Abstract
Mitochondria accumulate at neuronal and immunological synapses [1, 2] and yeast bud tips [3], and associate with the ER during phospholipid biosynthesis, calcium homeostasis and mitochondrial fission [4, 5]. We show that mitochondria are associated with cortical ER (cER) sheets [6, 7] underlying the plasma membrane in the bud tip, and confirmed that a deletion in YPT11, which inhibits cER accumulation in the bud tip [8], also inhibits bud tip anchorage of mitochondria [9]. Time-lapse imaging reveals that mitochondria are anchored at specific sites in the bud tip. Mmr1p, a member of the DSL1 family of tethering proteins, localizes to punctate structures on opposing surfaces of mitochondria and cER sheets underlying the bud tip, and is recovered with isolated mitochondria and ER. Deletion of MMR1 impairs bud tip anchorage of mitochondria, without affecting mitochondrial velocity or cER distribution. Deletion of the phosphatase PTC1 results in increased Mmr1p phosphorylation, mislocalization of Mmr1p, defects in association of Mmr1p with mitochondria and ER, and defects in bud tip anchorage of mitochondria. These findings indicate that Mmr1p contributes to mitochondrial inheritance as a mediator of anchorage of mitochondria to cER sheets in the yeast bud tip, and that Ptc1p regulates Mmr1p phosphorylation, localization and function.
Results and Discussion
A role for cER in anchorage of mitochondria in the bud tip
In budding yeast, mitochondria must be transported to and retained in the bud tip for mitochondrial quantity and quality control during inheritance and for control of daughter cell lifespan [3, 10]. We obtained evidence that mitochondria are anchored to ER in the yeast bud tip.
By EM, cER is resolved as flattened sacs underlying 28±5% of the plasma membrane (Fig. 1A). 92% of mitochondrial profiles at the cell cortex by EM are closely apposed to or are contacting cER. Thus, apposition of mitochondria to cER occurs 3.3-fold more frequently than would be expected by chance based on the coverage of plasma membrane perimeter by ER. In the bud tip, 71% of mitochondria are in close proximity to cER.
Fig. 1. Interaction of mitochondria with cER sheets at sites of accumulation of mitochondria in the yeast bud tip.
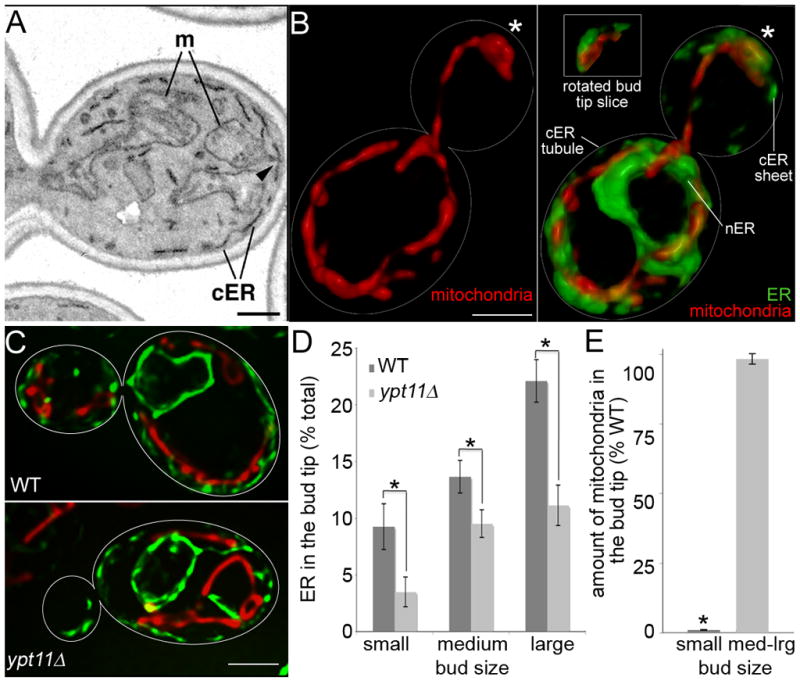
(A) Transmission electron micrograph of the bud and part of the mother of a wild-type cell (BY4741). m, mitochondria; cER, cortical ER. Arrowhead: example of mitochondria under tension at its site of contact with cER in the bud tip. Bar,1 μm. (B) Volume rendering of SIM images of HcRed-labeled mitochondria (red) and Sec63-GFP-labeled ER (green) in wild-type cells (CZY036). Asterisks: bud tip. Inset: slices through the bud tip, rotated to illustrate apposition of mitochondria and cER sheets. Bar, 1 μm. (C) Volume rendering of HcRed-labeled mitochondria and GFP-labeled ER in WT (CZY036) and ypt11Δ (JCY007) cells. Bar,1 μm. (D) Percentage of total cellular ER in the bud tip as a function of bud size in WT and ypt1Δ, as assessed by measuring the fluorescence of Sec63p-GFP in volume renderings of deconvolved images. Small, medium and large buds are <33%, 33–50%, or >50% of the diameter of the mother cell, respectively. Asterisks indicate statistically significant differences between strains (p = 0.028, 0.047 and 0.0002 for small, medium and large buds, respectively). n = 47 (WT) and 54 (ypt11Δ). Error bars are standard error of the mean. (E) The percent of mitochondria in the bud tip of ypt11Δ cells and wild-type cells was determined as for Fig. 1D, using mitochondria-targeted HcRed. The asterisk indicates a statistically significant difference with WT (p = 0.043). n = 47 (WT) and 54 (ypt11Δ). Error bars are standard error of the mean.
