Abstract
Although polymers, polyplexes, and cells are exposed to various extracellular and intracellular pH environments during polyplex preparation and polymeric transfection, the impact of environmental pH on polymeric transfection has not yet been investigated. This study aims to understand the influence of environmental pH on polymeric transfection by modulating the pH of the transfection medium or the culture medium. Changes in the extracellular pH affected polymeric transfection by way of complex factors such as pH-induced changes in polymer characteristics (e.g., proton buffering capacity and ionization), polyplex characteristics (e.g., size, surface charge, and decomplexation), and cellular characteristics (e.g., cellular uptake, cell cycle phases, and intracellular pH environment). Notably, acidic medium delayed endocytosis, endosomal acidification, cytosolic release, and decomplexation of polyplexes, thereby negatively affecting gene expression. However, acidic medium inhibited mitosis and reduced dilution of gene expression, resulting in increased transfection efficiency. Compared to pH 7.4 medium, acidic transfection medium reduced gene expression 1.6~7.7-fold whereas acidic culture medium enhanced transfection efficiency 2.1~2.6-fold. Polymeric transfection was affected more by the culture medium than by the transfection medium. Understanding the effects of extracellular pH during polymeric transfection may stimulate new strategies for determining effective and safe polymeric gene carriers.
1. Introduction
A great effort for developing effective polymeric vectors has focused primarily on cellular receptor targeting [1-3], endosomal escape [4-6], cytosolic transport [7-9], nuclear import [8, 10, 11], and decomplexation [12-17]. The effects of the transfection environment with respect to proteins [18-20], ions [3, 18, 21-23], pH [13, 16, 17, 24, 25], reduction/oxidation potentials [14, 15, 26], and hypoxia [14, 27-29] have also been investigated in relation to polyplex preparation and polymeric transfection. Among these effectors, we focused on pH because the solution (or medium) pH, extracellular pH, and intracellular pH can all modify characteristics of polymers, polyplexes, and cells.
When dissolving polycations and pDNA or preparing polycation/pDNA complexes, buffer solutions, saline, and/or deionized water have all been routinely used. These solutions can be artificially modulated by adjusting the pH, which can affect the ionization of polycations and pDNA. A change in ionization influences the physicochemical characteristics (e.g., particle size and surface charge) and complexation/decomplexation behavior of polyplexes [30].
The aforementioned solution can be described as the “extracellular medium,” especially when the medium surrounds cells in vitro and in vivo. The extracellular medium used for laboratory cell cultures can be modulated by adding or removing various components and by adjusting the pH to fit specific purposes. However, in vivo extracellular environments are predominantly affected by pathological differences. The extracellular pH of healthy organs is close to pH 7.4 (e.g., pH 7.4 for normal blood, pH 7.2 for brain [31], and pH 7.5 for heart [32]). Under certain pathological conditions, the extracellular pH can become acidic (e.g., approximately pH 6.4-6.8 for solid tumors [33], pH 6.4 for brain ischemia [31], and pH 6.8 for heart ischemia [32]). The extracellular pH can modulate various biological functions, such as gene expression [34], growth rate [35], viability [36], cellular uptake [37], endocytosis [37, 38], exocytosis [38], and lysosomal trafficking [39]. However, the effects of pH on polymeric transfection have rarely been studied.
The intracellular environment is not fixed at a specific pH value. Subcellular compartments such as endosomes (pH 5-7), lysosomes (pH 4-5), the cytosol (pH 6.7-7.1), and the nucleus (pH 7.1-7.2) have separate pH environments [40-42]. Upon endosomal formation, the pH drops with maturation from early to late forms. In particular, the late endosomal and lysosomal pHs are quite distinctive depending on a cell’s drug resistance and/or sensitivity (e.g., approximately pH 6.0 and pH < 5.8, respectively, for drug-resistant MCF7 cells and pH 6.5 and pH > 5.8, respectively, for drug-sensitive MCF7 cells) [40, 41]. Regarding intracellular pH, polymers for polymeric vectors have been designed primarily to target endolysosomal pathways by either disrupting endolysosomal membranes [4-6] or degrading polycations [16, 17].
As described, the pH environment affects characteristics of polymers, polyplexes, and cells. However, after polyplexes or cells are exposed to certain medium or extracellular pH values, it is unknown how the changed pH environments influence polymers, polyplexes, or cells during cellular internalization and intracellular trafficking of polyplexes. Thus, this research aims to understand how extracellular pH affects polyplexes and polymeric transfection. This study examines whether the effects of extracellular pH on transfection efficiency are caused by polyplex/polymer characteristics (e.g., pH-induced changes in surface charge, particle size, and decomplexation of polycation/pDNA complexes and proton buffering capacity of polymers) and/or cellular characteristics (e.g., cellular uptake of polyplexes, cell cycle phases, and cell viability). Branched polyethyleneimine (PEI) and poly(L-lysine) (PLL) were selected as model polymers due to their different degrees of ionization in response to environmental pH changes. Four pHs (i.e., pHs 7.4, 7.0, 6.7, and 6.3) were selected between the physiological pH 7.4 and the pathological lowest possible acidic pH 6.3.
2. Materials and methods
2.1. Materials
PLL hydrobromide (Mw(viscosity) 27.4 kDa), branched PEI (Mw 25 kDa, Mn 10 kDa), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), RPMI1640 medium, Ca2+-free and Mg2+-free Dulbecco’s phosphate buffered saline (DPBS), fluorescein isothiocyanate (FITC), rhodamine B isothiocyanate (RITC), triethylamine (TEA), dimethyl sulfoxide (DMSO), 4-(2-hydroxy-ethyl)-1-piperazine (HEPES), 2-(N-morpholino)ethanesulfonic acid (MES), nigericin, monensin, glucose, sodium bicarbonate, propidium iodide (PI), doxorubicin (DOX) (or adriamycin (ADR)), Triton®X-100, recombinant human insulin, and paraformaldehyde (PFA) were purchased from Sigma-Aldrich Companies (St. Louis, MO). Plasmid DNA (pDNA) encoding firefly luciferase (gWiz-Luc or pLuc) was purchased from Aldevron, Inc. (Fargo, ND). Fetal bovine serum (FBS), penicillin-streptomycin antibiotics, trypsin-EDTA, RNase, and YOYO-1 were purchased from Invitrogen, Inc. (Carlsbad, CA). The luciferase assay kit and BCA™ protein assay kit were bought from Promega Corporation (Madison, WI) and Pierce Biotechnology, Inc. (Rockford, IL), respectively.
2.2. Cells and cell culture
MCF7 cells (a human breast adenocarcinoma cell line), MCF7/ADR-RES cells (a DOX-induced multidrug resistant subline of MCF7), and MES-SA cells (a human uterus sarcoma cell line) were used. The cells were cultured in RPMI1640 medium supplemented with glucose (2 g/L) and 10% heat-inactivated FBS under humidified air containing 5% CO2 at 37°C. Additionally, insulin (4 mg/L) was added to RPMI1640 medium for MCF7 and MCF7/ADR-RES cells. As previously reported [43], to maintain multidrug resistance (MDR) of MCF7/ADR-RES cells, DOX (400 ng/mL) was added once weekly.
2.3. Acid-base titration of polycations
Acid-base titration was performed to monitor the proton buffering capacity of polymeric gene carriers as previously reported [4]. PLL·HBr and PEI (10 mg) were dissolved in NaCl aqueous solution (150 mM; 10 mL) with 1 N NaOH (aq.) (100 μL). The polymer solution (1 mg/mL; 3 mL) was titrated with 0.1 N HCl at room temperature (RT). The pH changes of polymer solutions were monitored.
