Abstract
Tryptophan dioxygenase (TDO) and Indoleamine 2,3 dioxygenase (IDO) are the only two heme-proteins that catalyze the oxidation reaction of tryptophan (Trp) to N-formylkynurenine (NFK). While human IDO (hIDO) is able to oxidize both L and D-Trp, human TDO (hTDO) displays a major specificity towards L-Trp. In this work we aim to interrogate the molecular basis for the substrate stereoselectivity of hTDO. Our previous molecular dynamics simulation studies of Xanthomonas campestris TDO (xcTDO) showed that an H-bond between T254 (T342 in hTDO) and the ammonium group of the substrate is present in the L-Trp-bound enzyme, but not in the D-Trp bound enzyme. The fact that this is the only notable structural alteration induced by the change in the stereo structure of the substrate prompted us to produce and characterize the T342A mutant of hTDO to evaluate the structural role of T342 in controlling the substrate stereoselectivity of the enzyme. The experimental results indicate that the mutation only slightly perturbs the global structural properties of the enzyme, but it totally abolishes the substrate stereoselectivity. Molecular Dynamics simulations of xcTDO show that T254 controls the substrate stereoselectivity of the enzyme by (i) modulating the H-bonding interaction between the NH3+ group and epoxide oxygen of the ferryl/indole 2,3-epoxide intermediate of the enzyme, and (ii) regulating the dynamics of two active site loops, loop250–260 and loop117–130, critical for substrate-binding.
Tryptophan dioxygenase (TDO) is a heme-containing enzyme that catalyzes the conversion of L-Tryptophan (L-Trp) to N-Formyl kynurenine (NFK), which represents the first and rate-limiting step of the L-Trp catabolism through the kynurenine pathway.1 TDO is a ubiquitous enzyme found in bacteria, insects and mammals. In mammals, it is expressed mainly in the liver, where it is responsible for L-Trp processing that ultimately leads to the biosynthesis of NAD and NADP. 2, 3.
Indoleamine 2,3-dioxygenase (IDO) is also a heme-containing enzyme that catalyzes the same oxidation reaction of L-Trp. Contrary to TDO, in mammals IDO is expressed in all tissues other than the liver. Recently, significant efforts have been put forth to unravel the reaction and inhibition mechanisms of IDO and TDO, as promoted by the discovery that IDO plays a pivotal role in cancer immune escape.4, 5, 6,7, 8 Along this line, it has been demonstrated that the combination of a IDO inhibitor, 1-methyl Trp, and cytotoxic chemotherapy leads to significant tumour regression in mouse model systems.4 In this context, it is important to develop potent IDO inhibitors that do not interfere with normal TDO function.
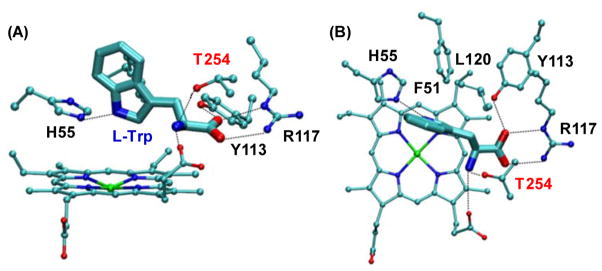
The crystal structure of mammalian TDO is not known; nonetheless those of two bacterial isoforms of TDO, from Xanthomonas campestris (xcTDO, PDB code: 2NW8)9 and Ralstonia metallidurans (rmTDO, PDB code: 2NOX),10 have been published in 2007. In the case of xcTDO, the enzyme was crystallized in both substrate-free and L-Trp-bound forms. In the structure of the L-Trp bound form, several tight contacts between L-Trp and the enzyme are evident (Fig. 1), including H-bonding interactions between the carboxylate group of L-Trp and Y113/R117, the ammonium group of L-Trp, T254 and the propionate A group of the heme, and the indoleamine group of L-Trp and H55, as well as the hydrophobic interactions between L-Trp, F51, L120 and F116 (it is noted that F116 is not shown in Fig. 1 for clarity). Structure-based sequence alignment data suggest that most of the key interactions between the substrate and the enzyme observed in xcTDO are conserved in human IDO (hIDO).9, 11, 12 Consistently, our hybrid Quantum Mechanics and Molecular Mechanics (QM-MM) studies13, 14 showed that xcTDO and hIDO carry out the Trp dioxygenation reaction with a similar ferryl-based mechanism. Based on this mechanism, the heme iron bound dioxygen is first inserted into the C2 atom of Trp to generate a ferryl/indole 2,3-epoxide intermediate. The subsequent ring opening reaction of the epoxide, catalyzed by a proton transfer from the ammonium group to the epoxide, triggers the insertion of the ferryl oxygen to the C2 atom to generate the NFK product. In this scenario, the proper positioning of the ammonium group of the substrate, as well as the electronic environment surrounding it, are believed to be critical for the catalysis.
Figure 1.
Frontal (A) and top (B) views of the active site of L-Trp-bound xcTDO.9 The key residues stabilizing the bound L-Trp are highlighted and labelled in black and red.
