Abstract
The Gram-positive bacterium Bacillus subtilis can initiate the process of sporulation under conditions of nutrient limitation. Here, we review some of the last five years of work in this area, with a particular focus on the decision to initiate sporulation, DNA translocation, cell-cell communication, protein localization and spore morphogenesis. The progress we describe has implications not just for the study of sporulation but also for other biological systems where homologs of sporulation-specific proteins are involved in vegetative growth.
Introduction
Some bacteria respond to nutrient limitation by forming an endospore, a morphologically distinct cell type (Fig. 1). This process is called sporulation and has been the subject of continuous microbiological investigation since the seminal 19th century reports of Robert Koch (Koch, 1876) and Ferdinand Cohn (Cohn, 1876). The endospore is a metabolically dormant, environmentally resistant cell, capable of surviving extremes of temperature, desiccation and ionizing radiation. Estimates of endospore longevity range from thousands to millions of years, although it is more likely on the lower end of that range; a number of factors are responsible for this robustness including dehydration of the spore core and compaction of chromosomal DNA (Nicholson, et al., 2000). The process by which these dormant cells reinitiate growth is called germination and it occurs in response to environmental signals including amino acids and cell wall peptidoglycan muropeptides derived from growing cells (Shah, et al., 2008). Neither germination nor the mechanisms underlying spore resistance will be further addressed here although the reader is directed to recent reviews (Setlow, 2006, Paredes-Sabja, et al., 2011). Endospore formation, sensu stricto, is reserved for Bacilli and Clostridia although many Streptomyces species produce spores with some limited similarity. Recently, endospore production by Mycobacterium marinum and M. bovis was reported (Ghosh, et al., 2009, Singh, et al., 2010); however, that observation has not been independently confirmed (Traag, et al., 2010).
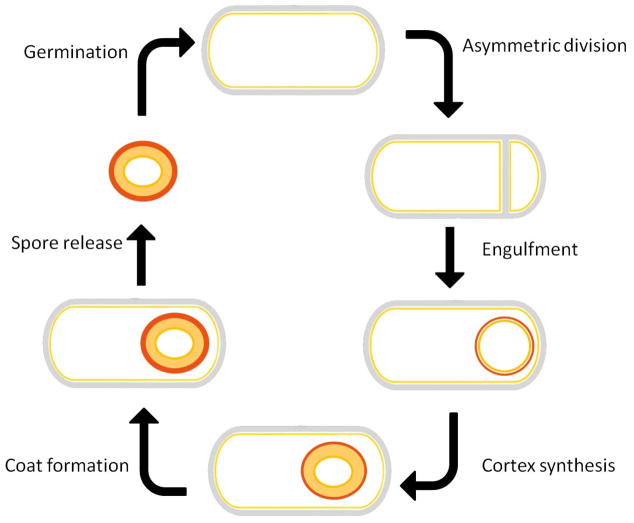
Figure 1. B. subtilis sporulation.
Upon nutrient limitation, B. subtilis ceases growth and initiates sporulation. An asymmetric cell division generates a smaller cell and a larger cell. Following completion of DNA segregation, the larger cell (the mother cell) proceeds to engulf the smaller cell (the forespore). During the initiation of spore metabolic dormancy and compaction of the spore DNA, the mother cell mediates the development of the forespore into the spore through the production of the spore cortex and the inner and outer coat. Upon completion of this process, the mother cell lyses, releasing the mature spore. When the dormant spore encounters an appropriate environmental stimulus, it initiates the process of germination that can result in the re-initiation of vegetative growth if sufficient nutrients are present.
Spores look very different from growing cells. This morphological differentiation initiates with an asymmetric division near to one pole of the cell, resulting in the formation of a smaller cell, the forespore and a larger cell, the mother cell. Recent evidence indicates, in contrast with earlier reports, that this septation can occur near either the “old” or the “new” pole (Veening, et al., 2008). The sporulation septum is similar, but not identical to the normal mid-cell division septum, and contains a thinner layer of peptidoglycan separating the two compartments. Numerous proteins are specifically localized to this septum, some of which are present on only one side of the septum. Following completion, the septum begins to curve and, eventually, the smaller forespore becomes wholly contained within the mother cell (Fig. 2A). We discuss recent progress in characterizing the mechanism underlying this process in Engulfment and in particular the role of specific protein-protein contacts that mediate the interactions between the forespore and the mother cell membrane. Given that these morphogenetic processes occur at a particular place in the cell, a critical question is how the proteins involved in these processes are properly targeted. We discuss recent progress in identifying mechanisms responsible for this targeting in Protein localization. Finally, during the later stages of spore development, the inner and outer proteinaceous layers of the spore are assembled and the spore cortex, consisting of a thick layer of peptidoglycan contained between the inner and outer spore membranes, is synthesized. Progress in defining the organization of the spore coat and the biochemical processes underlying cortex synthesis is described in Spore morphogenesis.
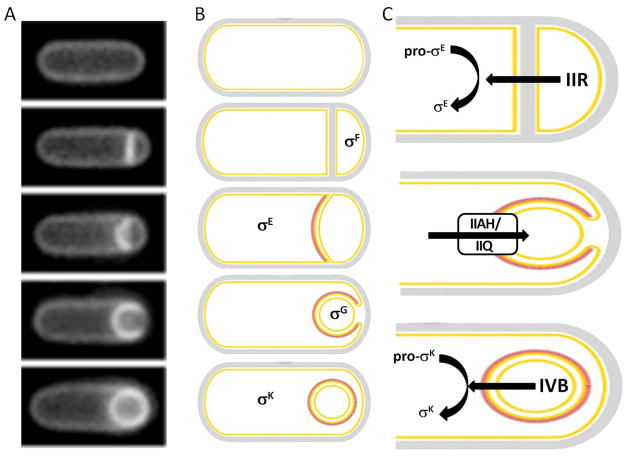
Figure 2. Morphological and genetic asymmetry during sporulation.
(A) Morphological asymmetry. Following entry into sporulation, the cells form an asymmetric septum, which then proceeds to curve out; it eventually becomes entirely distorted and surrounds the smaller cell. This process of engulfment is completed by the loss of attachment between the forespore and the mother cell. (B) Genetic asymmetry. These morphological changes are accompanied by activation of specific transcription factors in each compartment. Initially, σF is activated only in the forespore, followed by σE in the mother cell, σG in the forespore, and finally σK in the mother cell. (C) Intercellular signaling cascade underlying genetic asymmetry. (i) The SpoIIR protein that is under control of σF is produced and secreted by the forespore and activates proteolytic processing of pro-σE→σE in the mother cell. (ii) An as yet unknown signal, produced in the mother cell under control of σE, enters the forespore presumably via the SpoIIIAH/SpoIIQ pore and activates σG. (iii) The SpoIVB protein produced under the control of σG in the forespore activates pro-σK→σK proteolytic processing in the mother cell via an interaction with the SpoIVFA/SpoIVFB/BofA complex
Changes in gene expression underlie morphological differentiation in both the pre-divisional sporangium and later in the two compartments (Fig. 2B). SpoOA, a master transcription factor whose phosphorylation state governs its ability to bind to promoters and thereby regulate gene expression, drives these events (Fawcett, et al., 2000, Molle, et al., 2003). SpoOA phosphorylation is mediated by a phosphorelay composed of phosphatases and kinase-inhibitory proteins which transfers phosphates from several kinases that are assumed to respond to environmental cues (Jiang, et al., 2000). In Entry into sporulation, we describe some of the recent contributions to this literature, and in particular focus on the identification of possible environmental stimuli for kinase activity as well as the relationship between levels of SpoOA-P and the decision to initiate sporulation.
After septum formation, a “criss-cross” mechanism, beginning with following activation of the first compartment-specific transcription factor, σF in the forespore, governs the sequential activation of three other compartment-specific transcription factors (Kroos, 2007). Therefore the proper activation of σF is critical for all subsequent spore development and complex mechanisms are involved in this key checkpoint (Hilbert & Piggot, 2004). These mechanisms include volume differences between the forespore and the mother cell (Iber, et al., 2006, Igoshin, et al., 2006) and preferential localization to the forespore face of the sporulation septum of the SpoIIE membrane phosphatase that is key to the activation of σF (Guberman, et al., 2008). Furthermore, the formation of the polar sporulation septum results in a period of transient chromosomal asymmetry where the smaller forespore contains ~1/3 of the oriC-proximal part of the chromosome. This chromosome asymmetry transiently excludes the gene encoding the proteolytically unstable SpoIIAB anti-σF protein from the forespore, as this gene is located near the chromosome. Thus, genetic asymmetry directly contributes to specific activation of σF in the forespore (Dworkin, 2003) and the expression of ~50 genes under σF control (Wang, et al., 2006). In DNA translocation, we focus on recent progress in characterizing SpoIIIE, the polytopic membrane protein responsible for DNA segregation into the forespore.
Chromosomal asymmetry is also important for the correct activation of σE, the first mother-cell specific transcription factor. The forespore expressed SpoIIR protein crosses the first membrane of the polar septum and leads to the activation of σE and the subsequent expression of ~300 genes (Eichenberger, et al., 2004) (Fig. 2C, top). The spoIIR gene is located very near the origin of replication and this position plays an important role in appropriate σE activation (Khvorova, et al., 2000, Zupancic, et al., 2001). This mechanism was exploited in a recent study in which the timing of spoIIR expression was perturbed by placing it at ectopic chromosomal loci, resulting in the production of cells that contained “twin” spores (Eldar, et al., 2009). While activation of σG in the forespore requires a signal generated in the mother cell (Fig. 2C, middle), recent evidence, described later in Cell-cell communication, extends our understanding of this interaction and, further, has begun to characterize how the mother cell “nurses” the forespore. Finally, σG is responsible for the proper activation of σK, the final mother cell transcription factor (Fig. 2C, bottom), and we discuss recent advances in characterizing this inter-compartmental signaling cascade. Interestingly, the organization of the compartment-specific sigma-factor regulatory network and the genes comprising this network are largely conserved in multiple endospore-forming species, in contrast with the structural sporulation genes (de Hoon, et al., 2010).
