Abstract
A flow-based method employing a reverse displacement immunoassay was combined with ultrafast immunoextraction and near-infrared fluorescence detection for the analysis of free drug fractions, using phenytoin as a model analyte. Factors considered in the design of this method included the sample application conditions, the design of the immobilized drug analog column, the utilization of antibodies or Fab fragments as labeled binding agents, and the label application and column regeneration conditions. In the final method, sample injections led to the displacement of labeled binding agents from an immobilized phenytoin analog column. This displacement peak appeared within 20–30 s of sample injection and was proportional in size to the free phenytoin concentration in the sample. It was possible with this method to regenerate the column by using only the application of additional label between sample injections. This method was used to measure clinically-relevant concentrations of free phenytoin in serum and drug/protein mixtures and gave good correlation with ultrafiltration, while also being faster to perform and requiring significantly less sample. This technique was not limited to free phenytoin measurements but could be adapted for other drugs or analytes through the use of appropriate columns and binding agents.
INTRODUCTION
Many drugs bind reversibly with serum proteins, creating both a protein-bound fraction and a free fraction in the circulation. The free fraction is often thought to represent the active form of a drug because it is capable of crossing membranes and interacting with receptors.1–3 Although a drug’s total concentration is often used in pharmaceutical testing, the relationship between the total concentration and free fraction of a drug can be affected by factors such as illness, trauma, surgery, or age.3 Problems that arise because of these effects have created an ongoing need for rapid and accurate methods that can directly measure free drug fractions.1,4
Phenytoin is a common antiepileptic drug that is highly bound (e.g., 90%) in blood to human serum albumin (HSA)3–7 (see Refs. [3] and [5] for more details on the binding sites that are involved in this interaction and drugs or other solutes that may affect this binding). Equilibrium dialysis and ultrafiltration are often used to isolate free phenytoin fractions from clinical samples, but these methods tend to have long analysis times, problems with non-specific binding, and require a separate technique (e.g., HPLC) to measure the free drug fraction.1,4,6,7 Restricted access media (RAM) columns have also been used to isolate free phenytoin fractions from drug/protein mixtures but have not been used for this purpose with real clinical samples.8