2.4. Preparation and physicochemical characteristics of polyplexes
As previously reported [3, 43], polyplexes were prepared using pDNA and polycations (i.e., PEI and PLL) in HEPES buffer (20 mM, pH 7.4) supplemented with 5% glucose (HBG). After mixing pDNA and polycations using predetermined complexation conditions, the polyplexes (20 μL for 1 μg pDNA) were incubated for 30 min at RT. Complexation ratios of polyplexes were calculated by counting the amines (N) of polycations and the phosphate groups (P) of pDNA.
Particle size and surface charge of polyplexes were monitored under different medium pHs to understand whether medium pH affects these polyplex characteristics. The polyplex solution was added to HBG with different pHs (i.e., pHs 7.4, 7.0, 6.7 and 6.3). The concentration of pDNA in the polyplex solution was 2.5 μg/mL for surface charge measurements and 5 μg/mL for particle size measurements. Surface charge and particle size of polyplexes were measured using a Zetasizer 3000HS (Malvern Instrument, Inc., Worcestershire, UK) at a wavelength of 677 nm with a constant angle of 90° at RT.
The pH-induced dissociation kinetics of polyplexes under different pH environments were evaluated with a dye-dequenching method. PLL/pDNA and PEI/pDNA complexes were prepared with YOYO-1-intercalated pDNA (YOYO-1:pDNA = 1 molecules:5 base pair). After adding polyplexes (20 μL; 1 μg pDNA) into different pH RPMI1640 media (180 μL; adjusted to pHs 7.4, 7.0, 6.7, and 6.3), the fluorescence intensity of YOYO-1 in the polyplexes was monitored at 491 nm (excitation) and 509 nm (emission) every 5 min for 4 hr. To evaluate the time-dependent fluorescence change of each polyplex exposed to different pH media, the relative fluorescence units (RFU) of each polyplex at each pH were measured and t=0 was set to 100%.
2.5. In vitro transfection
MCF7, MCF7/ADR-RES, and MES-SA cells were used for in vitro transfection studies. As reported previously [1, 18, 43], transfections were performed in 6-well plates, and cells were seeded at a density of 5×105 cells/well. The seeded cells were cultured for 24 hr prior to adding polyplexes. One hour before transfection, the complete culture medium was replaced with serum-free and insulin-free medium. After dosing the polyplexes (20 μL; 1 μg pDNA), the cells were incubated with transfection mixtures for 4 hr, followed by an additional 44 hr incubation in complete culture medium. After transfection, the cells were rinsed twice with DPBS and then lysed using a reporter lysis buffer. Measurements of relative luminescence units (RLU) and protein content of transfected cells were performed per the manufacturer’s instructions.
To investigate the effects of extracellular pH on polymeric transfection, four different pHs, pH 7.4, 7.0, 6.7, and 6.3, were used. Transfection procedures were separated into two periods (i.e., the 4 hr transfection period and 44 hr incubation period) as follows:
Condition A: 4-hr transfection period at different pHs (pH 7.4, 7.0, 6.7, and 6.3) followed by the 44-hr incubation period fixed at pH 7.4.
Condition B: 4-hr transfection period at pH 7.4 followed by the 44-hr incubation period at different pHs (pH 7.4, 7.0, 6.7, and 6.3).
Condition AB: 48-hr transfection period and incubation period both at different pHs (7.4, 7.0, 6.7, and 6.3).
2.6. In vitro metabolic activity
The MTT-based metabolic activity of polyplex-transfected cells was assessed using MCF7, MCF7/ADR-RES, and MES-SA cells. Cells were seeded in 12-well plates at a density of 2.5×105 cells/well and cultured for 24 hr prior to polyplex addition. The experimental procedure was the same as previously described for in vitro transfection except for the polyplex loading dose (10 μL; 0.5 μg pDNA). After the 48-hr transfection procedure, MTT solution (0.1 mL; 5 mg/mL) was added to the cells in 1 mL of culture medium. After 4 hr, the MTT-containing medium was removed. Living cells produced formazan crystals that were dissolved in DMSO; crystal absorbance was measured at 570 nm with a microplate reader.
2.7. Cellular uptake of polyplexes
As previously described for in vitro transfection, cells were prepared in 6-well plates. Polyplexes (20 μL; 1 μg pDNA) prepared using YOYO-1-intercalated pDNA were added to the cells. After a 4-hr incubation under 4 different pH environments (pH 7.4, 7.0, 6.7, and 6.3), the cells were detached and then fixed using 4% PFA solution. The cells containing fluorescent polyplexes were monitored using flow cytometry (FACScan Anaylzer, Becton-Dickinson, Franklin Lakes, NJ) with a primary argon laser (488 nm) and fluorescence detector (530±15 nm) for YOYO-1. Polyplex uptake was analyzed using a gated population containing at least 5,000 cells.
2.8. Cell cycle phases
The cell-cycle phases of MCF7, MCF7/ADR-RES, and MES-SA cells incubated in different pH media were assessed. Cells were seeded in 6-well plates at a density of 5×105 cells/well and cultured for 24 hr prior to treatment with different pH media. Then, cells were exposed to 4 different pH transfection media (i.e., pH 7.4, 7.0, 6.7 and 6.3) for 4 hr and then 4 different pH culture media (i.e., pH 7.4, 7.0, 6.7 and 6.3) for 44 hr. During the transfection process, cells were sampled at predetermined time points (i.e., 0, 2, 4, 12, 24, 36, and 48 hr post-transfection). The cells were rinsed twice with DPBS and detached with trypsin-EDTA solution. The rinsed and suspended cells were fixed with cold 70% (v/v) ethanol (aq.). Fixed samples were rinsed twice with DPBS and incubated with 1 mL of a PI-containing DPBS solution (50 μg/mL PI, 0.1% Triton®X-100 solution, and 15 μg/mL RNase) for 30 min at RT. The stained samples were analyzed by flow cytometry with a primary argon laser (488 nm) and fluorescence detector (668 nm long pass); at least 15,000 cells were counted per condition.
2.9. Intracellular pH measurement of polyplexes
The intracellular pH environment of polyplexes was monitored using fluorescent dye-labeled polymers as previously reported [43]. PLL and PEI were labeled with pH-sensitive FITC and pH-insensitive RITC dyes using a simple coupling reaction. PLL and PEI were each labeled with both FITC and RITC, creating FITC-PLL-RITC (2.3 mol% (based on l-lysine units) FITC; 1.2 mol% RITC), and FITC-PEI-RITC (1.6 mol% (based on amines) FITC; 0.4 mol% RITC), respectively [43].