1. Entry into sporulation
Kinases
Phosphorylation of the transcription factor SpoOA governs the decision to initiate sporulation. The extent of SpoOA phosphorylation determines a range of physiological outcomes from the development of biofilms and cannibalism (when SpoOA is phosphorylated at lower levels) to sporulation (when SpoOA is phosphorylated at higher levels). Here, we focus primarily on the role of SpoOA in triggering sporulation (Fig.1) and direct the reader to a recent review that describes its role in other post-exponential processes (Lopez, et al., 2009).
KinA is the major kinase responsible for initiation of sporulation and KinA (or KinB) overexpression during exponential growth is sufficient to induce entry into sporulation (Fujita & Losick, 2005). In fact, inducing KinA synthesis beyond a certain level leads to entry into sporulation regardless of nutrient availability (Eswaramoorthy, et al., 2010). KinA contains three PAS domains: PAS-A, PAS-B, and PAS-C (from amino to carboxyl terminus). PAS domains sense a variety of stimuli including oxygen, redox potential, and light (Taylor & Zhulin, 1999). The KinA PAS-A domain binds ATP, which led to the hypothesis that KinA directly senses shifts in the available ATP pool (Stephenson & Hoch, 2001). However, recent studies have shown that no single KinA PAS domain is essential to induce sporulation (Eswaramoorthy, et al., 2009, Eswaramoorthy & Fujita, 2010, Nguele, et al., 2010). Furthermore, the loss of PAS-A has an essentially negligible effect on sporulation induction while the loss of either of the other two domains has a greater impact (Eswaramoorthy & Fujita, 2010). While KinA can clearly initiate sporulation (Eswaramoorthy, et al., 2010), the specific physiological signal(s) that stimulate KinA remain unclear.
The effect of a kinC mutation on sporulation is weaker than that of kinA or kinB. KinC responds to small-molecule-directed potassium leakage; this response depends upon its PAS sensory domains (Lopez, et al., 2009) and leads directly to SpoOA phosphorylation (Lopez, et al., 2009c). While a kinC mutant has a mild sporulation delay/defect, stimulation of KinC has not been shown to induce to sporulation. In contrast to KinA, B, and C, KinD was recently shown to delay the onset of sporulation under certain conditions as a kinD mutant proceeds to sporulation more quickly than a wild-type cell (Aguilar, et al., 2010). It is not known how KinD affects SpoOA though it has been hypothesized to function as a phosphatase and thereby inhibit the initiation and/or progression of sporulation.
Kinase activity is antagonized by regulatory factors, specifically interactions with Sda (see Cell cycle cues) and KipI. The C-terminal region of KinA is comprised of a dimerization/histidine-phosphotransfer (DHp) domain, containing the active auto-phosphorylated histidine residue, and a catalytic ATP-binding (CA) domain. Overexpression of KipI blocks KinA autophosphorylation but does not affect phosphate transfer to the phosphorelay in vitro (Wang, et al., 1997). KipA, whose gene is located in the same operon as kipI, inhibits KipI. Since KipI/KipA homologs are widespread in bacteria, they may function as general histidine kinase regulators. A recent crystallographic analysis of a native Thermus thermophilus KipI-KipA fusion protein suggests that the interaction of KipA with KipI blocks the site with which KipI recognizes KinA (Jacques, et al., 2011).
Cell cycle cues
For over forty years, it has been known that entry into sporulation is dependent upon cell cycle-derived cues. A key player in this regulation is Sda, a small protein (46 aa) that inhibits KinA (Burkholder, et al., 2001). NMR studies and mutational analysis revealed how one of its helices interacts with the DHp domain of KinA (Rowland, et al., 2004). Initial low-resolution structural analysis suggested that binding of the small Sda protein to KinA would not likely be able to directly block transfer of phosphate from the KinA CA domain to the active site histidine in its DHp domain though more recent experiments have suggested otherwise (Whitten, et al., 2007, Bick, et al., 2009). The larger KipI antikinase mentioned above binds to the same region as well as neighboring residues due to its greater size (Jacques, et al., 2008). Structural studies implicate possible allosteric effects for both Sda and KipI since KinA appeared “more compact” when bound to either inhibitor (Whitten, et al., 2007, Jacques, et al., 2008). However, inconsistent with such an allosteric mechanism, Sda is capable of preventing the CA domain from phosphorylating the DHp histidine when CA and DHp are divided into separate peptides (Cunningham & Burkholder, 2009). In addition to blocking autophosphorylation, Sda directly inhibits the ability of KinA to transfer phosphate to its target, SpoOF (Cunningham & Burkholder, 2009). A similar Sda-kinase structural interaction and reduction in the ability of the kinase to transfer phosphate downstream was recently observed in studies of Sda and KinB (Bick, et al., 2009).
Expression of sda is induced in order to prevent sporulation under conditions when initiation of DNA replication has been compromised (Burkholder, et al., 2001). Recently, it was shown that pulsed bursts of sda expression occur and are always correlated with the initiation of replication (Veening, et al., 2009), suggesting that Sda is more than a checkpoint for replication initiation. Instead, it inhibits initiation of sporulation during the period when the cell contains actively replicating chromosomes, thereby preventing the formation of inviable polyploid spores. Sda-mediated sporulation inhibition is relieved by the ClpXP protease, a process which depends upon the uncharged residues at the C-terminus of Sda (Ruvolo, et al., 2006). A corresponding mechanism regulating chromosome replication during sporulation is the sporulation-specific SirA (YneE) protein that displaces the replication initiation factor DnaA from the replication origin and thereby inhibits new rounds of replication (Rahn-Lee, et al., 2009, Wagner, et al., 2009).
Phosphatases
While kinase inhibition is one mechanism of phosphorelay regulation, dephosphorylation of kinase substrates also occurs (Fig. 3). The Rap proteins mediate this process and share six 34-residue (tetratricopeptide; TTP) repeats involved in protein-protein interactions (Perego, et al., 1994). RapA, RapB, RapE, and RapH (Smits, et al., 2007) all dephosphorylate SpoOF and thereby reduce the level of SpoOA~P. The recently reported RapH-SpoOF co-crystal structure identifies residues that mediate this interaction (Parashar, et al., 2011). Corresponding biochemical and genetic studies indicate that RapH is not only capable of dephosphorylating SpoOF, but can also sterically block phosphate transfer to and from SpoOF. Indirect evidence suggests that other Raps, notably RapJ and RapK, may be interacting with the phosphorelay as well (Auchtung, et al., 2006). Rap function is mediated by extracellular pentapeptides derived from the products of the phr genes, found adjacent to the rap genes. Phr proteins are cleaved, secreted from the cell, and accumulate in the culture supernatant (Pottathil & Lazazzera, 2003). Following import, the Phr-derived pentapeptides inhibit the activity of their cognate Raps. The rapA-phrA operon is heterogeneously induced in sporulating microcolonies, suggesting that phrA expression may play a role in regulating the heterogeneity underlying the decision to initiate sporulation (Bischofs, et al., 2009).
Figure 3. Activation of SpoOA.
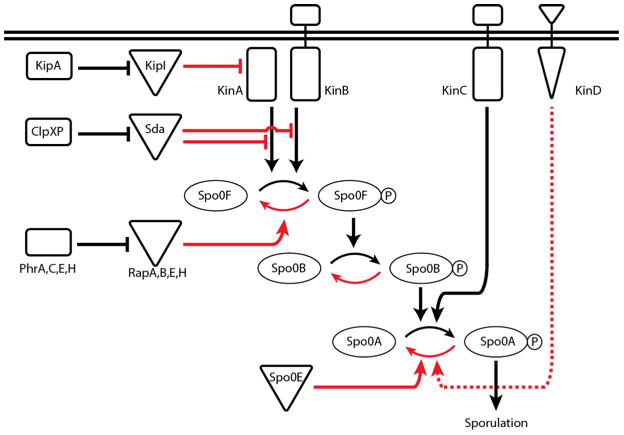
SpoOA phosphorylation is controlled by a phosphorelay comprised of several kinases and numerous intermediaries. Interactions that ultimately work against SpoOA phosphorylation appear in red while interactions that lead towards SpoOA~P are presented in black. At the beginning of the phosphorelay, the KinA and KinB kinases respond to as yet unknown signals by autophosphorylating and subsequently transferring phosphate to SpoOF. SpoOF phosphorylates SpoOB, which passes phosphate on to SpoOA. A third kinase, KinC, bypasses the phosphorelay (i.e. SpoOF and SpoOB) and acts directly on SpoOA. A number of phosphatases (RapA,B,E,&H and SpoOE) and kinase inhibitors (Sda and KipI) act at various steps in the pathway to directly or indirectly antagonize SpoOA phosphorylation. These inhibitors are subject to their own regulation. KinD has been shown to work against SpoOA phosphorylation though it has not yet been determined at which step this regulation occurs (it is therefore graphed as a dashed line). This entire network of interactions is ultimately represented by the phosphorylation state of SpoOA.