In the image shown, a mitochondrion that is closely apposed to cER in the bud tip is deformed into a thin tubular extension from its point of contact with cER (Fig. 1A, arrowhead), implying tension at the point of contact. In such cases, it is clear that mitochondria are not just in close proximity to, but are physically associated with cER. These findings indicate that mitochondria could accumulate in the bud tip by binding, not to the plasma membrane, but rather to cER.
cER forms tubules and sheets [11]. Super-resolution structured illumination microscopy (SIM) reveals mitochondria that are not associated with ER near the bud neck and in the center of the mother cell. SIM also reveals that mitochondria are closely apposed to cER sheets in the bud tip (Fig. 1B).
There is also a functional link between mitochondria and cER in the bud tip. Ypt11p is a Rab-like protein that localizes to cER in the bud and is required for localization of cER at that site [8]. Deletion of YPT11 has no obvious effect on cER morphology (Fig. S1). We and others have described defects in accumulation of ER and mitochondria in ypt11Δ mutants [8, 9, 12]. Here, we directly show defects in both ER and mitochondrial accumulation in the tips of the same small buds (Fig. 1C–E). Reduced bud tip accumulation of ER is observed in buds of all sizes in ypt11Δ cells. In contrast, only small buds of ypt11Δ cells show defective accumulation of mitochondria. The greater severity of the effect on cER suggests that the ER effect is primary. In addition, these results indicate that the absolute amount of ER is not the only factor determining mitochondrial accumulation, as the mitochondrial accumulation occurs in medium- and large-sized buds of ypt11Δ cells. We suggest that the ER contains a limited number of binding sites for mitochondria and that once a sufficient amount of ER is transferred to the bud tip then mitochondria will be able to accumulate to the same degree as in wild-type cells.
A role for Mmr1p in accumulation of mitochondria in the bud tip
Myo2p, a type V myosin, drives movement of numerous cargos from mother cells to buds using actin cables as tracks [13]. Mutation of MYO2 also results in defects in mitochondrial distribution [9, 12, 14, 15]. Indeed, Myo2p that is targeted to mitochondria as an artificial fusion protein can promote mitochondrial motility [16]. Nonetheless, mutations in MYO2 that block or severely impair its force-generating function inhibit anchorage of mitochondria in the bud tip but have no effect on the velocity or frequency of mitochondrial motility [12, 16]. Myo2p also does not a play a direct role in retention of mitochondria in the bud tip [12]. Thus, while it is clear that Myo2p has some role in mitochondrial distribution, the mechanism underlying this process is not well understood.