Although TDO and IDO exhibits high structural similarity and follow a similar dioxygenase mechanism, they exhibit several intriguing differences in the substrate stereoselectivity: hTDO shows comparable affinities towards L-Trp and D-Trp, but the kcat for L-Trp is 10-fold higher with respect to D-Trp;12 in contrast, hIDO displays ~170-fold higher affinity towards L-Trp with respect to D-Trp15, but the kcat are similar for the two stereoisomers. The structural factors leading to the differences in the substrate stereoselectivity of the two enzymes are unclear. Nonetheless, our previous molecular dynamics (MD) simulation data16 suggest that most of the interactions between the substrate and the enzyme in xcTDO are conserved in the L-Trp and D-Trp-bound complexes, except that the strong H-bond between the ammonium group of L-Trp and the OH group of T254 (equivalent to T342 in hTDO) is significantly weakened in D-Trp-bound enzyme, implying that T254 plays a critical role in controlling the substrate stereoselectivity of xcTDO.
To define the role of T342 in the substrate stereoselectivity of hTDO, we have produced and characterized the T342A mutant by using resonance Raman and optical absorption spectroscopies, and determined its activity towards L-Trp and D-Trp. Our data show that the T342A mutation only causes minor changes to the global structure of the enzyme, but it totally abolishes the substrate stereoselectivity. To unveil the structural perturbations to the enzyme underlying its absence of substrate stereoselectivity, the experimental work was complemented with classical MD simulations. As the crystal structure of hTDO is not available, the simulations were carried out with xcTDO. The simulation data of the L- and D-Trp bound O2 complexes, as well as the corresponding ferryl/indole 2,3-epoxide intermediates, indicate that T254 controls the substrate stereoselectivity of xcTDO by modulating the H-bonding interaction between the ammonium group and epoxide oxygen of the ferryl/indole 2,3-epoxide intermediate, and by regulating the interactions between two critical loops, loop250–260 and loop117–130, that sequester substrate in the active site.
Materials and Methods
Expression and Purification of the wild type hTDO
The details of protein expression and purification are described elsewhere.12, 17 Briefly, the hTDO protein, with N- and C-terminal tails truncated, plus a 6X-His tag extension at the C-terminus, was over-expressed in E. coli BL21 Star (DE3) cells by using the pET30b vector (Stratagene, La Jolla, CA). The transformant was selected from a single colony on a LB kanamycin agar plate and was used to inoculate a LB medium, supplemented with 50 μg/mL kanamycin. This starter culture was grown overnight at 37 °C, and was subsequently used to inoculate a 1 L of LB medium. The resulting E. coli culture was grown at 37 °C in a shaker at 250 rpm until the optical density at 600 nm reached ~0.8. The expression of hTDO was induced by Isopropyl β-D-1-thiogalactopyranoside (IPTG), with a final concentration of 1 mM. An aliquot of hemin, with a final concentration of 8–10 μM, was added to the culture to ensure the complete incorporation of the heme prosthetic group into the recombinant protein. The culture was grown at 25 °C for additional 6–8 hrs in a shaker at 150 rpm. The cells were harvested by centrifugation and stored at −20 °C until use.
The recombinant protein was purified by affinity chromatography with a Ni-NTA column (Novagen). The protein was eluted with 250 mM imidazole (Sigma) in 50 mM phosphate buffer (pH 7.8) and 250 mM KCl. To stabilize the protein, 10 mM L-Trp and 5% glycerol was present throughout the purification procedure. To ensure that the protein was in its ferric state, it was oxidized with potassium ferricyanide, and subsequently passed through a sephadex G25 column to remove the ferricyanide, L-Trp, imidazol and glycerol. The purity of the protein was confirmed by SDS PAGE analysis as shown in Fig. S2. The protein thus collected was stored in 100 mM phosphate buffer (pH 7.5) with 150mM KCl at −80°C until use.
The T342A mutant was created by using the QuikChange II kit (Stratagene) with the following primers: sense primer: 5′-CAGCAAAGCTGGCGCCGGTGGTTCCTC-3′ and antisense primer: 5′-GAGGAACCACCGGCGCCAGCTTTGCTG-3′. The sequence was verified by DNA sequence analysis. The mutant was cloned, expressed, and purified by using the same protocol as that described for the wild type protein.
Steady-State Activity Assay
For the activity assay, the ferric hTDO was rapidly mixed with sodium ascorbate (100-fold excess with respect to the protein) in the presence of a desired amount of L-Trp in pH 7.0 phosphate buffer (100 mM). The temperature was controlled with a water bath at 25 °C. The final concentration of hTDO was 0.5 μM. The reaction rate was followed by monitoring the product formation at 321 nm (ε = 3750 M−1cm−1 for N-formyl kynurenine).12, 18 Two representative kinetic traces obtained with 8 mM L- and D-Trp are shown in Fig. S3. The initial product formation rate was plotted as a function of L-Trp concentration; the data were analyzed by Michealis-Menten curve fitting with Origin 6.1 software (Microcal Software, Inc., MA).
Spectroscopic Measurements
The optical absorption spectra were taken on a spectrophotometer, UV2100, from Shimadzu Scientific Instruments, Inc. (Columbia, MD) with a spectral slit width of 1 nm. The resonance Raman spectra were taken on the instrument described elsewhere.19 Briefly, the 413.1 nm excitation from a Kr ion laser (Spectra Physics, Mountain View, CA) was focused to a 30 μm spot on a spinning quartz cell rotating at 1000 rpm. The scattered light, collected at a right angle to the incident laser beam, was focused on the 100 μm-wide entrance slit of a 1.25 m Spex spectrometer equipped with a 1200 grooves/mm grating (Horiba Jobin Yvon, Edison, NJ), where it was dispersed and then detected by a liquid nitrogen-cooled CCD detector (Princeton Instruments, Trenton, NJ). A holographic notch filter (Kaiser, Ann Arbor, MI) was used to remove the laser line. The Raman shift was calibrated by using indene (Sigma) as a reference. The laser power was kept at ~5 mW for all measurements, except that used for the CO complexes, in which it was kept at <1 mW to avoid photodissociation of the heme-bound CO. The acquisition time was 30 min for all except that of the super high frequency spectra of the CO-bound complexes in which the acquisition time was 180 min. The ferrous derivative was prepared by reducing the ferric protein, prepurged with N2 gas, with sodium dithionite under anaerobic conditions. The CO-bound ferrous complexes were obtained by gentle purging CO gas on the surface of the solution containing the ferrous form of the enzyme under anaerobic conditions. The concentration of the protein samples used for the Raman measurements was 50 μM in pH 7.0 phosphate buffer (100 mM).