Other regulatory mechanisms
The ABC transporter FtsEX also delays sporulation (Garti-Levi, et al., 2008). FtsEX activity is surmountable by an active mutant of SpoOA or by high induction of the phosphorelay (Garti-Levi, et al., 2008). Since FtsEX is a transporter it may function like Rap-Phr circuits in incorporating extracellular cues into the decision to sporulate. Another protein, CodY, senses excess nutrients and prevents sporulation from occurring under these conditions (Ratnayake-Lecamwasam, et al., 2001). It binds GTP as well as branched-chain amino acids (isoleucine, leucine, and valine) to repress the transcription of ~200 genes (Handke, et al., 2008, Villapakkam, et al., 2009) including kinB as well as the operons that encode RapA and RapE and their cognate Phr-repressor peptides (Molle, et al., 2003).
The role of cyclic nucleotides in bacterial cell-cycle control has been the subject of increasing attention in a number of organisms (Duerig, et al., 2009). B. subtilis DisA, which monitors chromosome integrity during entry into sporulation (Bejerano-Sagie, et al., 2006), is a cyclic diadenosine monophosphate (c-di-AMP) synthase (Witte, et al., 2008). The level of ci-di-AMP rises at the onset of sporulation but is reduced by DNA-damaging agents in a DisA-dependent fashion (Oppenheimer-Shaanan, et al., 2011). The rise in ci-di-AMP levels therefore seems to act as a secondary messenger that couples chromosome integrity with progression of sporulation.
SpoOA
The phosphorelay culminates in the phosphorylation of SpoOA. While unphosphorylated SpoOA is inactive, progressive increases in both SpoOA phosphorylation and abundance result in phenotypic differentiation including biofilm development and/or nutrient scavenging (i.e. cannibalism) (Hamon & Lazazzera, 2001, Gonzalez-Pastor, et al., 2003) at moderate levels and sporulation at higher levels (Fujita, et al., 2005). SpoOA is comprised of a phospho-acceptor (receiver) domain and a DNA-binding (effector) domain. SpoOA uses an ‘aromatic-switch’ mechanism of activation where phosphorylation reorients a phenylalanine residue in the receiver domain allowing the effector domain to become active (Muchova, et al., 2004).
SpoOA directly affects expression of ~120 genes with different subsets of genes affected at low and at high levels of SpoOA~P (Molle, et al., 2003, Fujita, et al., 2005). Binding sites of genes that require high amounts of SpoOA~P for induction typically have low affinities whereas many genes induced at low levels of SpoOA~P have binding sites with high affinities. A large number of indirectly induced genes (at low SpoOA~P levels) are regulated via SpoOA~P-mediated inactivation of the repressor protein AbrB (Fujita, et al., 2005). SpoOA inhibits AbrB by two mechanisms including the direct transcriptional repression of abrB as well as the induction of the abbA gene encoding AbbA which inhibits AbrB protein function (Banse, et al., 2008).
Phosphorelay heterogeneity
Important bimodal development decisions are often maintained via a number of feedback mechanisms that reinforce a state of bistability as occurs in B. subtilis competence (Dubnau & Losick, 2006). The phosphorelay has a number of feedback mechanisms, such as the Raps, that would facilitate bistability (Veening, et al., 2005, Smits, et al., 2007). This proposal, however, has been challenged in experiments showing that artificial manipulation of feedback loop components does not increase the percentage of sporulating cells (Chastanet, et al., 2010). Such increases would be expected if any of the feedback loops were functioning to reinforce cells in a non-sporulating state. Furthermore, if the scenario were bistable, a bimodal distribution of SpoOA~P would be expected; however its distribution was always seen as a heterogeneous continuum (Chastanet, et al., 2010). The cellular heterogeneity in the output of the phosphorelay, ultimately resulting in the production of a variety of physiological states (Lopez, et al., 2009), could arise from stochastic fluctuations in individual cells in the levels of components of the phosphorelay (Eswaramoorthy, et al., 2010) or in the transcription of their genes (de Jong, et al., 2010).
2. DNA translocation
A hallmark of sporulation is the uneven distribution of chromosomal DNA between the mother cell and the forespore. Specifically, the asymmetric sporulation septum is almost completed before chromosome segregation is finished, resulting in the forespore initially containing only the origin-proximal one-third of one of the chromosome. The chromosome is attached to the cell pole through the action of the RacA protein (Ben-Yehuda, et al., 2003, Wu & Errington, 2003) that interacts with both the pole-associated protein DivIVA and with specific DNA sequences that are enriched in the origin-proximal region of the chromosome (Ben-Yehuda, et al., 2005). Loss of RacA only results in ~2-fold reduction in sporulation efficiency (Ben-Yehuda, et al., 2003), suggesting that the bacterium is able to orient its chromosomes reasonably well even in the absence of this mechanism. The relatively mild sporulation defect of a racA mutant strain and the fact that racA was first identified as part of a genomic screen of genes under control of SpoOA (Fawcett, et al., 2000) provides further evidence that traditional genetic approaches to sporulation (Piggot & Coote, 1976), while undeniably powerful, can miss relevant genes.
In order for sporulation to proceed, the distal two-thirds of the chromosome must be pumped into the forespore. SpoIIIE, a large polytopic membrane protein belonging to the FtsK family of DNA transporters, is responsible for this translocation. Members of this family have diverse physiological roles including chromosome partitioning during vegetative growth and conjugative transfer of plasmid DNA and are thought to transfer a single double-stranded DNA molecule at a time. Here, we will discuss recent work that has provided new mechanistic detail of how SpoIIIE acts to mediate DNA pumping into the forespore. In addition, SpoIIIE plays a second role during engulfment described in more detail in the Engulfment section below. Although genetic experiments had previously unambiguously demonstrated a role for SpoIIIE in DNA pumping into the forespore, several issues remained: (1) How is the directionality of pumping established and maintained; (2) How are the two arms of the chromosome be pumped simultaneously; and (3) How is the terminus translocated given the topological constraints of a closed circle?
Previously published experiments indicated that the selective assembly of SpoIIIE complexes on the mother-cell side of the septum determined the polarity of translocation (Sharp & Pogliano, 2002). However, alterations in chromosome architecture switched SpoIIIE assembly to the forespore, suggesting that the DNA was dominant (Becker & Pogliano, 2007). The SpoIIIE homolog FtsK is a reversible translocase whose orientation is governed by its DNA substrate, in particular by the presence of specific chromosomal sequence motifs called KOPS (Touzain, et al., 2011). The KOPS motif distribution is highly skewed towards the origin of replication to the terminus facilitating movement of the terminus toward the septum. Analogously, recognition/binding of the KOPS-like sequence SRS is necessary for directional DNA translocation by SpoIIIE (Ptacin, et al., 2008). Thus, the directionality of pumping is likely a direct result of the asymmetric chromosomal distribution of SRS sequences. Additional SpoIIIE studies revealed that it is able to strip all bound proteins off a chromosome during translocation, suggesting that it makes a robust and continuous interaction with DNA beyond the SRS sequences (Marquis, et al., 2008).
Previous models of the SpoIIIE hexameric complex suggested that it formed a single aqueous channel through which both arms of the chromosome were translocated. However, the width of the channel as predicted by the pore size of the ATPase domain seen in the crystal structure of the SpoIIIE homolog FtsK would only be sufficient to allow passage of a single double strand of DNA (Massey, et al., 2006) and considerable evidence exists that the two arms of the chromosome are translocated simultaneously (Becker & Pogliano, 2007, Burton, et al., 2007). The suggestion that there are two SpoIIIE channels (Fig. 4A) offers a possible resolution to this dilemma (Burton, et al., 2007) although this has not been demonstrated.
Figure 4. DNA translocation mediated by SpoIIIE.
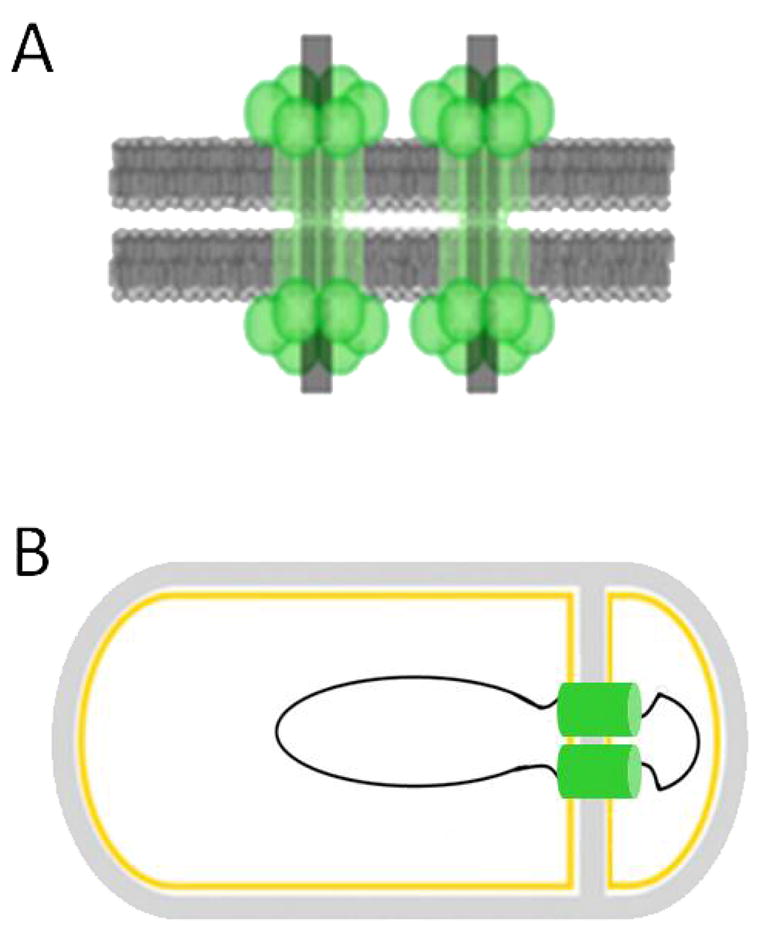
(A) The origin proximal one-third of the chromosome is contained within the forespore immediately following asymmetric septation. The polytopic membrane protein SpoIIIE is necessary for the translocation of the remaining two-thirds of the chromosome into the forespore. (B) SpoIIIE may be organized as a double-barreled DNA translocase, with each pore containing a single arm of the chromosome. Figures adapted from (Becker & Pogliano, 2007, Burton, et al., 2007).
