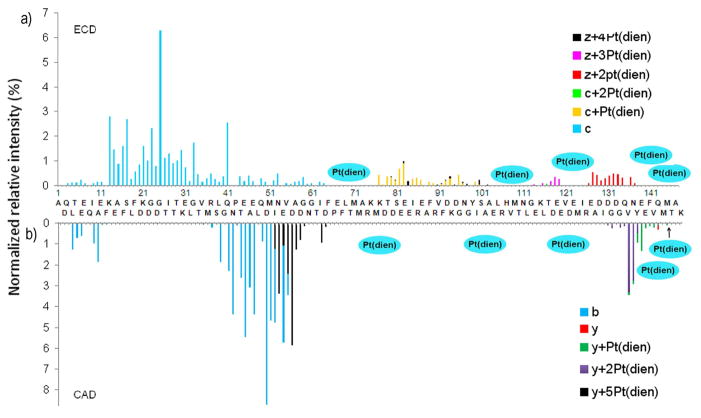
As clearly demonstrated in Figure 5a&b, by combination of the ECD and CAD results, the binding sites for five {Pt(dien)}2+ fragments to CaM are in the regions of CaM(64-76), CaM(102-113), CaM(120-125), CaM(138-144), and Met145. In view of these multiple potential binding sites, bottom-up experiments were performed to localize them. Specific {Pt(dien)}2+ binding sites were identified as Met51, Met71, Met72, His107, Met109, Met124, Met144, Met145, Glu45 or Glu47, and Asp122 or Glu123 (Figure S-6).
Figure 5.
Site-specific yields of products from backbone fragmentation vs. backbone cleavage sites. a). ECD of [CaM+5Pt(dien)+10H]20+; b) CAD of [CaM+5Pt(dien)+10H]20+.
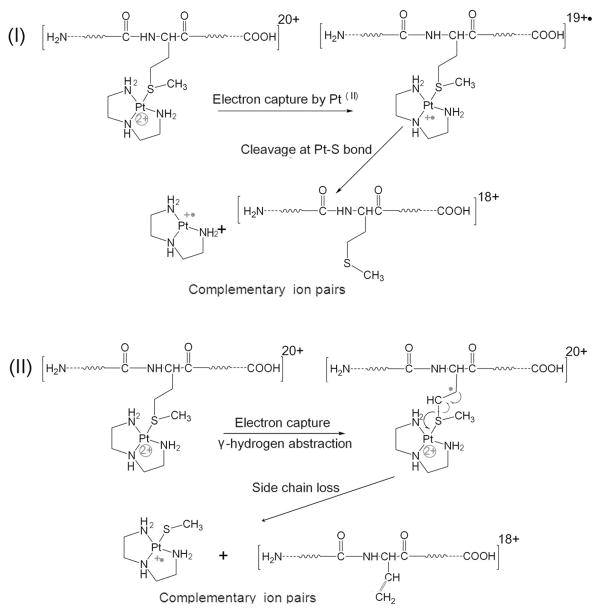
ECD studies of transition metal binding peptides have been previously reported by a number of groups.50–52 Different fragmentation behaviors were observed for different transition metal ions in these complexes. For ECD of multiply-charged metal-protein ions, numerous other dissociation channels exist. As can be seen in Figure 4, direct electron capture by platinum(II), side-chain losses, and backbone N-Cα bond cleavage leading to c/z· ions were all observed. Scheme 1(i) shows that direct electron capture by platinum(II) leads to the cleavage of Pt-S bonds, which is evidenced by the detection of Pt(dien)+· ions. Scheme 1(ii) illustrates abstraction of the γ-hydrogen, which leads to loss of a radical side chain fragment. Because of the two inherent positive charges of platinum(II), the side chain loss of Pt(dien)(SCH3)+• was detected, which otherwise is neutral. The side chain losses from Met residues have been reported as •C2H5S and C3H8S, the side chain loss of •SCH3 has rarely been observed for unmodified Met residues.53 It is likely due to the positive charge of platinum(II), therefore the side chain fragmentation pathway of Pt(II)-modified Met(S) is different. The side chain loss of Pt(dien)(SCH3)+• ions can be used as a signature ion which indicates the binding of platinum(II) to Met(S). Similarly, the observation of [Pt(dien)(CO)-H•]+ ions suggests the coordination of platinum(II) to the carboxyl group of Asp or Glu residues.