Starting structures for classical Molecular Dynamics Simulations
Due to the fact that the crystal structure of hTDO is not available, we have performed the simulations on the xcTDO structure. Sequence comparison of hTDO and xcTDO with 7 other TDOs from various organisms (Fig. S1) indicates that most of the residues involved in Trp binding are conserved, including H76 (55 in xcTDO), R144 (117 in xcTDO), F72 (51 in xcTDO), F143 (116 in xcTDO), L147 (120 in xcTDO), with the exception of Y113 of xcTDO that is replaced by a Phe residue in all the analyzed mammalian and insect sequences. Additionally, the loop250–260 and loop117–130 involved in the observed conformational change in the T254A mutant of xcTDO display high sequence homology, with all relevant residues involved in the interactions conserved. In this context, xcTDO structure provides a good model of the human protein justifying its use for the MD simulations.
The structure of the ternary complex of xcTDO was obtained based on the crystal structure of the ferrous L-Trp-bound xcTDO (PDB code: 2NW8)9, as described eleswhere.13, 16 Briefly, a one-subunit model of xcTDO was constructed by removing residues E19 to S35 from subunit A, and adding residues R21 to S35 of subunit B, which corresponds to a short helix that penetrates into the structure of the A subunit. The structure of D-Trp-bound xcTDO was obtained by a docking protocol described in reference.16 The structure of the ferryl/L or D-indole 2,3-epoxide intermediate of xcTDO was obtained from the QM-MM simulations of the corresponding ternary complexes.13, 20 Starting from the structures of the ternary complexes and the ferryl/indole 2,3-epoxide intermediates of the wild type enzyme, we constructed the corresponding T254A mutant derivatives in-silico. These structures were used for all the MD simulations.
Classical Molecular Dynamics Simulations
The starting structure for each complex, was immersed in a pre-equilibrated octahedral box of TIP3P water molecules. The standard protonation state at physiological pH was assigned to ionizable residues. Special attention was paid to the protonation states of histidines, which were assigned on the basis of the H-bonding patterns with neighboring residues. All simulations were performed at 300 K and pressure of 1 bar using Berendsen thermostat and barostat21. Periodic boundary conditions and Ewald sums (grid spacing of 1 Å) were used to treat long range electrostatic interactions. The SHAKE algorithm was used to keep bonds involving hydrogen atoms at their equilibrium length. A 2 fs time step was used for the integration of Newton’s equations. The Amber ff99SB force field22 was used for all residues but the heme, whose parameters were developed and thoroughly tested by our group in previous works23, 24. The parameters for the ferryl/indole 2,3-epoxide intermediate were obtained using standard Amber procedures, using ab-initio HF calculations and RESP22 method for determining the partial charges. All simulations were performed with the PMEMD module of the AMBER9 package.25 Equilibration consisted of an energy minimization of the initial structures, followed by a slow heating up to 300K. For each structure 20ns MD production runs were performed. Frames were collected at 2ps intervals, which were subsequently used to analyse the trajectories. Finally, in order to estimate the work required to produce the rotation of the Cα-Cβ bond in the ferryl/indole 2,3-epoxide intermediate of the wild type and T254A mutant of xcTDO and make a rough estimation of the free energy profile for the inter-conversion of the two extreme structures, we added a time-dependent potential that moves the system from the initial to the final configuration, resulting E′(r)=E(r)+k[(ξ−ξo)2]26. The constant k was 200 kcal/mol. K in all cases, and the coordinate ξ was chosen as the dihedral angle formed by the N- Cα-Cβ-Cγ atoms of the epoxide adduct (Fig. S9).
Results and Discussion
Optical absorption and resonance Raman spectra of the T342A mutant of hTDO
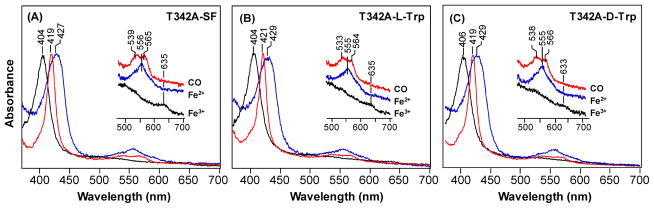
Fig. 2 shows the optical absorption spectra of the T342A mutant in the absence and presence of L-Trp. In the substrate-free form, the spectra of the ferric T342A shows a Soret band at 404 nm, and a charge transfer band at 635 nm, characteristic for a six-coordinate (6C) water-bound ferric heme, similarly to that reported for the wild type enzyme.12 The ferrous derivative has a Soret band at 427 nm, and a visible band at 556 nm, consistent with a five-coordinate (5C) high-spin heme. The CO-bound complex, on the other hand, shows a Soret band at 419 nm, and two visible bands at 539 and 565 nm, confirming the presence of a 6C low-spin heme. Like the wild type enzyme, the addition of L-Trp or D-Trp to the mutant does not introduce significant modifications to the spectra, as summarized in Table 1.