Separation of the termini during sporulation requires SpoIIIE (Bogush, et al., 2007) but it is not known whether SpoIIIE has the ability to translocate the final (closed) loop of the chromosome (Fig. 4B). Recent experiments on DNA translocation during vegetative growth indicate that SpoIIIE and SftA (a protein with significant homology in its DNA translocase domains with both SpoIIIE and FtsK) act coordinately to clear DNA away from the mid-cell (Biller & Burkholder, 2009, Kaimer, et al., 2009). SftA may be involved in chromosome dimer resolution but since it is not needed for sporulation (Biller & Burkholder, 2009), it cannot be responsible for this activity in sporulation. The B. subtilis RipX and CodV recombinases are necessary for vegetative dimer resolution (Sciochetti, et al., 2001) and may play a similar role in sporulation, perhaps by directly interacting with SpoIIIE. A precedent for such a possibility is the interaction of E. coli XerD recombinase and FtsK (Massey, et al., 2004).
3. Engulfment
During sporulation, the cell undergoes a diverse array of morphological changes. One prominent example is engulfment, the process by which the smaller of the two cells resulting from the asymmetric division becomes encased within the larger cell (Fig. 5A). Following the formation of the sporulation septum, the mother cell membranes move and eventually entirely encircle the forespore. While there are superficial similarities to membrane movements that occur during phagocytosis in eukaryotic cells, the analogy may be of limited relevance since in sporulating cells there is a layer of peptidoglycan (the germ cell wall) that surrounds the forespore and separates the two compartments. Initially, this peptidoglycan provides a rigid connection between the two cells. It must, however, be split in order for engulfment to continue past the initial septal bulging and to allow the final separation of the two cells. Thus, peptidoglycan must be modified so as to allow membrane movement. Here, we will describe the recent characterization of peptidoglycan-modifying proteins that are necessary for engulfment. We will also discuss a mechanism that drives membrane movements unidirectionally and appears to mediate the signaling necessary for the activation of the later steps in the developmental program.
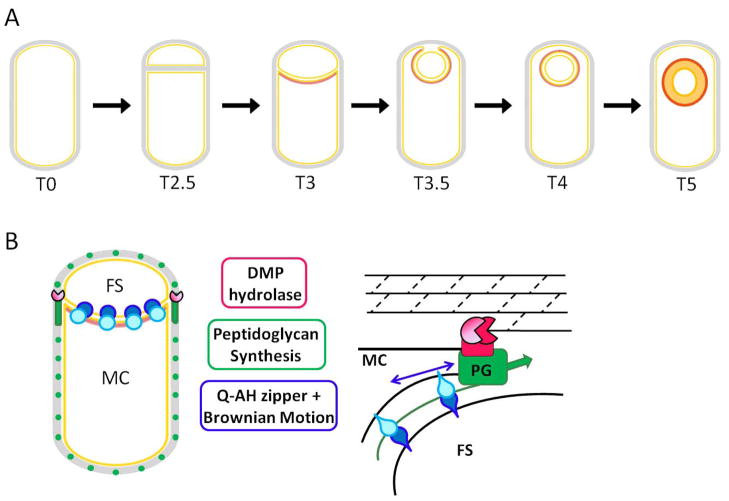
Figure 5. Engulfment.
(A) Following asymmetric septation, the polar septum begins to curve and bend, resulting in its surrounding the forespore. The forespore detaches from the mother cell at the last step of engulfment (T4), which is immediately followed by spore cortex (thick gray band) synthesis. Under each stage is the approximate time in hours since the onset of sporulation at 37oC. (B) Three partially redundant molecular mechanisms are responsible for the membrane movement that occurs during sporulation: (1) The ‘DMP’ machine (red) is responsible for membrane migration by hydrolyzing the peptidoglycan between the two membranes (black lines), thereby moving the mother-cell derived outer forespore membrane around the forespore; (2) the SpoIIQ-SpoIIIAH zipper (blue) may function as a ratchet to irreversibly drive engulfment; and (3) peptidoglycan synthesis (green) of the germ cell wall could provide a motive force for membrane movement during early engulfment as well as the final step of forespore detachment. Figure adapted from (Meyer, et al., 2010).
SpoIIQ and the ‘DMP’ machine
The proteins SpoIID (‘D’), SpoIIM (‘M’) and SpoIIP (‘P’) are located in the outer forespore membrane (facing the mother cell) and are necessary for engulfment. The demonstration that SpoIID is a peptidoglycan hydrolase led to the proposal that it acts as part of a so called ‘DMP’ complex, including SpoIIM and SpoIIP, to hydrolyze the peptidoglycan between the two membranes, thereby moving the mother cell membrane around the forespore (Abanes-De Mello, et al., 2002). Consistent with this model, the activity of SpoIID correlates with the rate of membrane migration. The peptidoglycan located between the membranes is joined with the peptidoglycan that surrounds the combined mother cell and forespore. This rigid connection peptidoglycan must be hydrolyzed in order for engulfment to occur. A candidate for this activity is SpoIIP (Chastanet & Losick, 2007), which is both an amidase and endopeptidase that removes the stem peptides from the cell wall and cleaves their cross-links (Morlot, et al., 2010). The interest in the peptidoglycan-modification activities that occur during engulfment was recently heightened by the observation that sites of active peptidoglycan synthesis track with the engulfing membrane all the way through the final detachment of the forespore into the mother cell cytosol. Compounds that block muropeptide synthesis or polymerization also block membrane fission at the completion of engulfment (Meyer, et al., 2010).
When the septal peptidoglycan of sporulating cells was enzymatically removed by lysozyme, the cells engulfed even in the absence of ‘DMP’ but instead required the forespore protein SpoIIQ and its mother cell partner SpoIIIAH. As described below in Protein Localization, the SpoIIQ and SpoIIIAH membrane proteins interact across the sporulation septum leading to the septal localization of SpoIIQ multimers that track the engulfing mother cell membrane (Rubio & Pogliano, 2004). Since photobleaching indicated that the SpoIIQ multimer does not freely diffuse, it was proposed that SpoIIQ and SpoIIIAH function as a ratchet to irreversibly drive engulfment (Broder & Pogliano, 2006). A model encompassing both this proposal as well as the observations about peptidoglycan hydrolysis suggests that several partially redundant mechanisms operate to ensure successful membrane movement (Fig. 5B). It remains unclear whether these mechanisms spatially and temporally overlap during normal engulfment and it is therefore possible that they participate in distinct steps of this process. For example, the ‘DMP’ machine may be necessary for detaching the septal peptidoglycan from the cell wall peptidoglycan and for initiating the early stages of membrane movement, SpoIIQ/SpoIIIAH may be necessary to drive the majority of membrane movements and peptidoglycan synthesis may be necessary for the final membrane fission event required to finally separate the forespore from the mother cell (Fig. 5B).
Finally, a subject of long-standing interest in sporulation has been the question of when and how the cell becomes committed to sporulation. Specifically, when is the forespore or the mother cell unable to resume growth even in the presence of nutrients? The forespore exhibits rod-like, longitudinal growth in the presence of nutrients only when SpoIIQ and SpoIIP are absent, whereas the mother cell can do so when SpoIIP alone is absent (Dworkin & Losick, 2005). These results indicate that once engulfment is initiated, a process that requires activities in both the forespore and the mother cell, differentiation is rendered irreversible.
SpoIIIE
Some mutations in SpoIIIE result in engulfment defects, and since SpoIIIE mediates chromosome translocation, it was suggested that engulfment and DNA translocation were coupled. However, approximately one-half of sporangia carrying a mutation in the SpoIIIE ATP binding site complete membrane fission during engulfment even though they are defective in DNA translocation (Sharp & Pogliano, 1999) so this coupling, even if present, is not absolute. In addition, a SpoIIIE mutant was identified that prevented the final steps of membrane fission but had no effect on DNA translocation (Liu, et al., 2006). Thus, an unresolved question was the nature of the SpoIIIE engulfment defects. The possibility that SpoIIIE had different roles in septal (early) and engulfment (late) fission was introduced. Specifically, DNA translocation only occurs during septal fission and a model was proposed where the assembly SpoIIIE complexes was the driving force for septal fission (Liu, et al., 2006). The quantitative relationship between the extent of septal membrane fission and the assembly of SpoIIIE complexes (Fleming, et al., 2010) is consistent with this proposal. The disassembly of these complexes was proposed to drive engulfment fission (Liu, et al., 2006) although how this mechanism, if at all, relates to the observed requirement for peptidoglycan synthesis remains unclear (Meyer, et al., 2010).
4. Cell-cell communication
Two distinct cells emerge following completion of asymmetric septation and DNA translocation. These cells will undergo dramatic morphological differentiation that results from unique patterns of gene expression. However, these events do not occur in isolation and a major theme in sporulation research is the characterization of the signaling pathways between the two cells that are required for proper developmental progression (Kroos, 2007). As mentioned in the introduction above, σF activation in the forespore is necessary for the expression of SpoIIR, a secreted protein that crosses one membrane into the space between the septal membranes and stimulates proσE →σE processing in the mother cell. Active σE is necessary for σG activation in the forespore, which, in turn, leads to σK activation in the mother cell. Here we will focus on two issues regarding this inter-cellular communication that have been the subject of much recent progress. First, the identity of the signal as well as the mechanism underlying σG activation has been mysterious. Activation is blocked in a σE mutant, indicating that a mother-cell specific gene product(s) is necessary. In the Feeding tube model, we describe recent progress in understanding the mother cell to forespore signaling in the activation of σG. Second, activation of the mother-cell specific transcription factor σK is dependent on a forespore-generated signal. A key point in this signaling is the processing of pro-σK to σK, its mature, active form, and we describe recent work below in σK processing.