Scheme 1.
Proposed mechanism for formation of Pt(dien)+• and Pt(dien)(CH3S)+• ions.
CONCLUSIONS
Top-down ECD mass spectrometry has been successfully applied to mapping the binding site of platinum complexes on calmodulin. Nearly 60% backbone cleavage efficiency (fraction of inter-residue bonds cleavage) was achieved in Pt-modified CaM, and over 90% backbone cleavage efficiency was achieved in CaM without Pt-modification. Due to the very high resolving power and mass accuracy of FTICR MS, differences in mass of one electron (0.55 mDa) differences are clearly differentiated, which leads to the confident assignment of Pt(dien)(SCH3)+• ions.
Multiple electron capture pathways were observed in the platinum-modified CaM ions, such as, direct electron capture by platinum(II), side chain losses, and normal backbone N-Cα bond cleavages leads to c/z· ions. In addition, the side-chain loss ions, Pt(dien)(SCH3)+• and [Pt(dien)(CO)-H•]+, can be used as markers to indicate the binding of Pt(dien) to Met(S), Asp(O), and Glu(O) groups.
The activity of platinum antitumor compounds is usually closely related to their binding to DNA.54 Therefore, the loss of all the ligands of cisplatin upon binding to Met-rich calmodulin could cause cisplatin to lose much of its antitumor activity due to a failure of reaching the target DNA.5–9 More importantly, the Met residues in calmodulin play an important role in the function of CaM by stabilizing the open conformation of Ca2+-CaM and providing a target binding interface.12,55–56 Therefore, the intramolecularly Pt-cross-linked CaM will lose its conformational flexibility to recognize calcium or target proteins, and thereby lose its function as a calcium sensor and a signal transducer. In addition, it has been widely reported that the oxidation of Met residues of CaM decreases the ability of CaM to activate target proteins.12,15–17 Particularly, the oxidation of Met144 and Met145 is largely responsible for the decrease in the activity of CaM to activate enzymes.15 Therefore, the direct binding of cisplatin or its analogues to either Met144 or Met145 may also decrease the activity of CaM to recognize other target proteins.
Supplementary Material
Acknowledgments
We thank the Warwick Postgraduate Research Scholarship (WPRS) and the Departmental Studentship for H. Li, and the WPRS for Y. Zhao. Financial support from NIH (NIH/NIGMS-R01GM078293), the ERC (247450), the Warwick Centre for Analytical Science (EPSRC funded EP/F034210/1), and EPSRC (BP/G006792) is gratefully acknowledged.
Footnotes
Supporting Information Available
References
- 1.Kelland L. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Guo Z. Anti-cancer Agents in Med Chem. 2007;7:19–34. doi: 10.2174/187152007779314062. [DOI] [PubMed] [Google Scholar]
- 3.Cohen SM, Lippard SJ. Prog Nucleic Acid Res Mol Biol. 2001;67:94–129. doi: 10.1016/s0079-6603(01)67026-0. [DOI] [PubMed] [Google Scholar]
- 4.Borst P, Rottenberg S, Jonkers J. Cell Circle. 2008;7:1353–1359. doi: 10.4161/cc.7.10.5930. [DOI] [PubMed] [Google Scholar]
- 5.Kroning R, Lichtenstein AK, Nagami GT. Cancer Chemother Pharmacol. 2000;45:43–49. doi: 10.1007/PL00006741. [DOI] [PubMed] [Google Scholar]