Another technique that has recently been developed for free drug measurements is an ultrafast immunoextraction/displacement assay (UFIDA).4,9 This flow-based approach uses an affinity microcolumn that contains immobilized antibodies for the drug of interest. These antibodies are loaded with a labeled drug analog, which is displaced by the free fraction of a drug during sample injection. This technique has been used with both near-infrared (NIR) fluorescent labels and chemiluminescent labels and provides good agreement with reference methods.4,9 One possible limitation of this approach is that the retained analyte does have to be eluted and the column regenerated on a regular basis. This requirement limits the sample throughput of this approach and can add a significant amount of time to the overall analysis.4,9,10
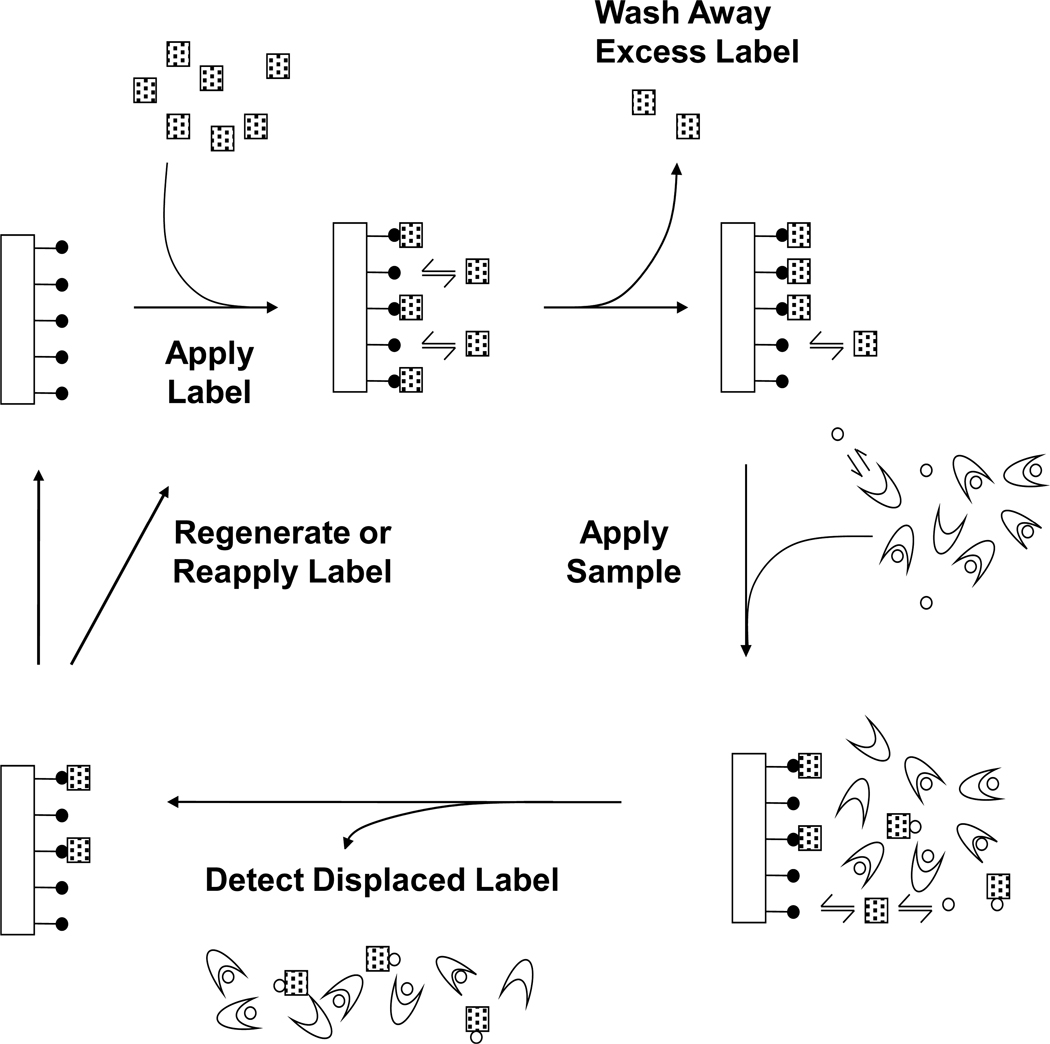
This report will explore a new flow-based approach for free drug measurements based on ultrafast affinity extraction and a reverse displacement immunoassay (RDIA). Phenytoin will be used as the model analyte in this work. The general scheme for this approach is given in Figure 1. First, a labeled binding agent (or “label”) will be applied to an affinity microcolumn containing an immobilized analog of the target drug, or a structurally-related species.10–12 After excess label has been washed away, a sample containing the analyte and proteins will be injected. This sample will be passed through the microcolumn on a sufficiently small timescale to minimize dissociation for the drug from binding proteins in the sample.4,9 Under these conditions, only the drug’s free fraction should displace the label from the column, giving a displacement peak that is proportional to the sample concentration of the free drug.
Figure 1.
General scheme for a reverse displacement immunoassay (RDIA). Symbols: ( ), immobilized drug analog; (
), immobilized drug analog; ( ), labeled monoclonal antibody or Fab fragments (i.e., the "label"); (○), drug or target analyte; (
), labeled monoclonal antibody or Fab fragments (i.e., the "label"); (○), drug or target analyte; ( ), serum protein or binding agent in the sample.
), serum protein or binding agent in the sample.
The RDIA method is similar to a one-site immunometric method, in which an immobilized analog of the analyte is also used to bind to the label.10–13 However, the one-site immunometric assay uses a relative large amount of the immobilized analog to extract excess label from a pre-incubated mixture of this label with the sample, with the remaining analyte-label complex in solution then being detected.10,13 In contrast to this, a signal is produced in an RDIA through the displacement of label by the analyte from a small immobilized analog column. This approaches means that no pre-incubation of the sample and label is required in the RDIA method, thus leading to shorter analysis times than a one-site immunometric assay. Another potential advantage of an RDIA over both a one-site immunometric method and the UFIDA is that no elution/regeneration step, other than the application of more label, should be needed between sample injections. The expected result is a relatively rapid method that can be employed for the direct analysis of free drug fractions or other analytes in complex samples.