As previously described for in vitro transfections [43], cells were prepared in 6-well plates. Polyplexes (20 μL; 1 μg pDNA) were prepared using FITC-PLL-RITC or FITC-PEI-RITC, and added to cells. At predetermined time points (i.e., 0.5, 1, 1.5, 2, 3, and 4 hr post-transfection), the cells were detached and then resuspended in DPBS 1% PFA solution. To create a pH calibration curve, the transfected cells were resuspended in 0.5 mL of pH clamp buffers (approximately pH 7.4, 6.8., 6.0, 5.0, and 4.0) that were prepared by mixing DPBS (pH 7.4) or MES (pH 4.0; 50 mM MES, 150 mM NaCl, 4 mM KCl, and 1 mM MgSO4). Monensin (20 μM) and nigericin (10 μM) were added into pH clamp buffers to ensure homogeneity of the pH environment for cells. The cells harboring fluorescent polyplexes were monitored using flow cytometry (FACScan Anaylzer, Becton-Dickinson) with a primary argon laser (488 nm) and fluorescence detectors (530±15 nm for FITC and 585±21 nm for RITC). The average intracellular pH of polyplexes was assessed by analyzing the ratio of FITC to RITC intensity from a gated population of at least 5,000 cells. In order to identify and assign the major intracellular compartments holding the polyplexes from the intracellular pH, the entire fluorescent cell population was divided into four areas based on the cellular pH calibration curve. The nucleus, and potentially the cytoplasm, was designated by pH values greater than pH 6.8, early endosomes were approximated between pH 6.0 and pH 6.8, late endosomes were between pH 5.0 and pH 6.0, and lysosomes were classified with a pH less than 5.0 [43].
3. Results and discussion
Prior to applying extracellular media of various pH values (i.e., culture medium and transfection medium) for in vitro polymeric transfection, the optimum conditions for PEI- and PLL-based transfection of MCF7, MCF7/ADR-RES, and MES-SA cells were determined using less toxic polymer/pDNA complexation ratios. For PEI/pDNA complexes, N/P 5 was applied to all cell lines used in this study because, in general, higher N/P values cause cytotoxicity [19, 22, 44]. For PLL/pDNA complexes, N/P 5 was used for MCF7 and MCF7/ADR-RES cells based on our previous report [43]. For MES-SA cells, N/P 10 was used as the optimum transfection condition based on results from a test of N/P values between 3 and 15 (Fig. S1).
3.1. Effects of extracellular pH on transfection efficiency
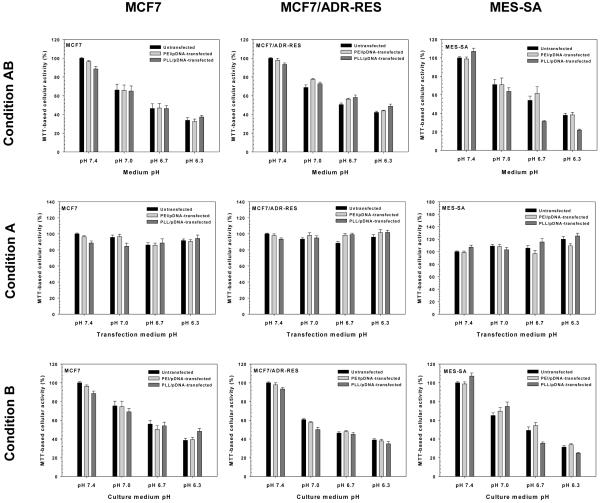
PEI- and PLL-mediated transfection efficiencies are shown in Fig. 1 for cells exposed to specific extracellular pHs. Luciferase expression of the cells transfected with 4 different medium pHs (i.e., transfection medium and culture medium) for 48 hr (Condition AB) was polyplex- and cell-dependent. The medium pH strongly influenced transgene expression of PEI/pDNA-transfected MCF7, MCF7/ADR-RES, and MES-SA cells (p<0.001, p<0.001, and p=0.004 by one-way ANOVA, respectively). Using pH 7.4 medium, PEI/pDNA-transfected MCF7 cells had approximately 2-fold to 3-fold higher transfection efficiency than those at pH 7.0, 6.7, and 6.3 (p=0.01, p=0.02, and p=0.09 by one-way ANOVA with Tukey HSD test, respectively). Interestingly, PEI/pDNA-transfected MCF7/ADR-RES and MES-SA cells showed increased transgene expression with decreasing medium pH values. Specifically, gene expression in medium pH 6.3 was approximately 2-fold higher than in medium pH 7.4 (p<0.001 for MCF7/ADR-RES cells and p=0.01 MES-SA cells by one-way ANOVA with Tukey HSD test).
Fig. 1.
Effects of extracellular pH (e.g., medium pH for Condition AB, transfection medium pH for Condition A, and culture medium pH for Condition B) on the transfection efficiency of PEI/pDNA- and PLL/pDNA-transfected MCF7, MCF7/ADR-RES, and MES-SA cells. (n≥8; mean±SEM)
PLL/pDNA-transfected cells experienced approximately 2-fold lower (for MES-SA cells) or higher (for MCF7 and MCF7/ADR-RES cells) gene expression with medium pH 7.4 than medium pH 6.3. However, PLL-mediated transfections were less sensitive to medium pH (p=0.27 for MCF7, p=0.07 for MCF7/ADR-RES, and p<0.05 for MES-SA cells by one-way ANOVA) than PEI-mediated transfections. Also, transfection results conducted at medium pH 7.4 and medium pH 6.3 showed less significant differences (p=0.03 for MES-SA, p=0.21 for MCF7, and p=0.06 for MCF7/ADR-RES cells by one-way ANOVA with Tukey HSD test) for PLL versus PEI.
These medium pH-induced polymeric transfection results were further analyzed to understand which steps of polymeric transfection are strongly affected by the extracellular pH. Thus, pH-controlled medium treatment was divided into transfection medium for 4-hr transfection periods and culture medium for 44-hr incubation periods. When applying Condition A (transfection media of various pHs) as shown in Fig. 1, acidic transfection media caused either decreased or nearly equal transfection efficiencies compared with neutral transfection medium. For PEI/pDNA-transfected MCF7 and MCF7/ADR-RES cells, transfection medium pH 6.3 reduced transfection efficiency by as low as 7.7-fold and 2.1-fold, respectively, compared with transfection medium pH 7.4 (for both, p=0.004 by one-way ANOVA with Tukey HSD test). Acidic transfection medium (pH 6.3) also caused approximately 25-35% reduced transgene expression of MES-SA cells compared to transfection medium pH 7.4, although transfection efficiency was less affected by medium pH (p=0.23 by one-way ANOVA).
PLL-mediated transfections were also influenced by transfection medium pH (p=0.04 for MCF7, p=0.02 for MCF7/ADR-RES, and p<0.001 for MES-SA by one-way ANOVA). MCF7/ADR-RES and MES-SA cells transfected with transfection medium pH 6.3 had 2.4-fold and 2.7-fold lower transfection efficiencies than those with transfection medium pH 7.4 (p<0.05 and p=0.001 by one-way ANOVA with Tukey HSD test, respectively). For PLL/pDNA-transfected MCF7 cells, transfection medium pH 7.0 caused the highest transfection efficiency, which was approximately 2-fold higher than those from other pH transfection media.
However, unlike the pH effects of transfection media, transfection efficiencies increased in acidic culture media (called as Condition B) (Fig. 1). The pH of the culture medium significantly influenced transfection efficiencies for PEI/pDNA-transfected MCF7/ADR-RES and MES-SA cells (for both, p<0.001 by one-way ANOVA), and their transfection efficiencies in culture medium pH 6.3 were 1.9-fold and 2.6-fold higher than those in culture medium pH 7.4 (for both, p<0.001 by one-way ANOVA with Tukey HSD test). For PEI/pDNA-transfected MCF7 cells, although the effect of culture medium pH on PEI-mediated transfection efficiency was not statistically significant (p=0.31 by one-way ANOVA), transfection efficiencies using culture medium pH 6.3 were 1.6-fold higher than those at culture medium pH 7.4 (p=0.49 by one-way ANOVA with Tukey HSD test).