Feeding tube model
Activation of σG in the forespore is dependent on a complex of eight proteins, encoded by the spoIIIAA-AH operon, seven of which are membrane proteins of previously unknown function produced in the mother cell under control of σE. Although each of these proteins is required for σG activation, the identification of an internal promoter in spoIIIAF that is necessary and sufficient for the expression of spoIIIAG and spoIIIAH (Guillot & Moran, 2007) suggested that SpoIIIAG and SpoIIIAH might play particularly important roles in σG activation. In fact, a suppressor mutation that allowed σG activation in the absence of all SpoIIIA proteins except for SpoIIIAH was identified (Camp & Losick, 2008).
The observation that SpoIIIAH is similar to a component of the Type III secretion apparatus led to the hypothesis that mother cell to forespore signaling occurs via a channel that links the two cells (Meisner, et al., 2008). The ability of the C-terminal extracellular domain of SpoIIIAH to be biotinylated by a biotin ligase expressed in the forespore cytoplasm is consistent with the presence of an aqueous channel (Meisner, et al., 2008). Several forespore-expressed anti-sigma factor proteins are known to bind σG (e.g. CsfB (Karmazyn-Campelli, et al., 2008) and SpoIIAB (Chary, et al., 2005)) and so it is possible that, like the export of the anti-σ28 factor FlgM during flagella synthesis in Salmonella (Chevance & Hughes, 2008), export of these proteins through the SpoIIIAH/SpoIIQ channel could result in σG activation. However, neither CsfB nor SpoIIAB are required for the coupling between σE and σG.
Finally, the forespore has been described typically as being “nursed” by the mother cell. It has been unclear, though, how the metabolic activity of the forespore is maintained even though transcription dependent on the primary sigma factor σA still occurs in the forespore at this later stage of sporulation (Steil, et al., 2005). The observation that transcription mediated by a heterologous RNA polymerase (phage T7) is dependent on the presence of the SpoIIIAH/SpoIIQ channel suggests that small molecules necessary for biosynthetic activity of the forespore are imported via this channel (Camp & Losick, 2009, Doan, et al., 2009). In fact, it is possible that these molecules are necessary for σG-dependent transcription, and this would explain the requirement of this channel for σG activation.
σK processing
The final sigma factor to be activated is σK in the mother cell. σK is present as a pro-protein and the processing of pro-σK is mediated by the membrane-embedded metalloprotease SpoIVFB which itself is held inactive by two other membrane proteins SpoIVFA and BofA. This processing has been challenging to study but it has recently been shown that the pro-σK cleavage occurs in a transmembrane segment and depends upon ATP (Zhou, et al., 2009). The forespore-produced protein SpoIVB cleaves the inhibitory protein SpoIVFA. SpoIVFB is thereby activated to process pro-σK (Campo & Rudner, 2006). The specific signal that SpoIVB responds to is unknown, but SpoIVB fails to accumulate when engulfment is perturbed suggesting that it serves as a checkpoint for the progression of engulfment (Doan & Rudner, 2007). This view is somewhat complicated by the report that a second forespore-produced protease CtpB is capable of cleaving SpoIVFA (Zhou & Kroos, 2005, Campo & Rudner, 2006) and can itself serve as a substrate for cleavage by SpoIVB (Campo & Rudner, 2007).
5. Protein localization
As discussed previously, during the process of engulfment, the forespore becomes internalized by the mother cell. A critical issue is how the proteins that mediate this process are targeted to the membranes surrounding the forespore as opposed to the rest of the cytoplasmic membrane. This targeting has been the subject of intensive investigation, and we describe some of the recent progress here, with particular focus on the targeting of the SpoIIIAH protein necessary for inter-compartmental signaling described above in the Feeding tube model (as well as during Engulfment). In addition, as the forespore matures into a spore (described below in Spore morphogenesis), proteins that will comprise the spore coat need to be targeted to the outer forespore membrane. How proteins are able to differentiate intracellular membranes is an issue of general biological relevance (Shapiro, et al., 2009) so we discuss the two different solutions to this problem employed by SpoIIIAH and SpoVM.
Two membranes are found between the mother cell and the forespore: the outer forespore membrane, which starts out as the mother cell side of the polar septum, and the inner forespore membrane, which starts out as the forespore side of the polar septum. Importantly, the outer forespore membrane is initially contiguous with the cytoplasmic membrane so that proteins expressed in the mother cell have access to both membranes. The mechanism of “diffusion and capture” is responsible for keeping at least some proteins in the polar septal membrane, but it has been unclear how the initial anchoring protein is directed to the septum (Rudner, et al., 2002). However, one distinguishing feature of the incipient outer forespore membrane is that it is adjacent to the inner forespore membrane and this feature is used to target the mother cell expressed polytopic SpoIIIAH membrane protein to this membrane. This localization is dependent on SpoIIQ, a forespore-expressed membrane protein, once engulfment happens (Blaylock, et al., 2004, Doan, et al., 2005). The extracellular domains of SpoIIIAH and SpoIIQ interact across the sporulation septum, resulting in the tethering of SpoIIIAH to the sporulation septum as well as its assembly with SpoIIQ into helical arcs and foci around the forespore (Blaylock, et al., 2004) (Fig. 6A). Thus, specific gene expression (σF-dependent spoIIQ) in one compartment leads to asymmetrical protein distribution in another compartment and is another example of how the intrinsic asymmetry of the chromosome is converted to asymmetry in protein localization (Dworkin, 2003).
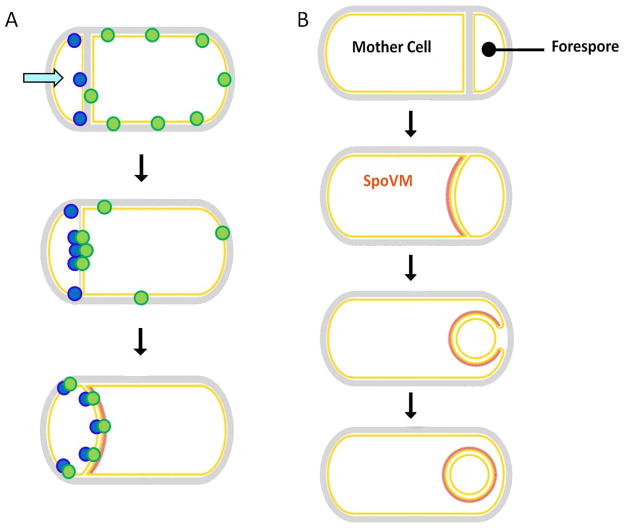
Figure 6. Protein localization during sporulation.
(A) The mother cell expressed protein SpoIIIAH (green) is initially present in the mother cell membrane but becomes progressively enriched at the sporulation septum through an interaction with the forespore expressed SpoIIQ protein (blue) that occurs in the space between the inner and outer sporulation membranes. Figure adapted from (Blaylock, et al., 2004). (B) The SpoVM protein is targeted to the outer forespore membrane because of its high negative curvature. Figure adapted from (Ramamurthi, 2010).
Proper spore coat assembly depends on SpoVM, a 26-amino-acid mother cell-produced peptide that forms an amphipathic helix. A SpoVM-GFP fusion targets exclusively to the outer forespore membrane via hydrophobic, amino acid side-chains on the hydrophobic face of the helix (van Ooij & Losick, 2003), suggesting that the SpoVM helix is oriented parallel to the membrane with the hydrophobic face buried in the lipid bilayer (Ramamurthi, et al., 2006). In addition, SpoVM-GFP produced after engulfment is still targeted to the forespore, even though this membrane was now topologically isolated (Ramamurthi, et al., 2009). One possible mechanism underlying this targeting is an anchoring complex in the outer forespore membrane containing the SpoIVA protein (see below in Spore coat). However, the very small size of SpoVM is problematic for this mechanism. Alternatively, membrane curvature (both positive and negative) has recently been proposed to be a basis for protein localization (Huang, et al., 2006, Mukhopadhyay, et al., 2008). Thus, given that the outer forespore membrane has positive curvature, SpoVM could be detecting this property. To examine this possibility, purified SpoVM was incubated with phospholipid vesicles similar in size to the forespore (Ramamurthi, et al., 2009). The binding of SpoVM was quantitatively related to the vesicle size (curvature) and was reduced by a mutation in the SpoVM amphipathic helix that also abolished localization of SpoVM to the outer forespore membrane in vivo but did not hamper membrane binding (Ramamurthi, et al., 2009) (Fig. 6B).
6. Spore morphogenesis
The B. subtilis spore is a complex structure. The spore core contains the chromosomal DNA that is maintained in a compact state by SASPs (small acid-soluble proteins). The original membrane that surrounded the forespore surrounds the core and the peptidoglycan rich cortex surrounds this membrane. Surrounding the cortex, the spore coat consists of ~80 proteins deposited by the mother cell arranged in inner and outer layers (Fig 7). In some spore-forming species, but not B. subtilis, the spore is surrounded by additional structures such as the exosporium and the S-layer.
Figure 7. Spore structure.