EXPERIMENTAL
Reagents
The phenytoin (99% pure), HSA (>96%, essentially fatty acid free) and commercial sample of pooled human serum (negative for hepatitis B or C and HIV; handled with standard precautions for bloodborne pathogens) were from Sigma-Aldrich (St. Louis, MO). Reagents for the bicinchoninic acid (BCA) assay were from Pierce (Rockford, IL). The 3-N-amino-5,5-diphenylhydantoin (ADPH) was from Ryan Scientific (Mount Pleasant, SC). The glutaraldehyde was obtained from Fisher Scientific (Fairlawn, NJ). The IRDye 800 CW N-hydroxysuccinimide (NHS) ester was from Li-Cor (Lincoln, NE). The monoclonal anti-phenytoin antibodies (clone 16302, protein A purified from ascites fluid) were purchased from QED Biosciences (San Diego, CA). The Nucleosil Si-1000 (1000 Å pore size, 5 µm particle size) and Nucleosil Si-300 (300 Å pore size, 7 µm particle size) were from Macherey Nagel (Düren, Germany). All aqueous solutions were prepared using water from a Nanopure system (Barnstead, Dubuque, IA). All other chemicals were reagent grade or better.
Apparatus
The Vivaspin 6 PES membrane concentrators (30 kDa MW cutoff) and 5702RH temperature controlled centrifuge were from VWR (West Chester, PA). The Zeba Desalt spin columns were from Pierce. Ultrafiltration was performed using Centrifree Micropartition devices (30 kDa MW cutoff, 0.15–1.5 mL sample capacity) from Amicon (Danvers, MA). Columns were packed using an Alltech 1666 slurry packer (Deerfield, IL).
The HPLC system contained two PU-2080 pumps, a CO-2057 column oven, and an AS-2057 autosampler from Jasco (Easton, MD). The autosampler was equipped with a 100 µL loop and operated in the partial loop injection mode. This system was controlled using a Jasco LC-Net II system controller and EZ Chrom SI software (Scientific Software, Pleasanton, CA). A Jasco UV-2075 absorbance detector was used with this system to examine the ultrafiltration samples, which were injected onto a 5 cm × 4.6 mm i.d. column containing HSA immobilized to Nucleosil Si-300.14,15 For the RDIA method, the HPLC system included a custom-built NIR fluorescence detector supplied by Li-Cor, as described previously.9,16 All peaks were integrated using PeakFit 4.12 (Systat Software, San Jose, CA); moments analysis was performed by using the residual option of this program and an EMG-GMG fit with a linear progressive baseline.