Figure 2.
Optical absorption spectra of the T342A mutant of hTDO in the substrate-free form (SF) (A), L-Trp-bound form (B) or D-Trp-bound form (C). Results for the ferric, ferrous and CO-bound forms are presented in black, blue and red, respectively. All the spectral intensities are normalized based on the Soret band.
Table 1.
The νFe-His, νFe-CO and νC-O modes (cm−1) and electronic transition bands (nm) of the ferric, ferrous and CO-bound ferrous derivatives of the wild type and T342A mutant of hTDO in the substrate free (SF), L-Trp and D-Trp bound forms.
| hTDO | νFe-His | νFe-CO | νC-O | AFerric | AFerrous | ACO | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Wild type | SF | 229 | 496 | 1958 | 406 633 |
432 556 |
420 538, 568 |
12 |
| +L-Trp | 229 | 488 | 1972 | 407 | 433 555 |
421 537, 567 |
12 | |
| +D-Trp | 228 | 490 | n.d. | 406 | 432 558 |
421 569 |
12 | |
| T342A | SF | 227 | 495 | 1963 | 404 635 |
427 556 |
419 539, 565 |
|
| +L-Trp | 227 | 488 | 1971 | 404 635 |
429 555 |
421 533, 564 |
||
| +D-Trp | 227 | 488 | n.d. | 406 633 |
429 555 |
419 538, 566 |
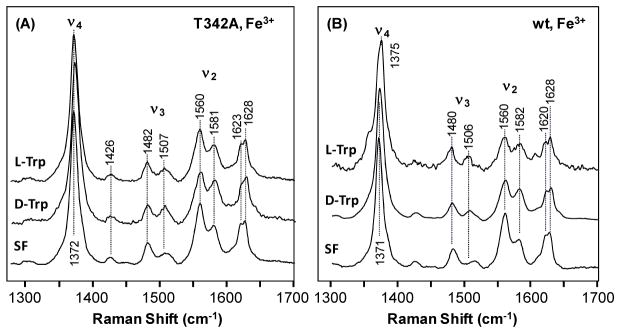
The resonance Raman spectra of the ferric derivatives of the T342A mutant are shown in Fig. 3A. The substrate-free enzyme has a ν4 mode at 1372 cm−1, and ν2/ν3 modes at 1560/1482 and 1581/1507 cm−1, indicating a 6C high-spin/low-spin mixed configuration, in good agreement with a water-bound ferric heme, as indicated by the optical absorption spectral data. The addition of L-Trp or D-Trp to the ferric enzyme does not perturb the spectral feature, except that the low-spin component is slightly increased, similar to that observed in the wild type enzyme (Fig. 3B).
Figure 3.
Resonance Raman Spectra of the ferric derivative of the T342A mutant (A) and wild type (B) hTDO in the substrate-free (SF), L-Trp-bound form or D-Trp bound form. The spectra of the SF and L-Trp-bound form of the wild type enzyme are taken from literature.12 The Trp concentration used for generating the substrate-bound enzyme was 20 mM.
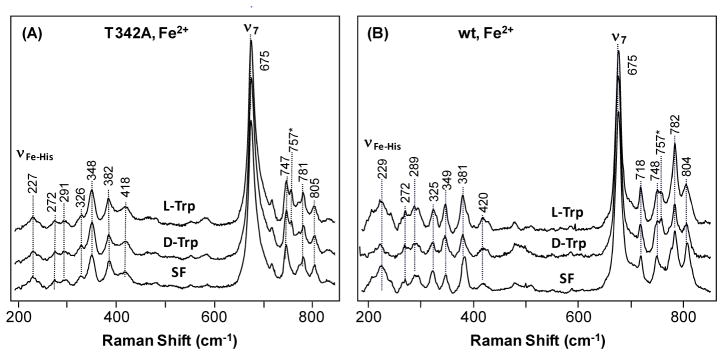
The ferrous derivative of the T342A mutant exhibits ν3 and ν4 modes at 1470 and 1355 cm−1, respectively, in the absence of substrate, indicating a 5C high-spin heme (data not shown), same as the reported data for the wild type enzyme.12 In the low frequency window of the spectrum, the Fe-His stretching mode (νFe–His) is identified at 227 cm−1 (Fig. 4A), similar to that of the wild type enzyme (Fig. 4B and Table 1),12 indicating that the mutation does not perturb the proximal heme environment. Nonetheless, the relative intensities of several in-plane and out-of-plane heme modes in the 250–450 cm−1 and 700–800 cm−1 region of the spectrum17, 20 are significantly perturbed by the mutation, suggesting conformational changes to the heme prosthetic group due to the mutation. L-Trp or D-Trp binding to the mutant does not introduce noticeable changes to the spectrum, in contrast to the small changes observed in the wild type enzyme (Fig. 4B), manifesting slightly weaker substrate-enzyme interactions in the mutant enzyme.
Figure 4.
Resonance Raman Spectra of the ferrous derivative of the T342A mutant (A) and wild type (B) hTDO, in the substrate-free (SF), L-Trp-bound form or D-Trp bound form. The spectra of the SF and L-Trp-bound form of the wild type enzyme are taken from literature.12 The Trp concentration used for generating the substrate-bound enzyme was 20 mM. The peak at 757 cm−1 indicated by an asterisk (*) is derived from Trp substrate.