When transfecting cells with PLL/pDNA using Condition B, there was no statistically significant influence of decreased culture medium pH values on transfection efficiencies (p=0.22 for MCF7, p=0.28 for MCF7/ADR-RES, and p=0.30 for MES-SA cells by one-way ANOVA). However, the culture medium pH 6.3 induced approximately 1.6-fold higher transfection efficiencies for PLL/pDNA-transfected MCF7 and MCF7/ADR-RES cells than culture medium pH 7.4 (p=0.33 and p=0.34 by one-way ANOVA with Tukey HSD test, respectively).
3.2. Effects of extracellular pH on MTT-based cellular activity
MTT-based cellular activity assays were applied to understand how the extracellular pH influences cell number, cell viability, metabolic activity, and the cell proliferation rate of polyplex-transfected cells. When different medium pHs were applied to untransfected cells (Condition AB), the cellular activities significantly decreased with decreasing pHs, regardless of cell type (for all cell lines, p<0.001 by one-way ANOVA) as shown in Fig. 2. These results are consistent with previous studies [35, 45]. However, regardless of the type of polyplex used, the cellular activity of most transfected cells was almost the same as untransfected cells at the same pH.
Fig. 2.
Effects of extracellular pH (e.g., medium pH for Condition AB, transfection medium pH for Condition A, and culture medium pH for Condition B) on MTT-based cellular activity of PEI/pDNA- and PLL/pDNA-transfected MCF7, MCF7/ADR-RES, and MES-SA cells. (n=6; mean±SEM)
In Fig. 2, 4-hr treatment of acidic transfection media (Condition A) did not significantly damage the cellular activities of untransfected or transfected cells versus neutral transfection media treatment. On the other hand, longer treatment (44 hr) of acidic medium (Condition B) induced similar cellular activities to Condition AB treatment. However, it is not clear whether acidic medium caused reduced cell viability and/or metabolic activity or inhibited cell proliferation (without cell death).
3.3. Effects of medium pH on polymers and polyplexes
When polymers and polyplexes are exposed to different pH environments, their chemical, physical and electrochemical properties can be changed. First, the proton buffering capacity of PEI and PLL were monitored by acidic titration. As shown in Fig. S2, a PLL solution had no proton buffering capacity like a NaCl aqueous solution (150 mM) because the primary amines of PLL stay protonated within the range of basic to acidic pHs. On the contrary, a PEI solution exhibited proton buffering throughout a broad pH range (approximately pH 3-10) due to continuous protonation of primary, secondary, and tertiary amines upon acidification. These results are consistent with previous reports [46].
During acidification, protonation of amines increases the net positive charge character, and the altered charge could affect electrostatic interactions between polymers and pDNA. In this way, medium pH (i.e., buffer pH) could influence complexation and decomplexation between polymers and pDNA as well as particle size and surface charge of polyplex. Nevertheless, Godbey et al., reported that PEI-mediated transfection efficiencies were not different when PEI/pDNA complexes were prepared in different pH solutions [47]. Thus, this study excluded the effects of medium pH on complexation. All polyplexes were prepared at pH 7.4.
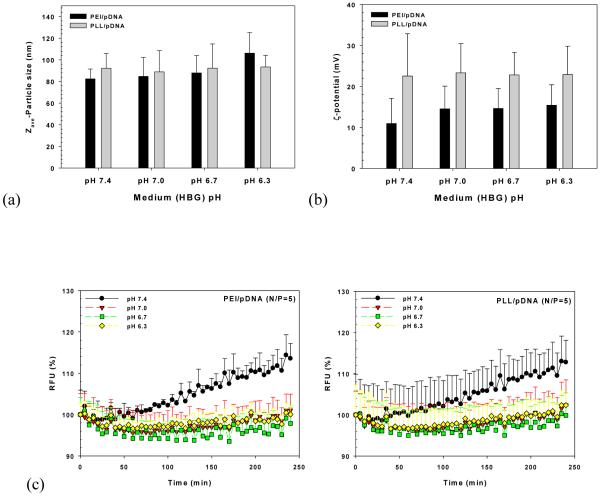
When polyplexes were exposed to different medium pHs, the changes of PEI/pDNA and PLL/pDNA complexes’ particle size, surface charge, and decomplexation were investigated. As expected, the particle size and surface charge of PLL/pDNA complexes were not affected by HBG with different pHs (p=0.88 and p=0.96 by one-way ANOVA, respectively) (Fig. 3(a) and 3(b)). These results may be attributed to unaltered primary amine protonation of PLL within the pH range of 6.3 to 7.4; the side chain of l-lysine has pKa 8.95. In the case of PEI/pDNA complexes (Fig. 3(a) and 3(b)), particle sizes were around 80-90 nm in solutions pH 6.7-7.4, although the size at pH 6.3 (106±19 nm) was somewhat increased compared to those at other pHs. Surface charges ranged between 10-15 mV without statistical significance related to pH effects.
Fig. 3.
Effects of medium pH on (a) particle size, (b) surface charge, and (c) decomplexation of PEI/pDNA (N/P=5) and PLL/pDNA (N/P=5) complexes. (n=3; mean±SD)
The effects of medium pH on decomplexation were monitored over time as shown in Fig. 3(c). Medium pH 7.4 resulted in increased decomplexation (i.e., increased fluorescent intensity) with time, regardless of polyplex type. However, acidic medium (pH 6.3 to 7.0) compared to medium pH 7.4 showed decreasing or constant fluorescent intensity over time. These results may be caused by increasing (+/−) charge ratios of polyplexes because the phosphate groups of pDNA (approximately pKa 6.3) are less negatively charged at acidic pH than pH 7.4. This phenomenon may be similar to tight complexation of polyplexes at high (+/−) charge ratios.
Our findings suggest that polyplexes may be stable in acidic extracellular environments and during endosomal acidification, but could be dissociated in neutral pH environments such as the cytoplasm or the nucleus. If medium pH 7.4 can induce a weak attraction between polycations and pDNA in polyplexes within 4 hr, before cellular internalization, this may facilitate pDNA release from polyplexes after endosomal release, thereby enhancing polymeric transfection efficiency. That is, high pH-induced decomplexation could generate higher transfection efficiency than low pH-induced decomplexation.
3.4. Effects of medium pHs on polyplex uptake
In transfection experiments, transfection medium was applied for 4 hr and then replaced with culture medium. At 4 hr post-transfection, the polyplexes that were not endocytosed will be removed. Thus, only internalized polyplexes will be available for gene expression. During the first 4 hr post-transfection, the different pHs of transfection medium for Conditions AB and A could affect cellular polyplex uptake, whereas the same pH of transfection medium under Condition B could cause the same cellular uptake.
Thus, when different medium pHs were applied, cellular uptake 4 hr post-transfection was monitored by flow cytometry. As shown in Fig. S3, PEI/pDNA uptake in MCF7 cells in transfection medium pH 7.4 was somewhat lower than uptake under other transfection medium pHs. A similar amount of PLL/pDNA complexes was internalized into transfected MCF7 cells regardless of the transfection medium pH. However, MCF7/ADR-RES and MES-SA cells showed negligible effects of altered transfection medium pH for PEI/pDNA and PLL/pDNA uptake.