Preparation of affinity microcolumns
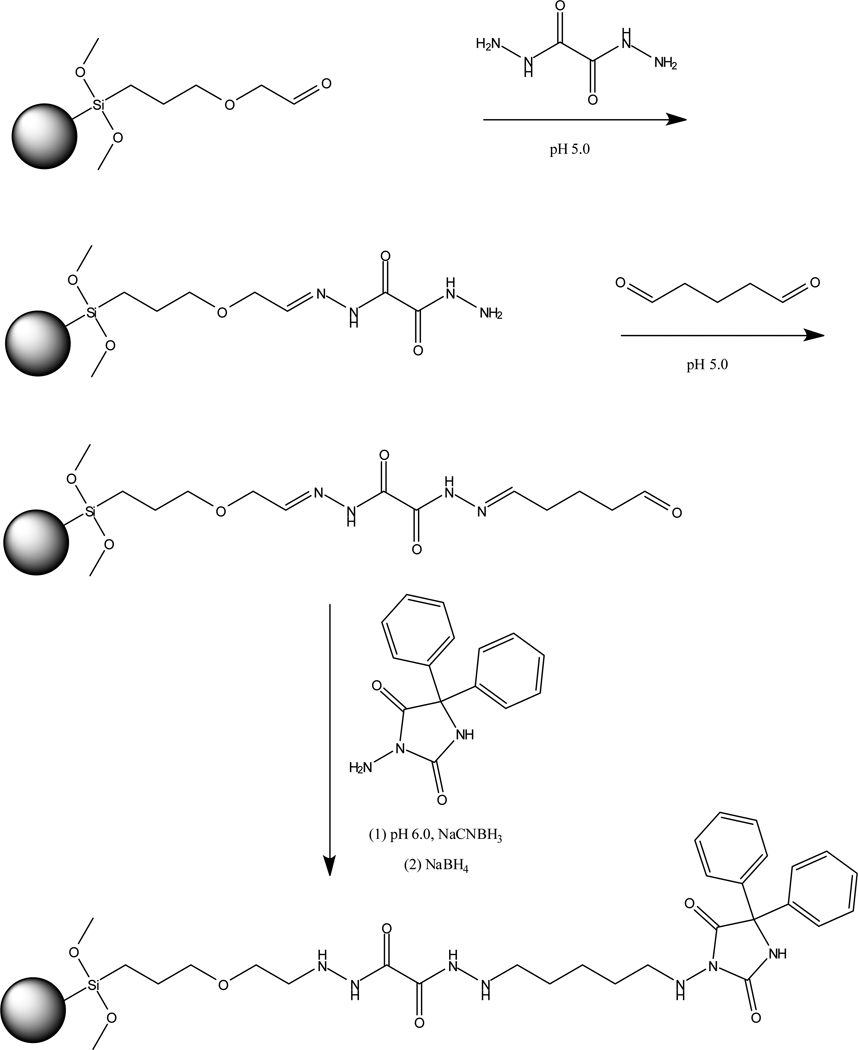
A support using immobilized ADPH as an analog of phenytoin was prepared using Nucleosil Si-1000 silica (see Supplemental Material for details). The silica was first converted into a diol form and oxidized with periodate to generate aldehyde groups,14,15 giving the starting material shown in Figure 2. This aldehyde-activated support was reacted with oxalic dihydrazide at pH 5.0.14 The dihydrazide silica was then combined at pH 5.0 with an excess of glutaraldehyde, followed by the addition of ADPH and sodium cyanoborohydride at pH 6.0. This support was later treated with sodium borohydride at pH 8.0 to remove any remaining aldehyde groups.14,15 The ADPH content of the final support was estimated with a BCA assay16 to be a maximum of 1–2 µmol ADPH/g silica. A control support was prepared in the same manner but with no ADPH being added during the immobilization step. The ADPH support and control support were packed into separate 1 mm × 2.1 mm I.D. columns at 3000 psi (20.7 MPa) using pH 7.4, 0.067 M phosphate buffer as the packing solution. All columns were stored at 4°C in pH 7.4, 0.067 M potassium phosphate buffer when not in use.
Figure 2.
Preparation of a silica support containing an immobilized analog of phenytoin.
Preparation of labeled antibodies and Fab fragments
The anti-phenytoin monoclonal antibodies were placed into pH 8.5, 0.10 M potassium phosphate buffer by using a Vivaspin concentrator and adjusted to a concentration of ~1 mg/mL. A 0.1–0.22 mg portion of IRDye 800 CW NHS ester was dissolved in 25 µL water and a 7.5 µL aliquot was combined with 1 mL of the pH 8.5 antibody solution. This mixture was shaken for 2 h in the dark at room temperature. A Zeba desalt spin column eluted with pH 7.4, 0.067 M potassium phosphate buffer was used to separate the labeled antibodies from the unreacted dye. Absorbance measurements at 780 and 280 nm were used to determine the dye/protein ratio and the approximate antibody concentration of this mixture. The labeled antibodies had a final concentration of 0.4–0.8 mg/mL and a dye/protein ratio of 0.5–1.5. These labeled antibodies were stored at 4°C in pH 7.4, 0.067 M phosphate buffer prior to further experiments.