CO has been demonstrated to be a useful probe for investigating active site structure of heme-proteins. The spectral features of the CO-complex of T342A mutant (Fig. S4) are similar to those reported for the wild type enzyme,12 and are not affected by the addition of L-Trp or D-Trp, expect that the Fe-CO stretching mode (νFe-CO) at 495 cm−1 found in the substrate-free enzyme shifted to 488 cm−1 in responding to L-Trp or D-Trp binding. The assignments of the νFe-CO modes at 495 and 488 cm−1, along with those of the associated C-O stretching modes (νC-O) at 1963 and 1970 cm−1, were confirmed by 12CO-13CO isotope substitution experiments (insert in Fig. S4). As listed in Table 1, in general, all the spectral features, as well as their responses to substrate binding, of the various derivatives of the T342A mutant are similar to those reported for the wild type enzyme,12 confirming that the mutation does not introduce major structural modifications to the enzyme.
Activity of the T342A mutant of hTDO towards L and D-Trp
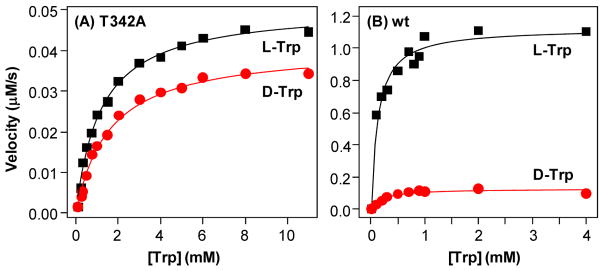
To determine how the T342A mutation affects the enzyme activity, NFK production activities of the wild type and T342A mutant were followed as a function L-Trp or D-Trp concentration. As shown in Fig. 5B, the activity of the wild type enzyme towards either L-Trp (black symbols) or D-Trp (red symbols) follows typical Michaelis-Menten behaviour with kcat/Km of 2.24±0.08 s−1/0.12±0.02 mM and 0.20±0.06 s−1/0.26±0.02 mM, respectively. The kcat/Km values are similar to those reported previously (2.1 s−1/0.19 mM and 0.2 s−1/0.18 mM for L- and D-Trp, respectively);12 the small differences are plausibly a result of the slight difference in the experimental temperatures, as the current data were obtained at 25 °C, while the previously reported data were acquired at ambient temperature. The data indicate that hTDO exhibits similar affinity towards L-Trp and D-Trp, but the kcat towards L-Trp is much higher than D-Trp. The T342 mutation leads to the elevation of the Km to 1.19±0.09 and 1.59±0.13 mM for L-Trp and D-Trp, respectively (Fig. 5A); at the same time the kcat is reduced to 0.101±0.002 and 0.082±0.002 s−1, respectively. As listed in Table 2, the T432A mutation results in similar reduction (~10-fold) in the substrate affinity towards L-Trp and D-Trp, but it leads to much higher reduction in kcat for L-Trp (~20-fold) as compared to D-Trp (~3-fold), indicating the abolishment of the substrate stereoselectivity (as manifested by the similar kcat/Km values of the mutant towards the two stereoisomers). Taken together the data confirm that T342 plays a pivotal role in controlling the substrate stereoselectivity of hTDO.
Figure 5.
Michaelis-Menten plots of the T342A mutant (A) and wild type (B) hTDO. The black and red symbols were obtained by using L-Trp and D-Trp as the substrate, respectively. The solid lines are best fit of the data with the Michaelis-Menten equation. The best fitted parameters are listed in Table 2. The enzyme concentration used for the measurements was 0.5 μM.
Table 2.
Michaelis-Menten parameters of the wild type and T254 mutant of hTDO. All parameters are taken from the data presented in Figure 5.
| hTDO | Substrate | Km (mM) | kcat (s−1) | kcat/Km |
|---|---|---|---|---|
| Wild type | L-Trp | 0.12±0.02 | 2.24 ± 0.08 | 18.7 |
| D-Trp | 0.20±0.06 | 0.26 ± 0.02 | 1.3 | |
|
| ||||
| T342A | L-Trp | 1.19±0.09 | 0.101±0.002 | 0.08 |
| D-Trp | 1.59±0.13 | 0.082±0.002 | 0.05 | |
Molecular dynamics simulations of the ternary complex of xcTDO
To understand the molecular basis for the abolished substrate stereoselectivity in the T342A mutant of hTDO, we performed 20 ns MD simulations of the ternary complex of the T254A mutant of xcTDO (equivalent to T342A mutant of hTDO), in which O2 and Trp were bound to the active site. The data show that the structures of both the L- and D-Trp bound complexes remained stable during the 20 ns simulations, with a RMSD<2.5Å with respect to the initial structure. (Figs. S5 and S6). In addition, the final averaged structures of the mutant show RMSD of 1.14 and 1.27 Å with respect to the L and D-Trp-bound wild type enzyme, respectively, indicating that the mutation does not introduce global structural changes to the enzyme, consistent with the conclusion drawn from the aforementioned spectroscopic studies.