The protein Mmr1p binds to the Myo2p tail, is required for normal mitochondrial distribution, is recovered with isolated yeast mitochondria and co-localizes with mitochondria in the bud [14, 17]. These findings raise the possibility that Mmr1p is a Myo2p receptor on mitochondria. However, evidence indicates that Mmr1p functions in the bud tip and not in the mother cell where mitochondria are highly motile. Specifically, MMR1 mRNA localizes to the bud tip, and is transported there using the She2p/She3p/Myo4p complex [18]. Moreover, MMR1 protein undergoes Myo2p-dependent localization to the bud tip [14].
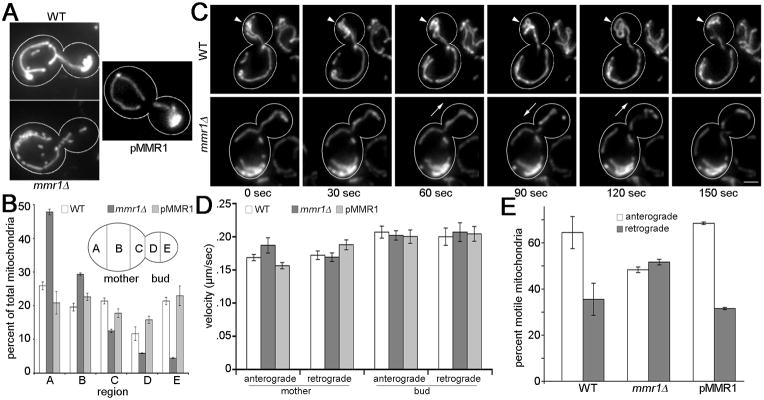
We confirm that mmr1Δ cells have defects in mitochondrial distribution and inheritance (Fig. 2A, Fig. S2). The greatest defect in mitochondrial distribution in mmr1Δ cells is a defect in accumulation of mitochondria in the bud tip (Fig. 2B). Mitochondria in mmr1Δ cells also exhibit an abnormal accumulation in the tip of the mother cell distal to the bud. Since expression of plasmid-borne wild-type MMR1 in mmr1Δ cells restores normal mitochondrial distribution, the bud tip accumulation defect observed in mitochondria of mmr1Δ cells is due to loss of Mmr1p.
Fig. 2. Deletion of MMR1 results in defects in anchorage of mitochondria in the bud tip without affecting the velocity of mitochondrial movement.
(A) GFP-labeled mitochondria in wild-type cells (CZY001), mmr1Δ cells (CZY002), and mmr1Δ cells bearing plasmid-borne MMR1 (pMmr1) (CZY096). Bar = 1 μm. (B) Mitochondrial distribution in different regions of wild-type and mmr1Δ cells was assessed by measuring the integrated intensity of mitochondria-targeted GFP fluorescence in budded cells. Error bars are standard error of the mean. n >100 cells for each strain. (C) Still frames from time-lapse series obtained at 3 sec intervals for 3 min in wild-type (WT) (CZY001) and mmr1Δ (CZY002) cells expressing mitochondria-targeted GFP. Arrowheads: a cortical site in the bud tip that remains associated with a mitochondrial cluster in a wild-type cell. Arrows: changes in the position of mitochondria in the bud tip in an mmr1Δ cell. Numbers shown are times after initial image acquisition. Bar = 1 μm. (D) Anterograde and retrograde mitochondrial velocity in the mother cell and bud in wild-type cells, mmr1Δ mutants and mmr1Δ mutants expressing pMmr1. n > 100. (E) Relative amounts of anterograde and retrograde movement of mitochondria in the bud tip. Error bars are standard error of the mean. n > 100 cells for each strain.