Fab fragments were generated from the anti-phenytoin monoclonal antibodies by using a mouse IgG1 Fab and F(ab’)2 preparation kit from Pierce (Rockford, IL). The Fab fragments were separated from the Fc fragments and undigested antibodies by collecting the non-retained fraction of the digest that passed through a protein A column, as supplied with the kit. The Fab fragments were stored in pH 8.5, 0.1 M potassium phosphate buffer at −80°C prior to labeling. The labeling of the Fab fragments with IRDye 800 CW NHS ester, and the purification and characterization of the labeled Fab fragments, was carried out in the same manner as described for the labeled antibodies. This preparation had a dye/protein ratio of 4.2 and a final Fab concentration of 0.13 mg/mL. The labeled Fab fragments were stored at 4°C in pH 7.4, 0.067 M phosphate buffer.
RDIA and ultrafiltration
All experiments were performed in duplicate or triplicate at 37°C. In the RDIA method, pH 7.4, 0.067 M potassium phosphate buffer was used for sample or label injection and pH 2.5, 0.067 M phosphate buffer was utilized for column regeneration. The final RDIA method used an injection of 100 pmol label at 0.1 mL/min, followed by a switch to 1.2 mL/min for the injection of a 20 µL sample. After the displacement peak had appeared, a switch was made to 0.1 mL/min and more label was applied prior to the next sample injection. For column regeneration, the flow rate was switched to 1 mL/min for the application of pH 2.5 elution buffer, followed by application of the pH 7.4 buffer at 1 mL/min and label at 0.1 mL/min.
In the ultrafiltration experiments, a 1 mL aliquot of each sample was added to a Centrifree micropartition device, which was then centrifuged at 37°C for 45 min at 1500 × g. To measure the free phenytoin concentration, the filtrate of each sample was collected and 20 µL was injected in duplicate at 0.5 mL/min onto a 5 cm × 4.6 mm i.d. HSA column in pH 7.4, 0.067 M potassium phosphate buffer and with detection at 205 nm, as described previously.9
RESULTS AND DISCUSSION
General method design and initial column development
The key components for the scheme shown in Figure 1 for the RDIA method can be represented by the following reactions.
| (1) |
| (2) |
Eq (1) represents the binding and dissociation of the immobilized analyte analog (I) with the labeled binding agent or “label” (L) in the affinity column, where this process creates a local pseudo-equilibrium between the free and bound states for L. The terms ka,LI and kd,LI represent the second-order association and first-order dissociation rate constants for this process. Eq (2) represents the binding that can then occur between an injected analyte (A) and the label that is present in a non-bound state in the mobile phase, where ka,LI is the second-order association rate constant for this reaction.
For the system in eqs (1–2), the labeled binding agent has already been applied to the column, allowed to bind to the immobilized analog, and excess label has been washed from the column. However, even after excess label has been removed, a small amount of the retained label will dissociate from the immobilized analog and either rebind to the column or interact with an injected analyte. The formation of a complex between the label and the analyte (L-A) is what leads to the formation of a displacement peak and the measured signal for this assay. If a small column and reasonably fast flow rate are used for this method, as employed in this report, the dissociation of L-A back to the analyte and non-bound label within the column can be assumed to be negligible on the time scale of the experiment.