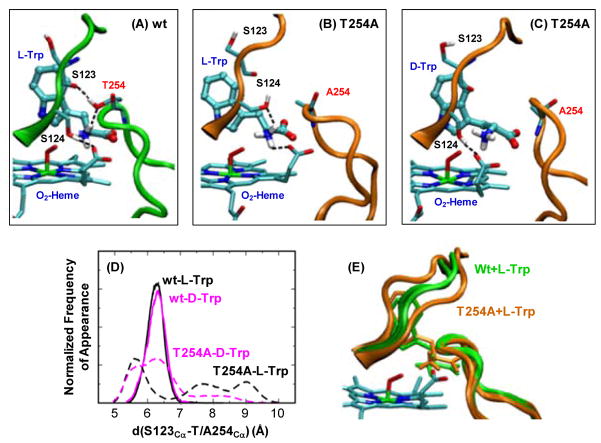
Visual inspection of the obtained MD trajectories, however, revealed important structural differences in the H-bonding network surrounding the NH3+ group of the substrate. In the L-Trp bound wild type enzyme, the substrate forms strong H-bonds with its surrounding environment, including (1) the indoleamine group and H55, (2) the COO− group and the sidechain groups of Y113/R117, and (3) the NH3+ group, the OH group of T254, the propionate A group of the heme and the OH group of S124 (Fig. 6A). The replacement of L-Trp with D-Trp leads to the interruption of the interaction between the NH3+ group and T254, while the rest of the H-binding interactions remain almost the same. It is important to note that T254 and S124 are located in two loop regions that are held close together by an H-bond between the OH group of T254 and the backbone carbonyl group of S123 (Fig. 6A). For clarity, the two loops are denoted as loop250–260 and loop117–130, respectively, hereafter.
Figure 6.
Comparison of the active site structures of the ternary complex of the wild type (A) and T254A mutant (B–C) of xcTDO, obtained from MD simulations. Panel (D) shows the histogram of the loop250–260-loop117–130 separation, defined by the distance between the Cα of S123 and the Cα of T/A254, obtained from MD simulations. The data associated with L- and D-Trp bound complexes are labelled in black and magenta, respectively; those associated with wild type and T254A are depicted in solid and dashed lines, respectively. Panel (E) shows superimposed two extreme structures of the ternary complex of the wild type (labelled in green) and T254A mutant (labelled in orange) of xcTDO, in which the loop250–260-loop117–130 separations are maximum and minimum.
In the L-Trp bound complex of the T254A mutant, the elimination of the H-bond between the NH3+ group of the substrate and T254 leads to only subtle modifications in the H-bonding interactions with H55, Y113 and R117. However, it significantly increases the fluctuations of the loop250–260 (Fig. 6E), as evident in the much wider distance distribution of the A254-S123 pair (Fig. 6D). The opening of the two loops results in a dynamic distal pocket with fluctuating interactions between the NH3+ group of the substrate and its surroundings. In addition, the OH group of S124 rotates to form an H-bond with the NH3+ group, instead of the heme propionate A (Fig. 6B versus 6A). When the L-Trp is replaced by D-Trp in the mutant complex, the opening of the two loops was also observed (Fig. 6D), in addition, the heme propionate A forms an H-bond with S124, instead of the NH3+ group (Fig. 6C versus 6B ).
Molecular dynamics simulations of the ferryl/indole 2,3-epoxide intermediate of xcTDO
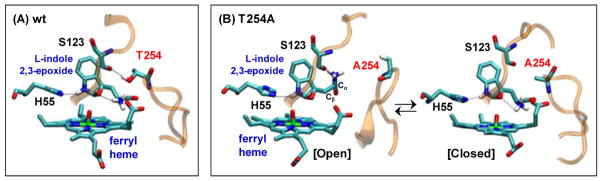
To gain insight into the role of T254 in controlling catalysis, we performed 20 ns MD simulations of the recently characterized ferryl/indole 2,3-epoxide intermediate of the wild type xcTDO.14, 27, 28 The data show that both the complexes with the L- and D-isomers remained stable during the 20 ns simulation, showing a RMSD<2.5 Å. (Figs. S7 and S8). In the previous studies we showed that, in the intermediate, the NH3+ group forms a strong H-bond with the epoxide oxygen (Fig. 7A). This H-bond was demonstrated to be critical for triggering the ring opening reaction of the epoxide that ultimately leads to product formation.28, 29 As observed in the ternary complex, in the epoxide intermediate of the wild type enzyme the NH3+ group is held in position by H-bonding to the heme propionate A and T254; in addition, the loop250–260 and loop117–130 are held together by an H-bond between T254 and S123 (denoted as “closed” conformation hereafter) (Fig. 7A). When L-indole 2,3-epoxide is replaced with the D-isomer, the H-bond between T254 and S123 is ruptured during the simulation, allowing the separation of the two loops (denoted as “open” conformation hereafter). Moreover, a spontaneous rotation of the Cα-Cβ bond of the epoxide intermediate brings the NH3+ group to a new position, thereby temporarily interrupting its H-bonding interaction with the epoxide.
Figure 7.
Representative snapshots of the ferryl/L-indole 2,3-epoxide intermediate of the wild type (A) and T254A (B) of xcTDO obtained from MD simulations.
In the ferryl/L-indole 2,3-epoxide intermediate of T254A, the structure fluctuates between open and closed conformation (Fig. 7B) due to the absence of the H-bond between T254 and S123. When the L-indole 2,3-epoxide is replaced with the D-isomer only open conformation was observed. In the open conformation of both L- and D-isomers, the H-bond between the NH3+ group and epoxide is broken, again due to the spontaneous rotation of the Cα-Cβ bond. Additional calculations show that the estimated free energy barrier for the rotation of the Cα-Cβ bond in the L-indole 2,3-epoxide intermediate of the wild type enzyme is very high (~39 kcal/mol), as a result of steric hindrance (Fig. S9). The steric hindrance is significantly reduced (~3 kcal/mol) for the D-isomer, and becomes barrierless for both isomer complexes of the T254A mutant (Fig. S9). Finally, we performed additional 6 ns MD simulations of the open conformations of the L- and D-intermediate of the wild type and T254A mutant, without restraints. It was found that, in the L-indole 2,3-epoxide intermediate of the wild type enzyme, the NH3+ group returned back to its initial position spontaneously; while in all other three cases, the NH3+ group remained in the open conformation during the time scale of the simulations, confirming that the conversion of the close to open conformation removes the steric hindrance, thereby allowing the rotation of the Cα-Cβ bond.