Role for Mmr1p in anchorage of mitochondria in the bud tip
In wild-type cells, bud tip mitochondria are resolved as dynamic clusters that move arbitrarily, but remain associated with a limited number of sites at the bud cortex throughout a 3 min imaging period (Fig. 2C). We find that deletion of MMR1 results in defects in anchorage of mitochondria in the yeast bud tip. Mitochondria in the bud tip of mmr1Δ cells do not remain associated with sites at the bud cortex or exhibit random movement during the 3-min time-course of imaging. Instead, they align along the mother-bud axis and oscillate to and from the bud tip (Fig. 2C).
The velocities of anterograde and retrograde movement in both the mother cell and bud are similar in wild-type cells, mmr1Δ cells and mmr1Δ cells expressing plasmid-borne MMR1 (Fig. 2D). Thus, Mmr1p, like Myo2p, is not required for normal mitochondrial motility in the mother cell or bud. Deletion of MMR1 also has no obvious effect on mitochondrial morphology or on cER ultrastructure or abundance in the bud (Fig. S1). However, the balance of anterograde and retrograde movement in the bud tip is altered in mmr1Δ cells (Fig. 2E). In wild-type cells and mmr1Δ cells expressing wild type MMR1, the ratio of anterograde to retrograde movement in the bud is ~65:35. Thus, for every 2 mitochondria that undergo anterograde movement into the bud tip, roughly one mitochondrion undergoes a retrograde movement out of the bud tip and one mitochondrion remains in the bud tip. In contrast, in mmr1Δ cells, the ratio of anterograde to retrograde mitochondrial movement in the bud is ~50:50. Thus, Mmr1p is required for anchorage of mitochondria in the bud tip and not for mitochondrial movement.
Mmr1p can associate with mitochondria and ER and localizes to opposing surfaces of mitochondria and cER in the bud tip
The Protein families database (Pfam; http://pfam.sanger.ac.uk/) was used to search for Mmr1p homologues. Mmr1p is a member of the DSL1 family of proteins (PF08505; e value: 3.0 e -196). There is 18.5% amino acid identity and 27.2% conserved or semi-conserved amino acid substitutions between Mmr1p and DSL1 (Fig. S3). The similarity between the proteins occurs over the entire sequence and is not limited to the predicted coiled-coil domain [19] near the N terminus of Dsl1p.
DSL1 and its mammalian homolog, syntaxin 18, are components of a multisubunit tethering complex (MTC) that links Golgi-derived COP-1 vesicles to ER during retrograde trafficking. ZW10, which links microtubules to the kinetochore, is also a DSL1 family protein [20]. Thus, Mmr1p has homology to tethering complex proteins. Indeed, previous studies revealed that mutation of Mmr1p in regions highly homologous to DSL1 results in defects in association of Mmr1p with mitochondria [14]. For example, residues 61–91 in Mmr1p, a region that is critical for association of Mmr1p with mitochondria, show 50% similarity to DSL1.
We tested whether Mmr1p localization is consistent with a role in tethering mitochondria to cER sheets in the bud tip. Previous studies indicate that Mmr1p localizes to the bud tip and to mitochondria in the bud [14, 18]. With the improved resolution of SIM, we find Mmr1p on opposing faces of mitochondria and cER sheets in the bud tip (Fig. 3A–B). Moreover, Mmr1p is recovered with mitochondria and ER by subcellular fractionation (Fig. 3C). Since there are no detectable mitochondrial marker proteins in the ER fraction, Mmr1p in the ER fraction is not due to mitochondrial contamination. Thus, Mmr1p has the capacity to associate with both mitochondria and cER and does so at sites where mitochondria are anchored to cER in the bud tip.
Fig. 3. Mmr1p can interact with mitochondria and ER and does so at sites of accumulation of mitochondria on cER in the bud tip.