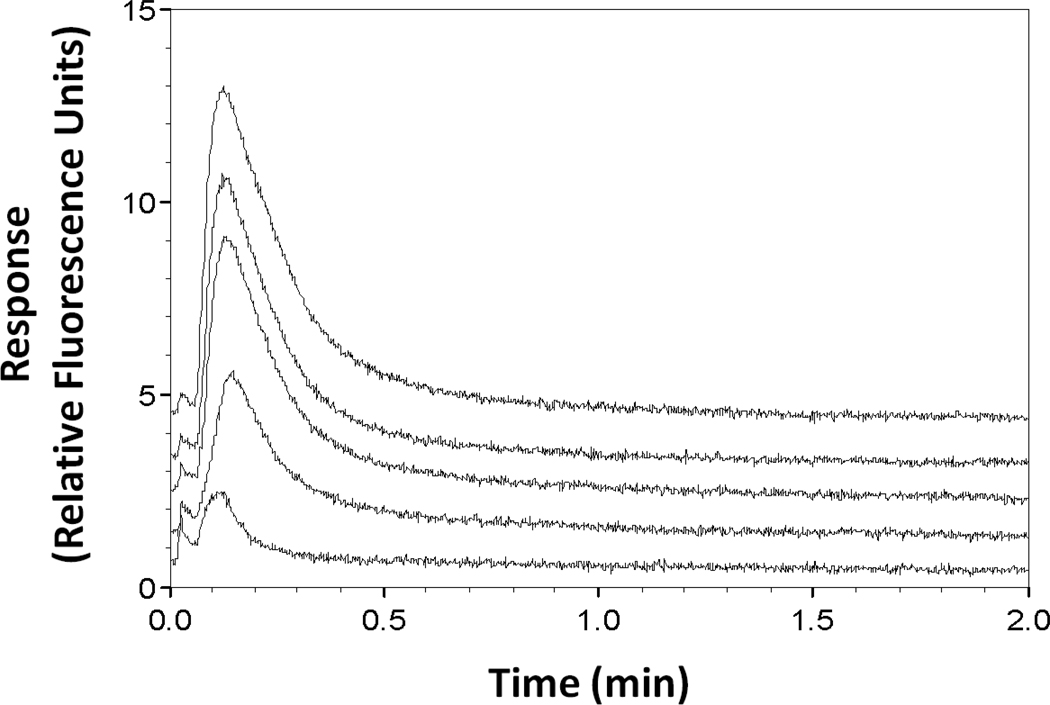
Two factors that were originally considered for the development of an RDIA method for free drug measurements were the column size and flow rate that were employed. It is known that the accurate measurement of free drug fractions by ultrafast affinity extraction requires that a sample be passed through the extraction column quickly enough to minimize dissociation of the protein-bound form of the drug in the sample.4,9,18 In a previous UFIDA method for measuring free phenytoin, an extraction time of 140 ms was found to be suitable for such work.9 The same extraction time was created in this current study by injected samples at a flow rate of 1.2 mL/min onto a 1 mm × 2.1 mm I.D. affinity microcolumn. The support that was placed within this column is shown in Figure 2 and contained an immobilized analog of phenytoin. The ability of this support to bind labeled anti-phenytoin antibodies was tested by injecting 100 pmol of this label at 0.1 mL/min, followed by an injection at 1.2 mL/min of a 100 µL sample of 50 µM phenytoin (i.e., a concentration ten-times that seen for free phenytoin in serum at therapeutic levels).9,19 As illustrated in Figure 3, a large displacement peak was observed under these conditions. However, little or no displacement peak was seen when a control column was used in place of the drug analog column or when only buffer was injected after the label onto the drug analog column (see Supplemental Material for examples).
Figure 3.
Typical displacement peaks obtained by the RDIA method for 100 µL sample injections of 50, 10, 5, 1, or 0.1 µM phenytoin (top-to-bottom). These displacement peaks were obtained at 1.2 mL/min and pH 7.4 with only re-application of label between samples being used for column regeneration.
In the RDIA method it was not necessary to restrict the extraction time for the label to the millisecond time domain. This meant that a lower application flow rate could be used for the label than for the sample, as has also been noted for the UFIDA method.9 The use of a slower flow rate for label application (i.e., 0.1 vs. 1.2 mL/min) was found to increase the signal of the displacement peaks by six- to seven-fold. This increase was believed to be due to the more efficient capture of the label as it was allowed more time for binding to the immobilized drug analog. A similar effect has been observed in other flow-based immunoassays and is typically related to the adsorption-limited kinetics that are often present for antibody-antigen interactions in the types of supports that were used in this study.4,9,10
Another factor found to affect the ability of the label to bind to the extraction column was the length of the spacer arm between the drug analog and the support. The reaction scheme in Figure 2 provided a seventeen-atom spacer between the drug analog and the support, which was the material used throughout the remainder of this report. Another support that was originally considered had the drug analog attached to aldehyde-activated silica through the Schiff base method, creating a six-atom spacer (see Supplemental Material for further details). This material was slightly easier and faster to make than the one in Figure 2, used the same site of attachment for the drug analog, and contained an equal or greater amount of the immobilized drug analog. Neither of these two materials or spacer arms gave any significant amount of non-specific binding to the label or measurable displacement peaks when used without the immobilized drug analog in the RDIA method. However, the support with the shorter spacer arm did result in a seven- to eight-fold smaller signal in the RDIA method than when the support with the larger spacer arm was utilized. The difference in these signals was determined to be mainly related to steric restrictions that affected the extent to which the labeled binding agent (e.g., antibodies) could interact with the immobilized drug analog as the spacer arm was decreased in length.