- 6.Arnesano F, Boccarelli A, Cornacchia D, Nushi F, Sasanelli R, Coluccia M, Natile G. J Med Chem. 2009;52:7847–7855. doi: 10.1021/jm900845t. [DOI] [PubMed] [Google Scholar]
- 7.Karotki AV, Vasak M. Biochemistry. 2008;47:10961–10969. doi: 10.1021/bi801253x. [DOI] [PubMed] [Google Scholar]
- 8.Knipp M, Karotki AV, Chesnov S, Natile G, Sadler PJ, Brabec V, Vasak M. J Med Chem. 2007;50:4075–4086. doi: 10.1021/jm070271l. [DOI] [PubMed] [Google Scholar]
- 9.Lau JK, Deubel D. Chem Eur J. 2005;11:2849–2855. doi: 10.1002/chem.200401053. [DOI] [PubMed] [Google Scholar]
- 10.Crivici A, Ikura M. Annu ReV Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 11.Nelson MR, Chazin WJ. Protein Sci. 1998;7:270–282. doi: 10.1002/pro.5560070206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vetter SW, Leclerc E. Eur J Biochem. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 13.O’Neil KT, DeGrado WF. Trends Biochem Sci. 1990;15:59–64. doi: 10.1016/0968-0004(90)90177-d. [DOI] [PubMed] [Google Scholar]
- 14.Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schoneich C, Williams TD, Squier TC. Biophysical J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartlett RK, Urbauer RJB, Anbanandam A, Smallwood HS, Urbauer JL, Squier TC. Biochemistry. 2003;42:3231–3228. doi: 10.1021/bi026956z. [DOI] [PubMed] [Google Scholar]
- 16.Vougier S, Mary J, Dautin N, Vinh J, Friguet B, Ladant D. J Biol Chem. 2004;279:30210–30218. doi: 10.1074/jbc.M400604200. [DOI] [PubMed] [Google Scholar]
- 17.Yao Y, Yin D, Jas GS, Kuczera K, Williams TD, Schoneih C, Squier T. Biochemistry. 1996;35:2767–2787. doi: 10.1021/bi951712i. [DOI] [PubMed] [Google Scholar]
- 18.Bigelow DJ, Squier TC. Biochimica et Biophysica Acta. 2005;1703:121–134. doi: 10.1016/j.bbapap.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Jarve RK, Aggarwal SK. Cancer Chemother Pharmacol. 1997;39:341–348. doi: 10.1007/s002800050581. [DOI] [PubMed] [Google Scholar]
- 20.Car SA, Annan RS. Curr Protoc Mol Biol. 1997;(unit):10.21.1–10.21.27. doi: 10.1002/0471142727.mb1021s38. [DOI] [PubMed] [Google Scholar]
- 21.Zhao T, King FL. J Am Soc Mass Spectrom. 2009;20:1141–1147. doi: 10.1016/j.jasms.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Gibson D, Costello CE. Eur Mass Spectrom. 1999;5:501–510. [Google Scholar]
- 23.Will J, Sheldrick WS. J Biol Inorg Chem. 2008;13:421–434. doi: 10.1007/s00775-007-0333-8. [DOI] [PubMed] [Google Scholar]
- 24.Allardyce CS, Dyson PJ, Coffey J, Johnson N. Rapid Commun Mass Spectrom. 2002;16:933–935. doi: 10.1002/rcm.662. [DOI] [PubMed] [Google Scholar]
- 25.Moreno-Gordaliza E, Canas B, Palacios MA, Gomez-Gomez MM. Anal Chem. 2009;81:3507–3516. doi: 10.1021/ac900046v. [DOI] [PubMed] [Google Scholar]
- 26.Hartinger C, Tsybin Y, Fuchser J, Dyson PJ. Inorg Chem. 2008;47:17–19. doi: 10.1021/ic702236m. [DOI] [PubMed] [Google Scholar]
- 27.Mandal R, Li X. Rapid Coummun Mass Spectrom. 2006;20:48–52. doi: 10.1002/rcm.2268. [DOI] [PubMed] [Google Scholar]
- 28.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J Am Chem Soc. 1999;121:806–812. [Google Scholar]
- 29.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FM. J Am Chem Soc. 1999;121:2857–2862. [Google Scholar]