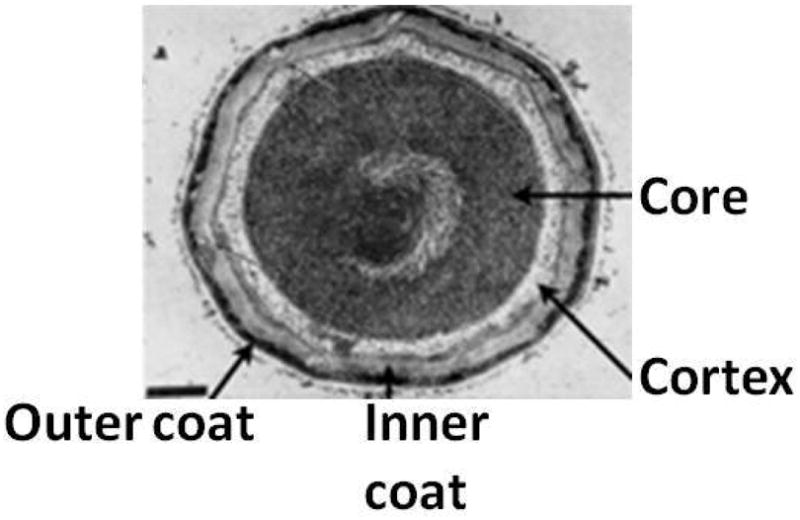
Multiple layers including the spore cortex composed of peptidoglycan and the inner and outer spore coats surround the spore core containing the compacted chromosome. Figure adapted from (Santo & Doi, 1974).
Spore coat
Two important questions regarding the spore coat have been: how is the coat assembled and what are the functions of the coat proteins? Coat proteins are generally not conserved between spore-forming species and their identification has traditionally been technically cumbersome. However, the use of fluorescent microscopy in combination with genomic approaches (Kim, et al., 2006, Takamatsu, et al., 2009) has greatly facilitated the identification of additional coat proteins. A recent extension of this work is the application of sophisticated image analysis to the localization of fluorescent fusions that allowed direct assignment of proteins to a particular coat layer as well as the identification of a previously undescribed layer (McKenney, et al., 2010). In addition, biochemical techniques such as gel filtration are facilitating the characterization of specific interactions in the spore coat (Krajcikova, et al., 2009). Here we will describe work reported since the most recent review on coat assembly(Henriques & Moran, 2007).
The assembling coat is synthesized in the mother cell and is targeted to the outer forespore membrane by SpoIVA (Wang, et al., 2009). SpoIVA binds and hydrolyzes ATP, allowing it to self-assemble into cable-like structures (Ramamurthi & Losick, 2008) that form a basement layer that serves as a platform for coat assembly. Other proteins involved in assembly are SpoVID that directly interacts with SpoIVA (Mullerova, et al., 2009, Wang, et al., 2009) and SafA, which is necessary for the encasement of the spore (Mullerova, et al., 2009). SafA was found to affect the localization of ~16 inner coat protein fusions (McKenney, et al., 2010) substantiating its central role in coat assembly.
The outer coat fails to assemble in a cotE mutant (Zheng, et al., 1988) and two previously identified spore coat proteins, CotC and CotU, were found to interact and assemble around the forming spore in a manner dependent on CotE (Isticato, et al., 2008, Isticato, et al., 2010). Expression of CotE later than normal led to defects in coat formation, but these spores were still lysozyme resistant, suggesting that CotE has functions outside of coat assembly (Costa, et al., 2007). Interestingly, B. anthracis CotE is necessary for exosporium assembly, but has little role in coat protein assembly, demonstrating functional variation of a well-conserved protein in different species (Giorno, et al., 2007).
Several enzymatic activities have been associated with particular coat proteins. For example, the coat protein LipC (YcsK) is a lipolytic enzyme, although its absence did not affect any spore phenotypes except germination in response to L-alanine (Masayama, et al., 2007). Four coat proteins (YtaA, CotS, YutH, YsxE) are members of a new family called “Bacterial Spore Kinases” (BSK) that have significant structural similarity but not homology at the level of primary sequence (Scheeff, et al., 2010) to bacterial eukaryotic-like Ser/Thr kinases (Pereira, et al., 2011). Although this enzymatic activity has not yet been demonstrated and may reflect similarities in substrate binding rather then function, this finding is intriguing and emphasizes the limitations of non-structural homology searches.
One issue in identifying the activity of a particular coat protein (with the exception of proteins necessary for overall coat structure) is that mutations in their respective genes often lack an observable phenotype. In fact, spores of a B. subtilis cotE gerE double mutant that almost entirely lack a spore coat germinated essentially normally and were resistant to wet heat, to mechanical disruption, and to treatment with detergents at an elevated temperature and pH (Ghosh, et al., 2008). One prospect in characterizing phenotypes is to expand the phenotypic assays, as has been done recently with spore digestion by the protozoan Tetrahymena thermophila (Klobutcher, et al., 2006) and the nematode Caenorhabditis elegans (Laaberki & Dworkin, 2008). Both of these studies revealed that the spore coat plays an important function in preventing spore degradation following host ingestion, presumably by preventing access of host muralytic enzymes to the spore cortex. These systems not only offer novel assays of spore structure, but also ecologically relevant assays which may have implications for understanding the biological role of spores.
Spore cortex
Bacterial endospores survive extremes of heat and desiccation because of the dehydrated state of the spore core that is in large part mediated by the cortex. This structure is composed of a form of peptidoglycan that is similar, although not identical, to vegetative peptidoglycan, with the presence of muramic δ–lactam residues resulting in fewer peptide side chains and a concomitant reduction in the cross-linking of the glycan strands (Popham, 2002). As a consequence of engulfment, two distinct membranes, separated by the germ cell wall, a thin layer of peptidoglycan, surround the forespore. The assembly of the spore peptidoglycan then occurs in the space between the two membranes, resulting from the action of genes expressed in the mother cell compartment including one encoding the SEDS (Shape, Elongation, Division and Sporulation) protein SpoVE. During sporulation, ΔspoVE mutants fail to form a cortex and they accumulate cytoplasmic peptidoglycan precursors (Vasudevan, et al., 2007), suggesting a defect at an early step in peptidoglycan polymerization. Consistent with its involvement in spore cortex synthesis, SpoVE localizes to the outer forespore membrane (Real, et al., 2008) and interacts closely both in vivo and in vitro with SpoVD, a σE-controlled penicillin-binding protein that is also required for spore cortex synthesis (Fay, et al., 2010). SpoVD is a target of the thioredoxin-like membrane protein StoA (Liu, et al., 2010), suggesting that the redox state of SpoVD may be a further level of regulation in cortex synthesis in addition to the compartment-specific gene transcription (Eichenberger, 2010).
The intimate interaction with SpoVD and the accumulation of peptidoglycan precursors are consistent with the proposed role of SpoVE (and its SEDS protein homologs FtsW and RodA) as a lipid II translocase. Recent direct biochemical evidence regarding the function of E. coli FtsW supports this conjecture (Mohammadi, et al., 2011). Another protein necessary for spore cortex synthesis is SpoVB, and it was recently proposed that the MurJ family of which SpoVB is a member is necessary for Lipid II translocation (Ruiz, 2008). However, B. subtilis strains lacking all four SpoVB orthologs (including SpoVB) display little or no phenotype during growth (Fay & Dworkin, 2009, Vasudevan, et al., 2009) suggesting that these proteins do not mediate the presumably essential process of Lipid II translocation in this organism.
Recently, B. subtilis spore-cortex peptidoglycan was found to be O-acetylated, a common peptidoglycan modification that reduces sensitivity to the innate immune anti-microbial lysozyme (Laaberki, et al., 2011). However, since lysozyme is unable to penetrate the outer coat (Driks, 1999), this modification would not appear to be useful. O-acetylation of B. anthracis peptidoglycan influences the formation of the S-layer by facilitating the anchoring of one of the major S-layer proteins to the peptidoglycan (Laaberki, et al., 2011), so O-acetylation of the spore cortex peptidoglycan may also mediate attachment of the spore coat to the cortex.
7. Conclusions
This review has highlighted some (but not all!) of the recent progress in understanding B. subtilis sporulation. Historically, experiments aimed at uncovering the processes underlying sporulation have identified important regulatory mechanisms that are also present in non-sporulating bacteria. These include the role of sigma factors in transcription (Sharma & Chatterji, 2010) and protein localization (Rudner & Losick, 2010). The success of these approaches is due to the genetic tractability of B. subtilis, in particular the non-essentiality of sporulation, as well as to the comparatively large size of the bacterium, which greatly facilitates microscopy. For example, a central question in bacterial cell biology is how proteins get targeted to different subcellular sites and the work described above with the membrane targeting of SpoVM will likely have implications for how this occurs in other bacteria(Ramamurthi, 2010). The development of sophisticated image analysis techniques to localize proteins to a particular spore coat layer (McKenney, et al., 2010) should be generally applicable as a method to investigate the organization of large proteinaceous complexes present in many bacteria in addition to more complex cells. Finally, the role of noise (‘stochastic fluctuations’) in various sporulation processes including compartment specific gene activation (Eldar, et al., 2009) has been the subject of intense recent experimental (Chastanet, et al., 2010) and theoretical efforts (Bischofs, et al., 2009); these should continue to serve as fertile grounds for developing synthetic biological approaches to single-cell decision making (Eldar & Elowitz, 2010).
Sporulation can be observed following inoculation of the mouse gut with B. subtilis (Tam, et al., 2006) suggesting that this process may play an important role in the colonization and ecology of the gut microbiota of which spore-forming bacteria are a significant (~50%) component (Ley, et al., 2008). In fact, the recent identification of a spore-forming Clostridia as a major player in the maintenance of immune homeostasis in the intestine (Atarashi, et al., 2011) and the ability of Clostridium difficile spores to survive antibiotic treatment due to their intrinsic lack of sensitivity to antimicrobials indicates that sporulation is a process with great implications for human health and disease (Nerandzic & Donskey, 2010).