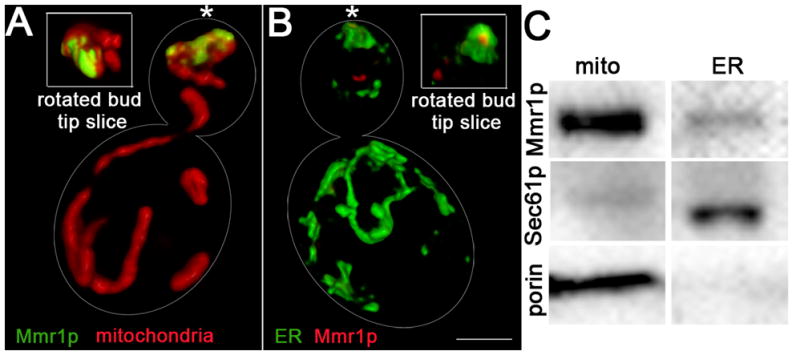
(A) Volume rendering of SIM images showing the localization of HcRed-labeled mitochondria (red) and myc-tagged Mmr1p, visualized by indirect immunofluorescence (green), in wild-type yeast cells (RMY017). Inset: rotated view of the outside of the bud tip. Bar,1 μm. (B) Volume rendering of SIM images of Sec63p-GFP-labeled ER and myc-tagged Mmr1p (red) (RMY016). Localization of Mmr1p in the cER sheet in the bud tip is not evident in the en face view. However, a punctate Mmr1p-containing structure that co-localizes with the medial surface of cER in the bud tip is evident when slices from the bud tip are rotated revealing the medial surface of the cER sheet. (C) Recovery of Mmr1p-GFP and mitochondrial (porin) and ER (Sec61p) marker proteins in subcellular fractions that are enriched in mitochondria (mito) and ER (ER) in wild-type cells with GFP-tagged Mmr1p (TSY4) assessed using western blots and antibodies against porin, Sec61p and GFP. Mmr1p is recovered in the mitochondria fraction and in the ER fraction that has no detectable mitochondrial marker protein.
Deletion of PTC1 results in mislocalization of Mmr1p and defects in accumulation of mitochondria in the bud tip
Ptc1p is a type 2C serine/threonine protein phosphatase that is activated by two mitogen-activated protein kinase (MAPK) pathways [21–23]. It has been implicated in the inheritance of mitochondria, vacuoles, peroxisomes and ER and in the distribution of secretory vesicles and mRNA [24–26]. Deletion of PTC1 also results in failure to localize Myo2p to the bud tip and a reduction in the steady-state levels of Vac17p and Inp2p, proteins that link vacuoles and peroxisomes to Myo2p [26].
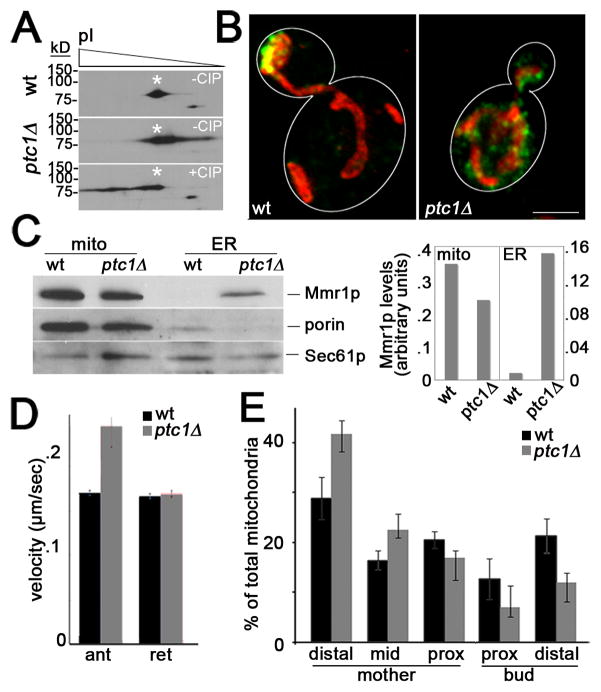
Since Mmr1p is a phosphoprotein [27] (Fig. S4), we tested whether Ptc1p affects Mmr1p phosphorylation, localization and function. We confirmed that deletion of PTC1 results in a 50% reduction in Mmr1p levels and that Mmr1p is a phosphoprotein (Fig. S4). 2D gel analysis also reveals that deletion of PTC1 results in increased phosphorylation of Mmr1p (Fig. 4A). Thus, Ptc1p either directly or indirectly regulates Mmr1p phosphorylation.