Method optimization
Several other factors were considered during the development of the RDIA method. As suggested by the scheme in eqs (1–2), one of these factors was the amount of label that was employed. To optimize this parameter, the quantity of applied label was varied while the resulting displacement peak was measured for a 100 µL sample of 50 µM phenytoin. As the amount of applied label was decreased below 100 pmol, a linear decrease in the size of the displacement peak was produced. However, the displacement peak began to show a nonlinear response as more than 100 pmol label was applied. Thus, a 100 pmol portion of labeled antibodies, or labeled Fab fragments, was used in all further experiments. A load of 100 pmol label represented at least a 30- to 60-fold excess versus the total estimated amount of immobilized drug analog in the affinity microcolumn; the actual excess was even higher because the label was able to bind to less than a third of the immobilized analog, as determined later in this section. This fact meant that only a small fraction of the label was binding to this column even at 0.1 mL/min, as confirmed experimentally. It should be possible in future work with the RDIA method to recapture and recycle this non-retained label for use in later cycles, thus helping to further reduce the amount of label that is needed for this approach.
The initial testing of the RDIA method used a pH 2.5 elution step to regenerate the drug analog column after label had been applied and a displacement peak for the sample had been detected (i.e., a process taking 10–15 min per cycle). Additional experiments examined whether multiple samples could be injected after one application of the label to the drug analog column. This was tested by applying 100 pmol of label followed by a series of 20 µL injections of 5 µM phenytoin (i.e., a typical concentration for free phenytoin in serum). When compared to the displacement peak for the first sample injection, the second, third and fourth injections gave displacement peaks that were 32%, 59%, and 63% smaller. The displacement peaks for further injections decreased even more in size. This was not surprising because the accumulated stoichiometric amount of injected phenytoin after four injections (i.e., 0.4 pmol) approached the estimated amount of retained label in the column (i.e., a maximum of 1.5–3 pmol). Although these results indicated that not all the label was displaced after a single sample injection, the amount of label that was displaced by one sample did affect the signal that was obtained for later samples if no additional label had been applied between injections. From this experiment it was estimated that 13–27% of the immobilized analog groups on the support were immunologically active and available for binding to the label under the given application conditions.
Further experiments were carried out in which a fixed amount of label was applied between samples while pH 7.4 buffer was continuously passed through the system. In this case, 100 pmol of label was applied between 100 µL injections of 50 µM phenytoin (i.e., 5 pmol analyte, or at least a 1.6- to 3.2-fold excess versus label that was adsorbed to the column). In this case, the displacement peak for the second sample showed an increase of 22.5% in its displacement peak when compared to the first sample injection. The next three injections showed only a variation of 4.7% in their displacement peaks, and the fourth and fifth injections gave peaks that varied by only 0.4%. These results indicated that the simple reapplication of label between sample injections could be used to regenerate the column. This approach added only a few minutes to the analysis and was much faster than the 10–15 min required for column regeneration when using both a pH 2.5 elution step and label application. Thus, the application of only label between sample injections was used in all further experiments unless otherwise indicated.