3.5. Effects of medium pH on cell cycle phases
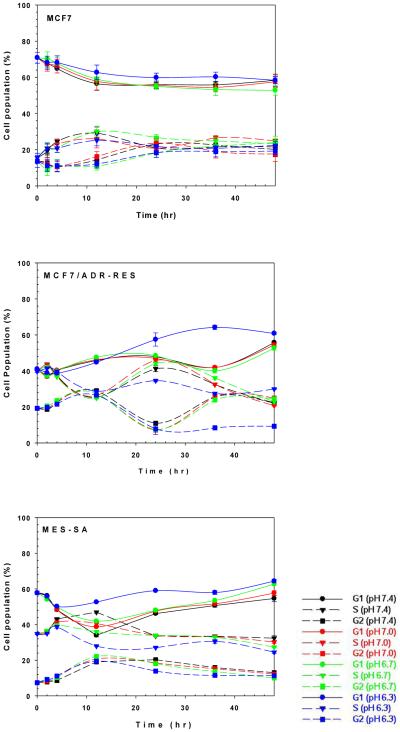
The cell-cycle phase of transfected cells affects cellular internalization, endocytosis, and transfection efficiency of gene complexes [48-51]. Cellular internalization of nonviral gene complexes during the G2/M phases was 1.5-fold lower than during late G1 [51]. Polymeric transfection initiated at the S or G2/M phases caused approximately 30-fold or more than 500-fold higher transgene expression than transfections beginning at G1, respectively [48]. Thus, we monitored the effect of medium pH on cell cycle phases. During the first 4 hr post-transfection, the G1, S, and G2 phases of cells from different medium pHs were not significantly different (Fig. S4). This may support the similar cellular internalization of polyplexes shown in Fig. S3. Under Condition A, the effects of different tranfection pHs on polymeric transfection efficiency may be caused by other effectors, but not the cell-cycle phase.
However, after the first 4 hr post-transfection, cell-cycle phases were indeed influenced by medium pH. Even though the impact of acidic medium is cell-dependent, the medium at pH 6.3 induced more G1 phase and less S and G2 phases than other pHs, regardless of the cell type (Fig. 4). In MCF7 cells, medium at pH 6.3 slightly inhibited cellular functions such as cell proliferation (G2 phase) and DNA duplication (S phase) compared with other medium pHs. Compared to MCF7 cells, MCF7/ADR-RES and MES-SA cells were strongly influenced by medium pH. In the case of MCF7/ADR-RES cells, medium at pH 6.7, 7.0 and 7.4 did not show any significant changes on cell cycle phases within the first 12 hr post-transfection. However, acidic (pH 6.3) medium-treated MCF7/ADR-RES cells had remarkably increased G1 phases and reduced S and G2 phases 24 hr post-transfection, unlike treatment with the other pH media. For MES-SA cells, acidic medium clearly showed higher G1 phases and lower S and G2 phases than neutral medium 4 hr post-transfection. These findings indicate that the cytoskeletal network for endocytosis of polyplexes may be maintained. Also, the lower proportion of mitotic cells in acidic medium could prevent dilution of gene expression per cell so long as the acidic medium does not damage cell viability. These two possibilities indicate that acidic culture media induces better transgene expression than neutral culture media under Condition B.
Fig. 4.
G1, S, and G2 phases of MCF7, MCF7/ADR-RES, and MES-SA cells after treatment with transfection media of varying pH (48 hr incubation).
3.6. Intracellular environment of polyplexes
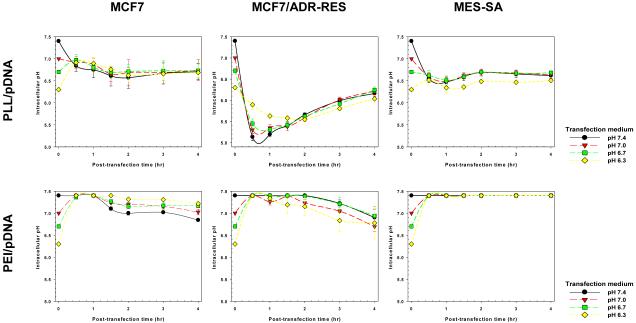
The intracellular location and pH of polyplexes or pDNA strongly affect polymeric transfection efficiency [43, 52]. Thus, we estimated how the medium pH influences the intracellular environment of polyplexes using flow cytometry as previously reported [43]. In cells transfected with PEI/pDNA and PLL/pDNA complexes, the average intracellular pH environment was monitored during the first 4 hr post-transfection (Fig. 5). Polyplexes continuously internalize until they are used up from the extracellular medium. Therefore, polyplexes could be exposed to different subcellular locations, and the transfected cells having these polyplexes may experience different intracellular pHs over time. Thus, as shown in Fig. 6 and Fig. 7, it was estimated how many polyplex-transfected cells had average intracellular pHs conducted from polyplexes exposed to the pH of subcellular compartments (e.g., the cytosol, the early and late endosomes, the lysosomes, and the nucleus) over time.
Fig. 5.
Average intracellular pH of PEI/pDNA- or PLL/pDNA-uptake in MCF7, MCF7/ADR-RES, and MES-SA cells within 4 hr after polymeric transfection (mean±SEM; n=3).
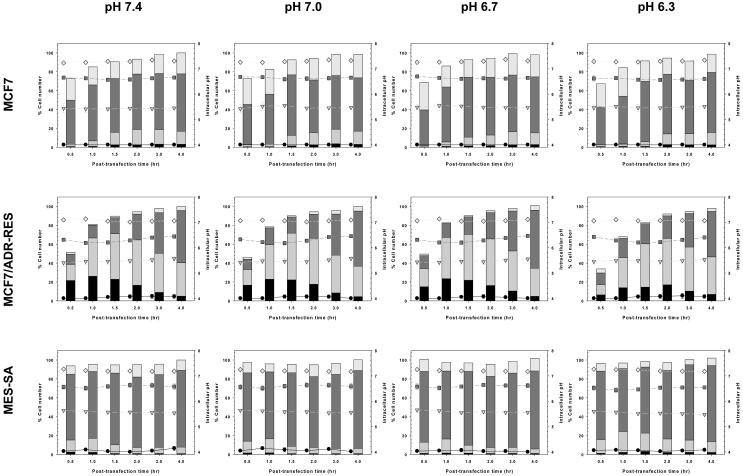
Fig. 6.
Subcellular pH distributions of PLL/pDNA-uptake in MCF7, MCF7/ADR-RES, and MES-SA cells within 4 hr after polymeric transfection. The pH of each subcellular compartment in polyplex-transfected cells and the number of cells in each subcellular compartment at a given time point following transfection are represented in the following dot plots and column plots, respectively. The intracellular pH for polyplex-transfected cells relevant to the pH of lysosomes (black circle, ●), late endosomes (gray inverse triangle,  ), early endosomes (dark gray square,
), early endosomes (dark gray square,  ), and the cytosol/nucleus (bright gray diamond,
), and the cytosol/nucleus (bright gray diamond,  ) are represented in dot plots. The corresponding % cell numbers are represented as
) are represented in dot plots. The corresponding % cell numbers are represented as  ,
,  ,
,  , and
, and  in the column plots. The cell number at 4 hr post-transfection was set to 100% (mean±SEM; n=3).
in the column plots. The cell number at 4 hr post-transfection was set to 100% (mean±SEM; n=3).
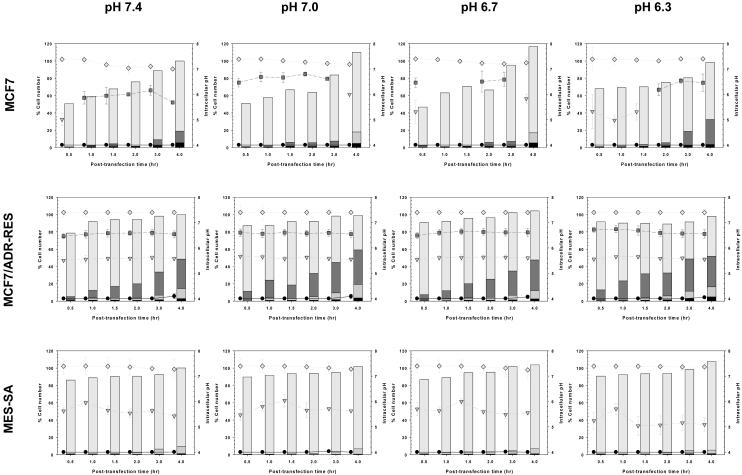
Fig. 7.