Fig. 4. Deletion of PTC1 results in mislocalization of Mmr1p, increased Mmr1p phosphorylation, increased velocity of anterograde mitochondrial movement and defects in anchorage of mitochondria in the bud tip.
(A) Mitochondria were isolated from wild-type cells (CZY057) or ptc1Δ cells (CZY106), each expressing myc-tagged Mmr1p, were analyzed by 2D gel electrophoresis. Deletion of PTC1 results in a shift of the isoelectric point of Mmr1p, consistent with increased phosphorylation of the protein. Treatment of mitochondrial extracts from ptc1Δ cells with calf alkaline phosphatase (CIP), as described in Fig. S4, dephosphorylates and alters the isoelectric point of Mmr1p. (B) Maximum projection of mitochondria (red) and Mmr1p (green) in a wild-type and a ptc1Δ cell. Cell outlines are shown in white. Bar, 1 μm. (C) Mitochondria and ER were isolated from wild-type (CZY057) and ptc1Δ (CZY106) cells expressing myc-tagged Mmr1p, as for Fig. 4. Left panel: The recovery of myc-tagged Mmr1p, mitochondrial (porin) and ER (Sec61p) marker proteins in fractions enriched in mitochondria (mito) and ER was determined as for Fig. 3C. Right panel: Quantitation of Mmr1p recovered in subcellular fractions. Values shown are arbitrary units normalized to recovery of a mitochondrial marker (porin) or an ER marker (Sec61p) for mitochondria and ER fractions, respectively. (D) ptc1Δ cells have increased anterograde mitochondrial velocity. The velocity of anterograde (ant) and retrograde (ret) mitochondrial movement in the mother cell and bud of wild-type (CZY001) and ptc1Δ (CZY089) cells was measured as for Fig. 2. Error bars are standard error of the mean. n >100 cells for each strain. * p<0.002. (E) Mitochondrial distribution in wild-type and ptc1Δ cells was determined as for Fig. 2.
Deletion of PTC1 results in mislocalization of Mmr1p to punctate structures throughout mother cells and buds in ptc1Δ cells (Fig. 4B). It also results in decreased association of Mmr1p with mitochondria, as assessed by visual analysis of Mmr1p in ptc1Δ cells and recovery of Mmr1p with mitochondria by subcellular fractionation (Fig. 4B–C). Interestingly, deletion of PTC1 also results in increased association of Mmr1p with ER, as assessed by recovery of Mmr1p with ER upon subcellular fractionation (Fig. 4C).
Deletion of PTC1, like deletion of MMR1, does not reduce the velocity of anterograde or retrograde mitochondrial movement (Fig. 4D). Indeed, there is a 40% increase in the velocity of anterograde mitochondrial movement. Since deletion of PTC1 impairs the targeting of Myo2p to various cargos [26], it is possible that the reduced density of Myo2p cargos on actin cables in ptc1Δ cells allows mitochondria a less restricted path for movement. Equally important, deletion of PTC1 results in defects in mitochondrial distribution that are similar to those observed in mmr1Δ cells: a decrease in accumulation of mitochondria in the bud tip and an increase in accumulation of mitochondria in the distal tip of the mother cell (Fig. 4E and Fig. S4).
These findings provide additional evidence that Mmr1p functions in the bud tip and has the capacity to bind to both mitochondria and ER. Moreover, it supports the model that PTC1-regulated phosphorylation of Mmr1p affects its localization to the bud tip and its association with ER and mitochondria, potentially to promote binding of mitochondria with ER in the bud tip and not in other areas in the cell.