- 30.Zubarev RA, Horn DM, Fridriksson EK, Keller NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FM. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 31.Xie Y, Zhang J, Yin S, Loo JA. J Am Chem Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 32.Breuker K, Jin M, Han X, Jiang H, McLafferty FM. J Am Soc Mass Spectrom. 2008;19:1045–1053. doi: 10.1016/j.jasms.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horn DM, Ge Y, McLafferty FW. Anal Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 34.Feketeova L, Ryzhov V, O’Hair RAJ. Rapid Commun Mass Spectrom. 2009;23:3133–3143. doi: 10.1002/rcm.4234. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Zhao Y, Phillips HIA, Qi Y, Lin TY, Sadler PJ, O’Connor PB. Anal Chem. 2011;83:5369–5376. doi: 10.1021/ac200861k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timerbaev AR, Hartinger CG, Aleksenko SS, Keppler BK. Chem Rev. 2006;106:2224–2248. doi: 10.1021/cr040704h. [DOI] [PubMed] [Google Scholar]
- 37.Dhara SC. Indian J Chem. 1970;8:193–194. [Google Scholar]
- 38.Annibale G, Brandolisio M, Pitteri B. Polyhedron. 1995;14:451–453. [Google Scholar]
- 39.Farrell N, Qu Y. Inorg Chem. 1989;18:3416–3420. [Google Scholar]
- 40.Caravatti P, Allemann A. Org Mass Spectrom. 1991;26:514–518. [Google Scholar]
- 41.Tsybin YO, Quinn JP, Tsybin OY, Hendrickson CL, Marshall AG. J Am Mass Spectrom. 2008;19:762–771. doi: 10.1016/j.jasms.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Gorshkov MV, Masselon CD, Nokolaev EN, Udseth HR, Pasa-Tolic L, Smith RD. Int J Mass Spectrom. 2004;234:131–136. [Google Scholar]
- 43.Mormann M, Peter-katalinic J. Rapid Commun Mass Spectrom. 2003;17:2208–2214. doi: 10.1002/rcm.1163. [DOI] [PubMed] [Google Scholar]
- 44.Kasherman Y, Sturup S, Gibson D. J Biol Chem. 2009;14:387–399. doi: 10.1007/s00775-008-0456-6. [DOI] [PubMed] [Google Scholar]
- 45.Crider SE, Holbrook RJ, Franz KJ. Metallomics. 2010;2:74–83. doi: 10.1039/b916899k. [DOI] [PubMed] [Google Scholar]
- 46.Wu Z, Liu W, Liang X, Yang X, Wang N, Wang X, Sun H, Lu Y, Guo Z. J Biol Chem. 2009;14:1313–1323. doi: 10.1007/s00775-009-0576-7. [DOI] [PubMed] [Google Scholar]
- 47.Oehlsen ME, Qu Y, Farrell N. Inorg Chem. 2003;42:5498–5506. doi: 10.1021/ic030045b. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez J, Gupta N, Smith RD, Pevzner PA. J Proteome Research. 2007;7:300–305. doi: 10.1021/pr0705035. [DOI] [PubMed] [Google Scholar]
- 49.Carpenter FH. Methods Enzymol. 1967;11:237. [Google Scholar]
- 50.Kleinnigenhuis AJ, Mihalca R, Heeren RMA, Heck AJR. Int J Mass Spectrom. 2006;253:217–224. [Google Scholar]
- 51.Liu H, Hakansson K. J Am Mass Spectrom. 2006;17:1731–1741. doi: 10.1016/j.jasms.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 52.Turecek F, Jones JW, Holm AIS, Panja S, Nielsen SB, Hvelplund P. J Mass Spectrom. 2009;44:707–724. doi: 10.1002/jms.1546. [DOI] [PubMed] [Google Scholar]
- 53.Moore BN, Julian RR. J Am Chem Soc. 2011;133:6997–7006. doi: 10.1021/ja1096804. [DOI] [PubMed] [Google Scholar]
- 54.Fuertes MA, Alonso C, Perez JM. Chem Rev. 2003;103:645–662. doi: 10.1021/cr020010d. [DOI] [PubMed] [Google Scholar]
- 55.Balog EM, Norton LE, Thomas DD, Fruen BR. Am J Physiol Heart Circ Physiol. 2006;290:H794–H799. doi: 10.1152/ajpheart.00706.2005. [DOI] [PubMed] [Google Scholar]
- 56.Vogel HJ, Zhang M. Mol Cell Biochem. 1995;149/150:3–15. doi: 10.1007/BF01076558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.