Subcellular pH distributions of PEI/pDNA-uptake in MCF7, MCF7/ADR-RES, and MES-SA cells within 4 hr after polymeric transfection (mean±SEM; n=3).
As shown in Fig. 5, the effects of transfection medium pH on the average intracellular pH of polyplex-transfected cells were cell- and polyplex-dependent. The average intracellular pH of PLL-mediated MCF7 transfected cells was not significantly influenced by the medium pH, whereas the intracellular pH for MCF7/ADR-RES and MES-SA transfections was affected by medium pH. In PLL/pDNA-transfected MCF7/ADR-RES cells, acidic medium induced a slow drop in intracellular pH within the first 2 hr post-transfection. Recovery of intracellular pH, which was caused by endosomal escape of polyplexes [43], was somewhat lower (approximately 0.1~0.2 pH units). Furthermore, the time points for intracellular pH recovery were delayed (2 hr post-transfection for medium pH 6.3 vs. 0.5-1 hr post-transfection for medium pH 7.4). Similarly, MES-SA cells transfected with PLL/pDNA complexes showed an acidic medium (pH 6.3)-mediated slow intracellular pH recovery (1.5 hr post-transfection vs. 0.5-1 hr post-transfection at medium pH 7.4) and low recovered intracellular pH (~ pH 6.5 vs. pH 6.7 at medium pH 7.4). These results are probably not attributed to polymer characteristics because PLL does not have proton buffering capacity in the pH range experienced during intracellular polyplex trafficking. One significant contribution could be the acidic medium-induced changes to cell characteristics; acidic extracellular medium lowered intracellular (cytoplasmic) pHs [37], and acidic cytosolic pHs are known to inhibit/delay the endocytosis of therapeutics such as proteins [38, 53-55].
Delayed endosomal acidification rates and lower intracellular recovery pHs of PLL/pDNA-transfected cells in acidic medium could be supported by the population data of cells with average intracellular pHs related to subcellular compartments. As shown in Fig. 6, it seems that medium pH does not significantly affect time-dependent polyplex uptake (column plots) or the average intracellular pH (dot plots) relevant to subcellular compartments. Regardless of the medium pH, PLL/pDNA-transfected MCF7 cells had an average intracellular pH between pH 7.25-7.35 (relevant to the cytosol/the nucleus), pH 6.55-6.70 (relevant to the early endosomes), pH 5.40-5.55 (relevant to the late endosomes), and ~ pH 4 (relevant to the lysosomes). The pH ranges of PLL/pDNA-transfected MCF7/ADR-RES cells were pH 7.01-7.13, pH 6.18-6.47, pH 5.40-5.57, and pH 4.00-4.10, and were lower than those of MCF7 transfected cells due to the fast endosomal acidification rates of MDR cells [43]. Also, PLL/pDNA-transfected MES-SA cells had pH 7.17-7.27, pH 6.44-6.64, pH 5.48-5.65, and pH 4.00-4.15. However, in PLL/pDNA-transfected MCF7/ADR-RES cells and MES-SA cells, the peak cell populations relevant to late endosomes and lysosomes (from column plots of Fig. 6) were delayed with acidic medium treatment. This acid medium-induced delayed acidification process could cause delayed cytosolic release of polyplexes. Also, the lower intracellular pH may slow down decomplexation rates. Together, these phenomena may influence the acidic transfection medium-induced decrease in polymeric transfection efficiency.
When applying PEI/pDNA complexes (having endosomal disruption capability) to the cells, the intracellular pH of transfected cells was higher (> ~pH 6.7) than those of PLL/pDNA-transfected cells (Fig. 5) because the proton buffering capacity of PEI can break endosomal membranes, quickly releasing PEI/pDNA into the cytoplasm. Like PLL/pDNA-transfected cells, PEI/pDNA-transfected cells were influenced by delayed endocytosis and endosomal acidification of polyplex-trapped endosomes when treated with acidic transfection medium. This was clearly demonstrated by the fast endosomal acidification rates (e.g., MCF7/ADR-RES cells in this study). As shown in Fig. 7, transfection medium pH 6.3 caused more cells to be exposed to early endosomal pHs than transfection medium pH 7.4. Also, the delayed endosomal acidification may negatively influence the endosomal release of PEI/pDNA complexes because the proton buffering capacity of PEI is strongly affected by decreasing pH.
Based on the transfection results of Conditions AB, A, and B (as summarized in Table 1), acidic transfection media decreased polymeric transfection efficiencies, whereas acidic culture media enhanced efficiencies. The effects of transfection media on transfection efficiency may be mediated by the delayed acidification rates of polyplex-trapped endolysosomes and the decomplexation rates during the transfection period. Delayed endosomal acidification caused by acidic transfection media resulted in delayed cytosolic release of polyplexes (i.e., sequestrated in the late endosomes and lysosomes). In addition, polyplexes in acidic extracellular environments and slightly acidic cytosol could be tightened and then slowly release pDNA. These reasons might explain why acidic transfection media decreased polymeric transfection efficiencies.
Table 1.
Summary of cell transfection enhancement or reduction with extracellular pH 6.3 compared to extracellular pH 7.4.
| MCF7 | MCF7 /ADR-RES |
MES-SA | ||
|---|---|---|---|---|
| PEI/pDNA- mediated transfection |
Condition AB | 2-fold ↓ | 2.2-fold ↑ | 2-fold ↑ |
| Condition A | 7.7-fold ↓ | 2.1-fold ↓ | 1.6-fold ↓ | |
| Condition B | 1.6-fold ↑ | 1.9-fold ↑ | 2.6-fold ↑ | |
|
| ||||
| PLL/pDNA- mediated transfection |
Condition AB | 1.7-fold ↑ | 1.8-fold ↑ | 1.5-fold ↓ |
| Condition A | 1-fold | 2.4-fold ↓ | 2.7-fold ↓ | |
| Condition B | 1.6-fold ↑ | 1.6-fold ↑ | 1.2-fold ↑ | |
↓ and ↑ mean lower and higher, respectively.
On the other hand, the culture medium affected the cell cycle phase and metabolic activity of transfected cells. Under acidic conditions, the metabolic activity of transfected cells was reduced. Although these results may have been caused by a reduction in viable cells, the metabolic functions of transfected cells could be limited by cellular arrest (i.e., increased G1 phase and decreased G2 and S phases) without cell death. Cellular arrest could delay mitosis, leading to less dilution of gene expression in transfected cells. As a result, acidic culture medium can enhance polymeric transfection efficiencies.
Thus, the impact of transfection media and culture media on cells may determine the effects of medium pH on transfection efficiency. Nevertheless, in regard to the effects of extracellular pH on polymeric transfection, the pH of the culture medium could be more influential than the pH of the transfection medium because transfected cells are exposed to culture medium longer (44 hr) than transfection medium (4 hr). The findings in this study may be helpful for developing effective polymeric vectors for solid tumors and ischemia because these cells pathologically feature acidic extracellular environments.