A model for anchorage of mitochondria in the bud tip in S. cerevisiae
We propose a model for Mmr1p localization and function that accounts for findings from all studies on Mmr1p and Myo2p (see Graphical Abstract). According to this model, Mmr1p localizes to punctate structures that undergo actin-, Myo2p- and Ptc1p-dependent localization to the bud. The movement of Mmr1p in living yeast cells has not been assessed. However, since Myo2p is a motor that drives cargo movement along actin cables in budding yeast, binds directly to Mmr1p, is required for localization of Mmr1p to the bud tip but is not required for normal velocities of mitochondrial movement, we favor the hypothesis that Myo2p plays a major role in mitochondrial distribution as a motor for Mmr1p movement to the bud tip.
Mmr1p in the bud tip mediates binding of mitochondria to cER sheets. This process is also regulated by Ptc1p. Since Mmr1p localizes to opposing surfaces on mitochondria and cER sheets in the bud tip, is recovered with isolated mitochondria and ER, and must be localized to the bud tip for anchorage of mitochondria at that site, Mmr1p has a direct role in binding of mitochondria to cER sheets, which leads to anchorage of mitochondria in the bud tip. Mmr1p is not required for conversion of PE to PS (data not shown). Thus, it is likely that Mmr1p mediates a specific mitochondria-cER interaction that leads to anchorage of mitochondria in the bud tip, and not a general mitochondria-ER interaction that is required for phospholipid biosynthesis.
Overall, these studies indicate that cytoskeletal function and organelle interactions contribute to mitochondrial inheritance through more complex mechanisms than previously appreciated. The cytoskeleton is required for movement of mitochondria and factors that mediate bud tip anchorage of mitochondria from mother cell to bud. Previous studies revealed a role for mitochondrial-ER interactions in phospholipid biosynthesis and calcium buffering. Our study reveals a new role for mitochondrial-ER interactions in anchorage of mitochondria in the yeast bud tip. Finally, proteins have been implicated in linking mitochondria to ER, including Mitofusin 2 in mammalian cells [28] and the ERMES/Mitochore protein complex (Mdm10p, Mmm1p, Mdm12p and Mdm34p) in yeast [29]. Interestingly, Mdm10p is also involved in mitochondrial protein import, morphology, motility, inheritance, DNA maintenance and bud tip accumulation [29–32]. Our study reveals a role for Mmr1p mitochondrial-ER interactions in the yeast bud tip. Since Mmr1p is part of the DSL1 family of tethering proteins, similar processes may contribute to accumulation of mitochondria at sites of polarized cell growth in neurons and cells of the immune system.
Materials and Methods
Analysis of mitochondrial motility and quantitation of the fluorescence of GFP-labeled mitochondria were carried out as described previously [33] and in the supplement.
Supplementary Material
Acknowledgments
We thank the members of the Pon laboratory for technical assistance and valuable discussions, and to Leonardo Peraza for model design. This work was supported by awards from the Amgen Scholars Program to CY, the Ellison Medical Foundation (AG-SS-2465) to LP, the National Institutes of Health (NIH) (GM45735, GM45735S1 and GM096445) to LP and (1 F31 AG034835) to JRMF. GM45735S1 and 1 F31 AG034835 were issued from the NIH under the American Recovery and Reinvestment Act of 2009.