Mechanistic implications
The simulation data of the ternary complex of xcTDO (with both L- and D-Trp) showed that the T254A mutation causes the opening of the loop250–260-loop117–130, leading to a more dynamic and open distal pocket, accounting for the much lower substrate affinity (i.e. higher Km) observed in the activity studies (Table 2). On the other hand, the simulation data of the ferryl/L-indole 2,3-epoxide intermediate indicate that the T254A mutation also results in the opening of the loop250–260- loop117–130. It leads to local reorganization of the H-bonding interactions surrounding the NH3+ group of the substrate, resulting in an open conformation, in which the H-bond between the NH3+ and the epoxide is temporarily lost. As the H-bond is critical for catalyzing the ring opening reaction of the epoxide during the dioxygenase chemistry,28, 29 the data account for the ~20-fold reduction of the catalytic activity of the enzyme (see kcat in Table 2). Similar result was observed in the complex of the mutant with the D-isomer, accounting for the similar kcat value observed for the two stereoisomers (Table 2). Intriguingly, similar open conformation is observed for the wild type complex with the D-isomer, accounting for its much lower kcat with respect to the L-isomer, confirming that T254 plays a crucial role in determining the stereoselectivity of xcTDO.
Conclusions
In the previous work, we have revealed that hTDO exhibits similar affinities towards L- and D-Trp, but the kcat for L-Trp is 10-fold higher with respect to D-Trp.12 The mutagenesis and MD simulation data obtained in this work demonstrate the critical role of T342 in hTDO (and the equivalent residue T254 in xcTDO) in controlling substrate binding, as well as substrate stereoselectivity of the enzyme, by modulating the H-bonding interaction between the NH3+ group and epoxide oxygen of the ferryl/indole 2,3-epoxide intermediate of the enzyme, and by regulating the dynamics of the loop250–260 and loop117–130. The crystallographic structural data show that substrate binding to xcTDO induces the closure/organization of the two loops to sequester and stabilize the bound substrate. Sequence alignment along with the structural data of the substrate-free hIDO (the only structure available for hIDO) reveal that the substrate induced loop closure also occurs in hIDO, in addition, the T254 and S124 are fully conserved in hIDO, but S123 is replaced by a Gly.
It is plausible that, due to the absence of the T254-S123 interaction, hIDO displays ~170-fold higher affinity towards L-Trp with respect to D-Trp, but the kcat are similar for the two stereoisomers,12 opposite to what observed in hTDO. Apparently, T254 (T342) is not the only player in determining the substrate stereoselectivity of TDO; we are in a process of evaluating the role of S123 in the stereoselectivity of xcTDO. In any case, the mutagenesis results presented here agree perfectly with the prediction made in our previous work, in which T254 in xcTDO was revealed as one of the key residues for controlling the stereoselectivity of the enzyme. In this sense, our new experimental work validates the computational methodology, and confirms the predictive power of this tool.
Supplementary Material
Acknowledgments
Funding: This work was supported by National Institutes of Health Grant GM086482 and National Science Foundation Grant 1026788 to S.-R.Y, and National Institute of Health Molecular Biophysics Training Grant GM008572 to A.L.-B. It was also partially supported by grants from Universidad de Buenos Aires 08-X625 to MAM and 08-X074 to DAE, ANPCYT 07-1650 to MAM, and 06-25667 and Conicet PIP 01207 to DAE. DAE and MAM are members of CONICET. Computer power was gently provided by Cecar at FCEN, UBA and Cluster MCG at UNC.
LC thanks CONICET, the Fulbright Foundation and the Bunge & Born Foundation for the fellowships that allowed her to perform this work.
Abbreviations
- TDO
Tryptophan dioxygenase
- IDO
Indoleamine 2,3-dioxygenase
- Trp
Tryptophan
- hIDO
human IDO
- NFK
N-formylkynurenine
- hTDO
human TDO
- rmTDO
Ralstonia metallidurans TDO
- xcTDO
Xanthomonas campestris TDO
- MD
Molecular Dynamics
- 6C
six-coordinate
- 5C
five-coordinate
Footnotes
Supplementary Information. Sequence alignment of various isoforms of TDO, SDS-PAGE data of the T324 mutant, kinetic trances for the product formation from the T254A mutant, Resonance-Raman spectra for the Trp-CO complexes of xcTDO, RMSD and Potential energy of the Trp-O2 complexes and ferryl/L-indole 2,3-epoxide intermediates of xcTDO as a function of the MD simulation time, and calculated work required for the transition between the closed and open conformations of the ferryl/L-indole 2,3-epoxide intermediate of xcTDO are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sono M, Roach MP, Coulter ED, Dawson JH. Heme-Containing Oxygenases. Chem Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 2.Takikawa O. Biochemical and medical aspects of the indoleamine 2,3-dioxygenase-initiated L-tryptophan metabolism. Biochem Biophys Res Commun. 2005;338:12–19. doi: 10.1016/j.bbrc.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 3.Knox WE, Mehler AH. The Conversion Of Tryptophan To Kynurenine In Liver: I. The Coupled Tryptophan Peroxidase-Oxidase System Forming Formylkynurenine. J Biol Chem. 1950;187:419–430. [PubMed] [Google Scholar]