4. Conclusion
In vitro polymeric transfection was strongly affected by the extracellular pH. Transfection media modulated both polymer/polyplex characteristics (e.g., proton buffering and decomplexation) and cellular characteristics (e.g., endocytic trafficking), whereas culture medium affected only cellular characteristics (e.g., cell proliferation, cell cycle phase, and mitosis). In conclusion, acidic transfection medium reduced and acidic culture medium enhanced polymeric transfection efficiency. When treating with a specific extracellular pH during polymeric transfection, the impact of transfection medium and culture medium may determine the effect of medium pH on transfection efficiency.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH GM82866.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Kang HC, Kim S, Lee M, Bae YH. Polymeric gene carrier for insulin secreting cells: Poly(l-lysine)-g-sulfonylurea for receptor mediated transfection. J Control Release. 2005;105:164–176. doi: 10.1016/j.jconrel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- [2].Kawakami S, Higuchi Y, Hashida M. Nonviral approaches for targeted delivery of plasmid DNA and oligonucleotide. J Pharm Sci. 2008;97:726–745. doi: 10.1002/jps.21024. [DOI] [PubMed] [Google Scholar]
- [3].Kang HC, Bae YH. Transfection of insulin-secreting cell line and rat islets by functional polymeric gene vector. Biomaterials. 2009;30:2837–2845. doi: 10.1016/j.biomaterials.2009.01.035. [DOI] [PubMed] [Google Scholar]
- [4].Kang HC, Bae YH. pH-Tunable endosomolytic oligomers for enhanced nucleic acid delivery. Adv Funct Mater. 2007;17:1263–1272. [Google Scholar]
- [5].Jones RA, Cheung CY, Black FE, Zia JK, Stayton PS, Hoffman AS, et al. Poly(2-alkylacrylic acid) polymers deliver molecules to the cytosol by pH-sensitive disruption of endosomal vesicles. Biochem J. 2003;372:65–75. doi: 10.1042/BJ20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kiang T, Bright C, Cheung CY, Stayton PS, Hoffman AS, Leong KW. Formulation of chitosan-DNA nanoparticles with poly(propyl acrylic acid) enhances gene expression. J Biomater Sci Polym Ed. 2004;15:1405–1421. doi: 10.1163/1568562042368112. [DOI] [PubMed] [Google Scholar]
- [7].Ng CP, Goodman TT, Park IK, Pun SH. Bio-mimetic surface engineering of plasmid-loaded nanoparticles for active intracellular trafficking by actin comet-tail motility. Biomaterials. 2009;30:951–958. doi: 10.1016/j.biomaterials.2008.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Choi JS, Ko KS, Park JS, Kim YH, Kim SW, Lee M. Dexamethasone conjugated poly(amidoamine) dendrimer as a gene carrier for efficient nuclear translocation. Int J Pharm. 2006;320:171–178. doi: 10.1016/j.ijpharm.2006.05.002. [DOI] [PubMed] [Google Scholar]
- [9].Drake DM, Pack DW. Biochemical investigation of active intracellular transport of polymeric gene-delivery vectors. J Pharm Sci. 2008;97:1399–1413. doi: 10.1002/jps.21106. [DOI] [PubMed] [Google Scholar]
- [10].Wagstaff KM, Jans DA. Nuclear drug delivery to target tumour cells. Eur J Pharmacol. 2009;625:174–180. doi: 10.1016/j.ejphar.2009.06.069. [DOI] [PubMed] [Google Scholar]
- [11].Park KM, Kang HC, Cho JK, Chung IJ, Cho SH, Bae YH, et al. All-trans-retinoic acid (ATRA)-grafted polymeric gene carriers for nuclear translocation and cell growth control. Biomaterials. 2009;30:2642–2652. doi: 10.1016/j.biomaterials.2009.01.025. [DOI] [PubMed] [Google Scholar]
- [12].Kang HC, Kang HJ, Bae YH. A reducible polycationic gene vector derived from thiolated low molecular weight branched polyethyleneimine linked by 2-iminothiolane. Biomaterials. 2011;32:1193–1203. doi: 10.1016/j.biomaterials.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim YH, Park JH, Lee M, Kim YH, Park TG, Kim SW. Polyethylenimine with acid-labile linkages as a biodegradable gene carrier. J Control Release. 2005;103:209–219. doi: 10.1016/j.jconrel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- [14].Christensen LV, Chang CW, Yockman JW, Conners R, Jackson H, Zhong Z, et al. Reducible poly(amido ethylenediamine) for hypoxia-inducible VEGF delivery. J Control Release. 2007;118:254–261. doi: 10.1016/j.jconrel.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30:5804–5814. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shim MS, Kwon YJ. Acid-transforming polypeptide micelles for targeted nonviral gene delivery. Biomaterials. 2010;31:3404–3413. doi: 10.1016/j.biomaterials.2010.01.019. [DOI] [PubMed] [Google Scholar]
- [17].Shim MS, Kwon YJ. Controlled cytoplasmic and nuclear localization of plasmid DNA and siRNA by differentially tailored polyethylenimine. J Control Release. 2009;133:206–213. doi: 10.1016/j.jconrel.2008.10.007. [DOI] [PubMed] [Google Scholar]
- [18].Kang HC, Bae YH. Polymeric gene transfection on insulin-secreting cells: sulfonylurea receptor-mediation and transfection medium effect. Pharm Res. 2006;23:1797–1808. doi: 10.1007/s11095-006-9027-0. [DOI] [PubMed] [Google Scholar]
- [19].Florea BI, Meaney C, Junginger HE, Borchard G. Transfection efficiency and toxicity of polyethylenimine in differentiated Calu-3 and nondifferentiated COS-1 cell cultures. AAPS PharmSci. 2002;4:E12. doi: 10.1208/ps040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dash PR, Read ML, Barrett LB, Wolfert MA, Seymour LW. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Ther. 1999;6:643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]
- [21].Spitnik P, Lipshitz R, Chargaff E. Studies in nucleoproteins. III. Deoxyribonucleic acid complexes with basic polyelectrolytes and their fractional extraction. J Biol Chem. 1954;215:765–775. [PubMed] [Google Scholar]
- [22].Kang HC, Bae YH. Transfection of rat pancreatic islet tissue by polymeric gene vectors. Diabetes Technol Ther. 2009;11:443–449. doi: 10.1089/dia.2008.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dautzenberg H, Karibyants N. Polyelectrolyte complex formation in highly aggregating systems. Effect of salt: response to subsequent addition of NaCl. Macromol Chem Phys. 1999;200:118–125. [Google Scholar]
- [24].Poon Z, Chang D, Zhao X, Hammond PT. Layer-by-Layer Nanoparticles with a pH-Sheddable Layer for in Vivo Targeting of Tumor Hypoxia. ACS Nano. 2011;5:4284–4292. doi: 10.1021/nn200876f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sethuraman VA, Na K, Bae YH. pH-responsive sulfonamide/PEI system for tumor specific gene delivery: In vitro study. Biomacromolecules. 2006;7:64–70. doi: 10.1021/bm0503571. [DOI] [PubMed] [Google Scholar]
- [26].Meng F, Hennink WE, Zhong Z. Reduction-sensitive polymers and bioconjugates for biomedical applications. Biomaterials. 2009;30:2180–2198. doi: 10.1016/j.biomaterials.2009.01.026. [DOI] [PubMed] [Google Scholar]