Footnotes
Supplemental data includes materials and methods, four figures, and one table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Quintana A, Schwindling C, Wenning AS, Becherer U, Rettig J, Schwarz EC, Hoth M. T cell activation requires mitochondrial translocation to the immunological synapse. Proc Natl Acad Sci U S A. 2007;104:14418–14423. doi: 10.1073/pnas.0703126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palay SL. Synapses in the central nervous system. J Biophys Biochem Cytol. 1956;2:193–202. doi: 10.1083/jcb.2.4.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264:545–553. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 5.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011 doi: 10.1126/science.1207385. published online Sept. 1, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buvelot Frei S, Rahl PB, Nussbaum M, Briggs BJ, Calero M, Janeczko S, Regan AD, Chen CZ, Barral Y, Whittaker GR, et al. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Mol Cell Biol. 2006;26:7299–7317. doi: 10.1128/MCB.02405-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh T, Watabe A, Toh-E A, Matsui Y. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:7744–7757. doi: 10.1128/MCB.22.22.7744-7757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA. Mitochondrial quality control during inheritance is associated with lifespan and mother–daughter age asymmetry in budding yeast. Aging Cell. 2011 doi: 10.1111/j.1474–9726.2011.00731.x. published online Aug, 7, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boldogh IR, Ramcharan SL, Yang HC, Pon LA. A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol Biol Cell. 2004;15:3994–4002. doi: 10.1091/mbc.E04-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 14.Itoh T, Toh-E A, Matsui Y. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 2004;23:2520–2530. doi: 10.1038/sj.emboj.7600271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altmann K, Frank M, Neumann D, Jakobs S, Westermann B. The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J Cell Biol. 2008;181:119–130. doi: 10.1083/jcb.200709099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Förtsch J, Hummel E, Krist M, Westermann B. The myosin-related motor protein Myo2 is an essential mediator of bud-directed mitochondrial movement in yeast. J Cell Biol. 2011;194:473–488. doi: 10.1083/jcb.201012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frederick RL, Okamoto K, Shaw JM. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 2008;178:825–837. doi: 10.1534/genetics.107.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andag U, Schmitt HD. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J Biol Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- 20.Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schonfisch B, Guiard B, Pfanner N, Meisinger C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin H, Flandez M, Nombela C, Molina M. Protein phosphatases in MAPK signalling: we keep learning from yeast. Mol Microbiol. 2005;58:6–16. doi: 10.1111/j.1365-2958.2005.04822.x. [DOI] [PubMed] [Google Scholar]
- 22.Warmka J, Hanneman J, Lee J, Amin D, Ota I. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol Cell Biol. 2001;21:51–60. doi: 10.1128/MCB.21.1.51-60.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mapes J, Ota IM. Nbp2 targets the Ptc1-type 2C Ser/Thr phosphatase to the HOG MAPK pathway. EMBO J. 2004;23:302–311. doi: 10.1038/sj.emboj.7600036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roeder AD, Hermann GJ, Keegan BR, Thatcher SA, Shaw JM. Mitochondrial inheritance is delayed in Saccharomyces cerevisiae cells lacking the serine/threonine phosphatase PTC1. Mol Biol Cell. 1998;9:917–930. doi: 10.1091/mbc.9.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Y, Walker L, Novick P, Ferro-Novick S. Ptc1p regulates cortical ER inheritance via Slt2p. EMBO J. 2006;25:4413–4422. doi: 10.1038/sj.emboj.7601319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y, Taylor Eves P, Tang F, Weisman LS. PTC1 is required for vacuole inheritance and promotes the association of the myosin-V vacuole-specific receptor complex. Mol Biol Cell. 2009;20:1312–1323. doi: 10.1091/mbc.E08-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep--a database of protein phosphorylation sites in model organisms. Nat Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 29.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang HC, Palazzo A, Swayne TC, Pon LA. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr Biol. 1999;9:1111–1114. doi: 10.1016/s0960-9822(99)80480-1. [DOI] [PubMed] [Google Scholar]
- 31.Boldogh IR, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meisinger C, Wiedemann N, Rissler M, Strub A, Milenkovic D, Schonfisch B, Muller H, Kozjak V, Pfanner N. Mitochondrial protein sorting: differentiation of beta-barrel assembly by Tom7-mediated segregation of Mdm10. J Biol Chem. 2006;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- 33.Fehrenbacher KL, Yang HC, Gay AC, Huckaba TM, Pon LA. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.