Acknowledgments
Work on sporulation in our laboratory is supported by the NIH (R01GM081368-04) and by an Irma T. Hirschl Scholar Award to JD. We thank Patrick Eichenberger (NYU) and Frederico Guieros-Filho (USP) for comments.
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD Is a Checkpoint Protein Linking Spore Formation to Extracellular-Matrix Production in Bacillus subtilis Biofilms. MBio. 2010;1 doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188:5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banse AV, Chastanet A, Rahn-Lee L, Hobbs EC, Losick R. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc Natl Acad Sci U S A. 2008;105:15547–15552. doi: 10.1073/pnas.0805203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EC, Pogliano K. Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol Microbiol. 2007;66:1066–1079. doi: 10.1111/j.1365-2958.2007.05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Fujita M, Liu XS, et al. Defining a centromere-like element in Bacillus subtilis by Identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17:773–782. doi: 10.1016/j.molcel.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Bick MJ, Lamour V, Rajashankar KR, Gordiyenko Y, Robinson CV, Darst SA. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J Mol Biol. 2009;386:163–177. doi: 10.1016/j.jmb.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Burkholder WF. The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Mol Microbiol. 2009;74:790–809. doi: 10.1111/j.1365-2958.2009.06893.x. [DOI] [PubMed] [Google Scholar]
- Bischofs IB, Hug JA, Liu AW, Wolf DM, Arkin AP. Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proc Natl Acad Sci U S A. 2009;106:6459–6464. doi: 10.1073/pnas.0810878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogush M, Xenopoulos P, Piggot PJ. Separation of chromosome termini during sporulation of Bacillus subtilis depends on SpoIIIE. J Bacteriol. 2007;189:3564–3572. doi: 10.1128/JB.01949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell. 2007;131:1301–1312. doi: 10.1016/j.cell.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Rudner DZ. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol Cell. 2006;23:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Campo N, Rudner DZ. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor sigmaK in Bacillus subtilis. J Bacteriol. 2007;189:6021–6027. doi: 10.1128/JB.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary VK, Meloni M, Hilbert DW, Piggot PJ. Control of the expression and compartmentalization of (sigma)G activity during sporulation of Bacillus subtilis by regulators of (sigma)F and (sigma)E. J Bacteriol. 2005;187:6832–6840. doi: 10.1128/JB.187.19.6832-6840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet A, Losick R. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol. 2007;64:139–152. doi: 10.1111/j.1365-2958.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- Chastanet A, Vitkup D, Yuan GC, Norman TM, Liu JS, Losick RM. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 2010;107:8486–8491. doi: 10.1073/pnas.1002499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn F. Untersuchungen ueber Bakterien. IV. Beitraege zur Biologie der Bacillen. Beitraege zur Biologie der Planzen. 1876;2:249–276. [Google Scholar]
- Costa T, Serrano M, Steil L, Volker U, Moran CP, Jr, Henriques AO. The timing of cotE expression affects Bacillus subtilis spore coat morphology but not lysozyme resistance. J Bacteriol. 2007;189:2401–2410. doi: 10.1128/JB.01353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KA, Burkholder WF. The histidine kinase inhibitor Sda binds near the site of autophosphorylation and may sterically hinder autophosphorylation and phosphotransfer to SpoOF. Mol Microbiol. 2009;71:659–677. doi: 10.1111/j.1365-2958.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr Biol. 2010;20:R735–745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong IG, Veening JW, Kuipers OP. Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. J Bacteriol. 2010;192:2053–2067. doi: 10.1128/JB.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Rudner DZ. Perturbations to engulfment trigger a degradative response that prevents cell-cell signalling during sporulation in Bacillus subtilis. Mol Microbiol. 2007;64:500–511. doi: 10.1111/j.1365-2958.2007.05677.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr, Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, et al. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J. Transient genetic asymmetry and cell fate in a bacterium. Trends Genet. 2003;19:107–112. doi: 10.1016/S0168-9525(02)00046-X. [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121:401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Eichenberger P. The red-ox status of a penicillin-binding protein is an on/off switch for spore peptidoglycan synthesis in Bacillus subtilis. Mol Microbiol. 2010;75:10–12. doi: 10.1111/j.1365-2958.2009.06963.x. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Fujita M, Jensen ST, et al. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Chary VK, Xenopoulos P, et al. Partial penetrance facilitates developmental evolution in bacteria. Nature. 2009;460:510–514. doi: 10.1038/nature08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P, Fujita M. Systematic domain deletion analysis of the major sporulation kinase in Bacillus subtilis. J Bacteriol. 2010;192:1744–1748. doi: 10.1128/JB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P, Guo T, Fujita M. In vivo domain-based functional analysis of the major sporulation sensor kinase, KinA, in Bacillus subtilis. J Bacteriol. 2009;191:5358–5368. doi: 10.1128/JB.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P, Dinh J, Duan D, Igoshin OA, Fujita M. Single-cell measurement of the levels and distributions of the phosphorelay components in a population of sporulating Bacillus subtilis cells. Microbiology. 2010;156:2294–2304. doi: 10.1099/mic.0.038497-0. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Duan D, Dinh J, Dravis A, Devi SN, Fujita M. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J Bacteriol. 2010;192:3870–3882. doi: 10.1128/JB.00466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Dworkin J. Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J Bacteriol. 2009;191:6020–6028. doi: 10.1128/JB.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Meyer P, Dworkin J. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J Mol Biol. 2010;399:547–561. doi: 10.1016/j.jmb.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TC, Shin JY, Lee SH, Becker E, Huang KC, Bustamante C, Pogliano K. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes Dev. 2010;24:1160–1172. doi: 10.1101/gad.1925210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator SpoOA. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the SpoOA regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garti-Levi S, Hazan R, Kain J, Fujita M, Ben-Yehuda S. The FtsEX ABC transporter directs cellular differentiation in Bacillus subtilis. Mol Microbiol. 2008;69:1018–1028. doi: 10.1111/j.1365-2958.2008.06340.x. [DOI] [PubMed] [Google Scholar]
- Ghosh J, Larsson P, Singh B, et al. Sporulation in mycobacteria. Proc Natl Acad Sci U S A. 2009;106:10781–10786. doi: 10.1073/pnas.0904104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Setlow B, Wahome PG, Cowan AE, Plomp M, Malkin AJ, Setlow P. Characterization of spores of Bacillus subtilis that lack most coat layers. J Bacteriol. 2008;190:6741–6748. doi: 10.1128/JB.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno R, Bozue J, Cote C, et al. Morphogenesis of the Bacillus anthracis spore. J Bacteriol. 2007;189:691–705. doi: 10.1128/JB.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: Noise and Asymmetry in Bacterial Division Revealed by Computational Image Analysis at Sub-Pixel Resolution. PLoS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot C, Moran CP., Jr Essential internal promoter in the spoIIIA locus of Bacillus subtilis. J Bacteriol. 2007;189:7181–7189. doi: 10.1128/JB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor SpoOA is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Handke LD, Shivers RP, Sonenshein AL. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol. 2008;190:798–806. doi: 10.1128/JB.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iber D, Clarkson J, Yudkin MD, Campbell ID. The mechanism of cell differentiation in Bacillus subtilis. Nature. 2006;441:371–374. doi: 10.1038/nature04666. [DOI] [PubMed] [Google Scholar]
- Igoshin OA, Price CW, Savageau MA. Signalling network with a bistable hysteretic switch controls developmental activation of the sigma transcription factor in Bacillus subtilis. Mol Microbiol. 2006;61:165–184. doi: 10.1111/j.1365-2958.2006.05212.x. [DOI] [PubMed] [Google Scholar]
- Isticato R, Pelosi A, De Felice M, Ricca E. CotE binds to CotC and CotU and mediates their interaction during spore coat formation in Bacillus subtilis. J Bacteriol. 2010;192:949–954. doi: 10.1128/JB.01408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato R, Pelosi A, Zilhao R, Baccigalupi L, Henriques AO, De Felice M, Ricca E. CotC-CotU heterodimerization during assembly of the Bacillus subtilis spore coat. J Bacteriol. 2008;190:1267–1275. doi: 10.1128/JB.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, Langley DB, Jeffries CM, Cunningham KA, Burkholder WF, Guss JM, Trewhella J. Histidine kinase regulation by a cyclophilin-like inhibitor. J Mol Biol. 2008;384:422–435. doi: 10.1016/j.jmb.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Jacques DA, Langley DB, Hynson RM, Whitten AE, Kwan A, Guss JM, Trewhella J. A novel structure of an antikinase and its inhibitor. J Mol Biol. 2011;405:214–226. doi: 10.1016/j.jmb.2010.10.047. [DOI] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Kaimer C, Gonzalez-Pastor JE, Graumann PL. SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis. Mol Microbiol. 2009;74:810–825. doi: 10.1111/j.1365-2958.2009.06894.x. [DOI] [PubMed] [Google Scholar]
- Karmazyn-Campelli C, Rhayat L, Carballido-Lopez R, Duperrier S, Frandsen N, Stragier P. How the early sporulation sigma factor sigmaF delays the switch to late development in Bacillus subtilis. Mol Microbiol. 2008;67:1169–1180. doi: 10.1111/j.1365-2958.2008.06121.x. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Chary VK, Hilbert DW, Piggot PJ. The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J Bacteriol. 2000;182:4425–4429. doi: 10.1128/jb.182.16.4425-4429.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hahn M, Grabowski P, et al. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol. 2006;59:487–502. doi: 10.1111/j.1365-2958.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Ragkousi K, Setlow P. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc Natl Acad Sci U S A. 2006;103:165–170. doi: 10.1073/pnas.0507121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch R. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beiträge zur Biologie der Pflanzen. 1876;2:277–231. [Google Scholar]