- 4.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nature medicine. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 5.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T Cell Proliferation by Macrophage Tryptophan Catabolism. The Journal of Experimental Medicine. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grohmann U, Fallarino F, Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends in Immunology. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 7.Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, Mellor AL, Munn DH, Antonia SJ. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int J Cancer. 2002;101:151–155. doi: 10.1002/ijc.10645. [DOI] [PubMed] [Google Scholar]
- 8.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, Van den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 9.Forouhar F, Anderson JLR, Mowat CG, Vorobiev SM, Hussain A, Abashidze M, Bruckmann C, Thackray SJ, Seetharaman J, Tucker T, Xiao R, Ma LC, Zhao L, Acton TB, Montelione GT, Chapman SK, Tong L. Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2007;104:473–478. doi: 10.1073/pnas.0610007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Kang SA, Mukherjee T, Bale S, Crane BR, Begley TP, Ealick SE. Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis. Biochemistry. 2007;46:145–155. doi: 10.1021/bi0620095. [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto H, Oda S-i, Otsuki T, Hino T, Yoshida T, Shiro Y. Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc Natl Acad Sci U S A. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batabyal D, Yeh SR. Human Tryptophan Dioxygenase: A Comparison to Indoleamine 2,3-Dioxygenase. J Am Chem Soc. 2007;129:15690–15701. doi: 10.1021/ja076186k. [DOI] [PubMed] [Google Scholar]
- 13.Capece L, Lewis-Ballester A, Batabyal D, Di Russo N, Yeh SR, Estrin DA, Marti MA. The first step in the dioxygenation reaction carried out by Tryptophan dioxygenase and Indoleamine 2,3- dioxygenase as revealed by QM-MM studies. J Biol Inorg Chem. 2010;15:811–823. doi: 10.1007/s00775-010-0646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capece L, Lewis-Ballester A, Yeh S-R, Estrin DA, Marti MA. The complete Reaction Mechanism of Indoleamine 2,3- Dioxygenase as revealed by QM/MM simulations. 2011. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C, Lin Y, Yeh SR. Inhibitory Substrate Binding Site of Human Indoleamine 2,3-Dioxygenase. J Am Chem Soc. 2009;131:12866–12867. doi: 10.1021/ja9029768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capece L, Arrar M, Roitberg AE, Yeh SR, Marti MA, Estrin DA. Substrate stereo-specificity in tryptophan dioxygenase and indoleamine 2,3-dioxygenase. Proteins: Structure, Function, and Bioinformatics. 2010;78:2961–2972. doi: 10.1002/prot.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batabyal D, Yeh SR. Substrate-Protein Interaction in Human Tryptophan Dioxygenase: The Critical Role of H76. J Am Chem Soc. 2009;131:3260–3270. doi: 10.1021/ja807969a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishimura Y, Nozaki M, Hayaishi O, Nakamura T, Tamura M, Yamazaki I. The Oxygenated Form of l-Tryptophan 2,3-Dioxygenase as Reaction Intermediate. J Biol Chem. 1970;245:3593–3602. [PubMed] [Google Scholar]
- 19.Egawa T, Yeh SR. Structural and functional properties of hemoglobins from unicellular organisms as revealed by resonance Raman spectroscopy. J Inorg Biochem. 2005;99:72–96. doi: 10.1016/j.jinorgbio.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Lewis-Ballester A, Batabyal D, Egawa T, Lu C, Lin Y, Marti MA, Capece L, Estrin DA, Yeh SR. Evidence for a Ferryl Intermediate in Heme-based Dioxygenases: Mechanistic Implications. Proc Natl Acad Sci U S A. 2009;106:17371–17376. doi: 10.1073/pnas.0906655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 22.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 23.Marti MA, Crespo A, Capece L, Boechi L, Bikiel DE, Scherlis DA, Estrin DA. Dioxygen affinity in heme proteins investigated by computer simulation. J Inorg Biochem. 2006;100:761–770. doi: 10.1016/j.jinorgbio.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Bidon-Chanal A, Martí MA, Estrin DA, Luque FJ. Exploring the nitric oxide detoxification mechanism of mycobacterium tuberculosis truncated haemoglobin N. NATO Science for Peace and Security Series A: Chemistry and Biology. 2009:33–47. [Google Scholar]
- 25.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, DeBolt S, Ferguson D, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and. Comput Phys Comm. 1995;91:1–41. [Google Scholar]
- 26.Marti MA, Capece L, Bidon-Chanal A, Crespo A, Guallar V, Luque FJ, Estrin DA. Nitric Oxide reactivity with Globins as investigated through computer simulation. Methods in enzymology. 2008;437:477–498. doi: 10.1016/S0076-6879(07)37024-9. [DOI] [PubMed] [Google Scholar]
- 27.Chung LW, Li X, Sugimoto H, Shiro Y, Morokuma K. Density functional theory study on a missing piece in understanding of heme chemistry: the reaction mechanism for indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase. J Am Chem Soc. 2008;130:12299–12309. doi: 10.1021/ja803107w. [DOI] [PubMed] [Google Scholar]
- 28.Capece L, Lewis-Ballester A, Yeh S-R, Estrin DA, Marti MA. The complete Reaction Mechanism of Indoleamine 2,3- Dioxygenase as revealed by QM/MM simulations. 2011. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung LW, Li X, Sugimoto H, Shiro Y, Morokuma K. ONIOM Study on a Missing Piece in Our Understanding of Heme Chemistry: Bacterial Tryptophan 2,3-Dioxygenase with Dual Oxidants. J Am Chem Soc. 2010;132:11993–12005. doi: 10.1021/ja103530v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.