- [27].Kim HA, Lee BW, Kang D, Kim JH, Ihm SH, Lee M. Delivery of hypoxia-inducible VEGF gene to rat islets using polyethylenimine. J Drug Target. 2009;17:1–9. doi: 10.1080/10611860802392982. [DOI] [PubMed] [Google Scholar]
- [28].Liu ML, Oh JS, An SS, Pennant WA, Kim HJ, Gwak SJ, et al. Controlled nonviral gene delivery and expression using stable neural stem cell line transfected with a hypoxia-inducible gene expression system. J Gene Med. 2010;12:990–1001. doi: 10.1002/jgm.1527. [DOI] [PubMed] [Google Scholar]
- [29].Kim HA, Lim S, Moon HH, Kim SW, Hwang KC, Lee M, et al. Hypoxia-inducible vascular endothelial growth factor gene therapy using the oxygen-dependent degradation domain in myocardial ischemia. Pharm Res. 2010;27:2075–2084. doi: 10.1007/s11095-010-0206-7. [DOI] [PubMed] [Google Scholar]
- [30].Ihm JE, Han KO, Hwang CS, Kang JH, Ahn KD, Han IK, et al. Poly (4-vinylimidazole) as nonviral gene carrier: in vitro and in vivo transfection. Acta Biomater. 2005;1:165–172. doi: 10.1016/j.actbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- [31].Sarantopoulos C, McCallum B, Sapunar D, Kwok WM, Hogan Q. ATP-sensitive potassium channels in rat primary afferent neurons: the effect of neuropathic injury and gabapentin. Neurosci Lett. 2003;343:185–189. doi: 10.1016/s0304-3940(03)00383-5. [DOI] [PubMed] [Google Scholar]
- [32].Hunjan S, Mason RP, Mehta VD, Kulkarni PV, Aravind S, Arora V, et al. Simultaneous intracellular and extracellular pH measurement in the heart by 19F NMR of 6-fluoropyridoxol. Magn Reson Med. 1998;39:551–556. doi: 10.1002/mrm.1910390407. [DOI] [PubMed] [Google Scholar]
- [33].Volk T, Jahde E, Fortmeyer HP, Glusenkamp KH, Rajewsky MF. pH in human tumour xenografts: effect of intravenous administration of glucose. Br J Cancer. 1993;68:492–500. doi: 10.1038/bjc.1993.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bumke MA, Neri D, Elia G. Modulation of gene expression by extracellular pH variations in human fibroblasts: a transcriptomic and proteomic study. Proteomics. 2003;3:675–688. doi: 10.1002/pmic.200300395. [DOI] [PubMed] [Google Scholar]
- [35].Bear MP, Schneider FH. The effect of medium pH on rate of growth, neurite formation and acetylcholinesterase activity in mouse neuroblastoma cells in culture. J Cell Physiol. 1977;91:63–68. doi: 10.1002/jcp.1040910107. [DOI] [PubMed] [Google Scholar]
- [36].Petronini PG, Alfieri R, Campanini C, Borghetti AF. Effect of an alkaline shift on induction of the heat shock response in human fibroblasts. J Cell Physiol. 1995;162:322–329. doi: 10.1002/jcp.1041620304. [DOI] [PubMed] [Google Scholar]
- [37].Davoust J, Gruenberg J, Howell KE. Two threshold values of low pH block endocytosis at different stages. EMBO J. 1987;6:3601–3609. doi: 10.1002/j.1460-2075.1987.tb02691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cosson P, de Curtis I, Pouyssegur J, Griffiths G, Davoust J. Low cytoplasmic pH inhibits endocytosis and transport from the trans-Golgi network to the cell surface. J Cell Biol. 1989;108:377–387. doi: 10.1083/jcb.108.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Glunde K, Guggino SE, Solaiyappan M, Pathak AP, Ichikawa Y, Bhujwalla ZM. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 2003;5:533–545. doi: 10.1016/s1476-5586(03)80037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Altan N, Chen Y, Schindler M, Simon SM. Defective acidification in human breast tumor cells and implications for chemotherapy. J Exp Med. 1998;187:1583–1598. doi: 10.1084/jem.187.10.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Simon SM. Role of organelle pH in tumor cell biology and drug resistance. Drug Discov Today. 1999;4:32–38. doi: 10.1016/s1359-6446(98)01276-8. [DOI] [PubMed] [Google Scholar]
- [42].Kang HC, Bae YH. Endolysosomolytically active pH-sensitive polymeric nanotechnology. In: Weissig V, D’Souza GGM, editors. Organelle-specific pharmaceutical nanotechnology. John Wiley & Sons, Inc.; Hoboken, NJ: 2010. pp. 247–262. [Google Scholar]
- [43].Kang HC, Samsonova O, Bae YH. Trafficking microenvironmental pH of gene vector polycation in drug-sensitive and multidrug-resistant MCF7 breast cancer cell. Biomaterials. 2010;31:3071–3078. doi: 10.1016/j.biomaterials.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Morimoto K, Nishikawa M, Kawakami S, Nakano T, Hattori Y, Fumoto S, et al. Molecular weight-dependent gene transfection activity of unmodified and galactosylated polyethyleneimine on hepatoma cells and mouse liver. Mol Ther. 2003;7:254–261. doi: 10.1016/s1525-0016(02)00053-9. [DOI] [PubMed] [Google Scholar]
- [45].Overgaard J. Influence of extracellular pH on the viability and morphology of tumor cells exposed to hyperthermia. J Natl Cancer Inst. 1976;56:1243–1250. doi: 10.1093/jnci/56.6.1243. [DOI] [PubMed] [Google Scholar]
- [46].Suh J, Paik HJ, Hwang BK. Ionization of poly(ethyleneimine) and poly(allylamine) at various pH’s. Bioorg Chem. 1994;22:318–327. [Google Scholar]
- [47].Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- [48].Brunner S, Sauer T, Carotta S, Cotten M, Saltik M, Wagner E. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- [49].Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol Ther. 2002;5:80–86. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- [50].Mortimer I, Tam P, MacLachlan I, Graham RW, Saravolac EG, Joshi PB. Cationic lipid-mediated transfection of cells in culture requires mitotic activity. Gene Ther. 1999;6:403–411. doi: 10.1038/sj.gt.3300837. [DOI] [PubMed] [Google Scholar]
- [51].Tseng WC, Haselton FR, Giorgio TD. Mitosis enhances transgene expression of plasmid delivered by cationic liposomes. Biochim Biophys Acta. 1999;1445:53–64. doi: 10.1016/s0167-4781(99)00039-1. [DOI] [PubMed] [Google Scholar]
- [52].Kang HC, Lee M, Bae YH. Polymeric gene carriers. Crit Rev Eukaryot Gene Expr. 2005;15:317–342. doi: 10.1615/critreveukargeneexpr.v15.i4.30. [DOI] [PubMed] [Google Scholar]
- [53].Smith RM, Baibakov B, Lambert NA, Vogel SS. Low pH inhibits compensatory endocytosis at a step between depolarization and calcium influx. Traffic. 2002;3:397–406. doi: 10.1034/j.1600-0854.2002.30603.x. [DOI] [PubMed] [Google Scholar]
- [54].Sandvig K, Olsnes S, Petersen OW, van Deurs B. Acidification of the cytosol inhibits endocytosis from coated pits. J Cell Biol. 1987;105:679–689. doi: 10.1083/jcb.105.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sandvig K, Olsnes S, Petersen OW, van Deurs B. Inhibition of endocytosis from coated pits by acidification of the cytosol. J Cell Biochem. 1988;36:73–81. doi: 10.1002/jcb.240360108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.