- Krajcikova D, Lukacova M, Mullerova D, Cutting SM, Barak I. Searching for protein-protein interactions within the Bacillus subtilis spore coat. J Bacteriol. 2009;191:3212–3219. doi: 10.1128/JB.01807-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu Rev Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- Laaberki M-H, Dworkin J. The role of spore coat proteins in the resistance of B. subtilis spores to C. elegans predation. J Bacteriol. 2008;190:6197–6203. doi: 10.1128/JB.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laaberki MH, Pfeffer J, Clarke AJ, Dworkin J. O-Acetylation of peptidoglycan is required for proper cell separation and S-layer anchoring in Bacillus anthracis. J Biol Chem. 2011;286:5278–5288. doi: 10.1074/jbc.M110.183236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NJ, Dutton RJ, Pogliano K. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol. 2006;59:1097–1113. doi: 10.1111/j.1365-2958.2005.05004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Carlsson Moller M, Petersen L, Soderberg CA, Hederstedt L. Penicillin-binding protein SpoVD disulphide is a target for StoA in Bacillus subtilis forespores. Mol Microbiol. 2010;75:46–60. doi: 10.1111/j.1365-2958.2009.06964.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis KA, Burton BM, Nollmann M, Ptacin JL, Bustamante C, Ben-Yehuda S, Rudner DZ. SpoIIIE strips proteins off the DNA during chromosome translocation. Genes Dev. 2008;22:1786–1795. doi: 10.1101/gad.1684008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masayama A, Kuwana R, Takamatsu H, Hemmi H, Yoshimura T, Watabe K, Moriyama R. A novel lipolytic enzyme, YcsK (LipC), located in the spore coat of Bacillus subtilis, is involved in spore germination. J Bacteriol. 2007;189:2369–2375. doi: 10.1128/JB.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey TH, Aussel L, Barre FX, Sherratt DJ. Asymmetric activation of Xer site-specific recombination by FtsK. EMBO Rep. 2004;5:399–404. doi: 10.1038/sj.embor.7400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eskandarian HA, et al. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol. 2010;20:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci U S A. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Gutierrez J, Pogliano K, Dworkin J. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol. 2010;76:956–970. doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi R, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011 doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. The SpoOA regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchova K, Lewis RJ, Perecko D, et al. Dimer-induced signal propagation in SpoOA. Mol Microbiol. 2004;53:829–842. doi: 10.1111/j.1365-2958.2004.04171.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullerova D, Krajcikova D, Barak I. Interactions between Bacillus subtilis early spore coat morphogenetic proteins. FEMS Microbiol Lett. 2009 doi: 10.1111/j.1574-6968.2009.01737.x. [DOI] [PubMed] [Google Scholar]
- Nerandzic MM, Donskey CJ. Triggering germination represents a novel strategy to enhance killing of Clostridium difficile spores. PLoS One. 2010;5:e12285. doi: 10.1371/journal.pone.0012285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguele JC, Eswaramoorthy P, Bhattacharya M, Ngou-Milama E, Fujita M. Genetic and biochemical analyses of sensor kinase A in Bacillus subtilis sporulation. Genet Mol Res. 2010;9:573–590. doi: 10.4238/vol9-1gmr758. [DOI] [PubMed] [Google Scholar]
- Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V, Mirouze N, Dubnau D, Neiditch MB. Structural Basis of Response Regulator Dephosphorylation by Rap Phosphatases. PLoS Biol. 2011;9:e1000589. doi: 10.1371/journal.pbio.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Sabja D, Setlow P, Sarker MR. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2011;19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Coote JG. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell Mol Life Sci. 2002;59:426–433. doi: 10.1007/s00018-002-8435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci. 2003;8:d32–45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Nollmann M, Becker EC, Cozzarelli NR, Pogliano K, Bustamante C. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat Struct Mol Biol. 2008;15:485–493. doi: 10.1038/nsmb.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn-Lee L, Gorbatyuk B, Skovgaard O, Losick R. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J Bacteriol. 2009;191:3736–3739. doi: 10.1128/JB.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS. Protein localization by recognition of membrane curvature. Curr Opin Microbiol. 2010;13:753–757. doi: 10.1016/j.mib.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell. 2008;31:406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real G, Fay A, Eldar A, Pinto SM, Henriques AO, Dworkin J. Determinants for the subcellular localization and function of a nonessential SEDS protein. J Bacteriol. 2008;190:363–376. doi: 10.1128/JB.01482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland SL, Burkholder WF, Cunningham KA, Maciejewski MW, Grossman AD, King GF. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol Cell. 2004;13:689–701. doi: 10.1016/s1097-2765(04)00084-x. [DOI] [PubMed] [Google Scholar]
- Rubio A, Pogliano K. Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. Embo J. 2004;23:1636–1646. doi: 10.1038/sj.emboj.7600171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci U S A. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo MV, Mach KE, Burkholder WF. Proteolysis of the replication checkpoint protein Sda is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis. Mol Microbiol. 2006;60:1490–1508. doi: 10.1111/j.1365-2958.2006.05167.x. [DOI] [PubMed] [Google Scholar]
- Santo LY, Doi RH. Ultrastructural analysis during germination and outgrowth of Bacillus subtilis spores. J Bacteriol. 1974;120:475–481. doi: 10.1128/jb.120.1.475-481.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeff ED, Axelrod HL, Miller MD, Chiu HJ, Deacon AM, Wilson IA, Manning G. Genomics, evolution, and crystal structure of a new family of bacterial spore kinases. Proteins. 2010;78:1470–1482. doi: 10.1002/prot.22663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciochetti SA, Piggot PJ, Blakely GW. Identification and characterization of the dif Site from Bacillus subtilis. J Bacteriol. 2001;183:1058–1068. doi: 10.1128/JB.183.3.1058-1068.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma UK, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev. 2010;34:646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci U S A. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002;295:137–139. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Ghosh J, Islam NM, Dasgupta S, Kirsebom LA. Growth, cell division and sporulation in mycobacteria. Antonie Van Leeuwenhoek. 2010;98:165–177. doi: 10.1007/s10482-010-9446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP, Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol. 2007;65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- Steil L, Serrano M, Henriques AO, Volker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- Stephenson K, Hoch JA. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:15251–15256. doi: 10.1073/pnas.251408398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu H, Imamura D, Kuwana R, Watabe K. Expression of yeeK during Bacillus subtilis sporulation and localization of YeeK to the inner spore coat using fluorescence microscopy. J Bacteriol. 2009;191:1220–1229. doi: 10.1128/JB.01269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam NK, Uyen NQ, Hong HA, et al. The intestinal life cycle of Bacillus subtilis and close relatives. J Bacteriol. 2006;188:2692–2700. doi: 10.1128/JB.188.7.2692-2700.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzain F, Petit MA, Schbath S, El Karoui M. DNA motifs that sculpt the bacterial chromosome. Nat Rev Microbiol. 2011;9:15–26. doi: 10.1038/nrmicro2477. [DOI] [PubMed] [Google Scholar]
- Traag BA, Driks A, Stragier P, et al. Do mycobacteria produce endospores? Proc Natl Acad Sci U S A. 2010;107:878–881. doi: 10.1073/pnas.0911299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, Popham DL. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol. 2007;65:1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x. [DOI] [PubMed] [Google Scholar]
- Vasudevan P, McElligott J, Attkisson C, Betteken M, Popham DL. Homologues of the Bacillus subtilis SpoVB protein are involved in cell wall metabolism. J Bacteriol. 2009;191:6012–6019. doi: 10.1128/JB.00604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Hamoen LW, Kuipers OP. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol Microbiol. 2005;56:1481–1494. doi: 10.1111/j.1365-2958.2005.04659.x. [DOI] [PubMed] [Google Scholar]
- Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapakkam AC, Handke LD, Belitsky BR, Levdikov VM, Wilkinson AJ, Sonenshein AL. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. J Bacteriol. 2009;191:6865–6876. doi: 10.1128/JB.00818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Marquis KA, Rudner DZ. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol Microbiol. 2009;73:963–974. doi: 10.1111/j.1365-2958.2009.06825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Isidro AL, Domingues L, et al. The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol. 2009;74:634–649. doi: 10.1111/j.1365-2958.2009.06886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Grau R, Perego M, Hoch JA. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ST, Setlow B, Conlon EM, et al. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Whitten AE, Jacques DA, Hammouda B, et al. The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J Mol Biol. 2007;368:407–420. doi: 10.1016/j.jmb.2007.01.064. [DOI] [PubMed] [Google Scholar]
- Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. RacA and the Soj-SpoOJ system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol. 2003;49:1463–1475. doi: 10.1046/j.1365-2958.2003.03643.x. [DOI] [PubMed] [Google Scholar]
- Zheng LB, Donovan WP, Fitz-James PC, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- Zhou R, Kroos L. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-sigmaK during Bacillus subtilis development. Mol Microbiol. 2005;58:835–846. doi: 10.1111/j.1365-2958.2005.04870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Cusumano C, Sui D, Garavito RM, Kroos L. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc Natl Acad Sci U S A. 2009;106:16174–16179. doi: 10.1073/pnas.0901455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupancic ML, Tran H, Hofmeister AE. Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol Microbiol. 2001;39:1471–1481. doi: 10.1046/j.1365-2958.2001.02331.x. [DOI] [PubMed] [Google Scholar]