Response of RDIA method
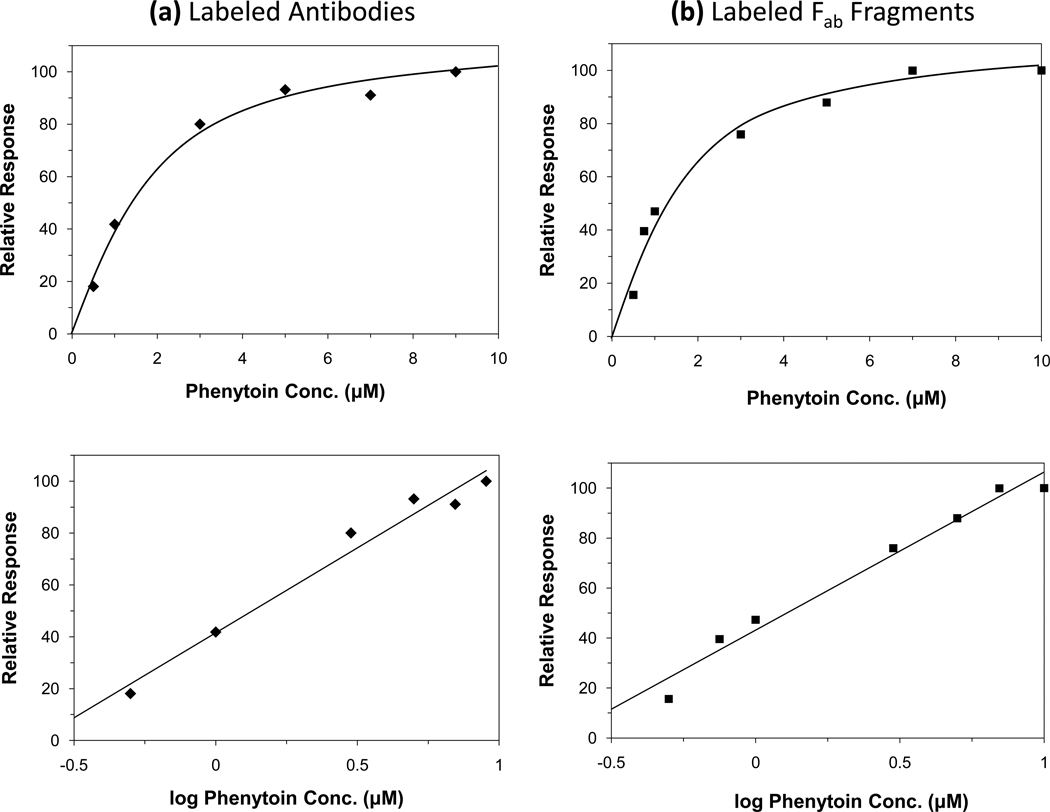
Some typical calibration curves for the RDIA method are shown in Figure 4. When these results were plotted as a function of analyte concentration, a non-linear change in response was observed. An essentially identical response and calibration range was obtained when using equal amounts of labeled antibodies or labeled Fab fragments. This information indicated that the curvature of these plots was not the result of using bivalent instead of monovalent binding agents as the labeled species (e.g., intact antibodies versus Fab fragments). An alternative explanation suggested by the model in eqs (1–2) and the results in the previous sections was that this curvature was at least partly a result of the kinetic processes occurring in the RDIA method.10,18 Thus, a second set of calibration curves were prepared in which the response of the assay was plotted as a function of the logarithm of the analyte concentration. The plots obtained for the assay response versus log(analyte concentration) appeared to follow a linear response over the range of phenytoin concentrations that were tested (correlation coefficient = 0.986–0.987, n = 6–7). This linear response was observed when using either labeled antibodies or labeled Fab fragments in the assay. Such a response suggested that the assay signal could be described, to an initial approximation, by using eq (2) and a pseudo-first order reaction between the injected analyte and labeled binding agent in the mobile phase. Further studies are being planned to examine the theoretical basis of this response in more detail.
Figure 4.
Calibration curves obtained by the RDIA method for phenytoin when using (a) labeled anti-phenytoin antibodies or (b) labeled anti-phenytoin Fab fragments. These results were obtained for 20 µL sample injections. Other conditions were the same as in Figure 3 or as given in the text. All of these measurements were made in duplicate or triplicate; the typical precisions that were obtained are summarized in the text. The use of six calibration standards in (a) instead of the seven used in (b) simply reflects that different amounts of each type of label that were available at the different times at which these studies were conducted; however, the overall range and response of the two curves are otherwise quite similar.
As noted for other flow-based assays involving similar supports, the response of this method was found to be related to the moles of injected analyte.10,18 One practical consequence of this relationship was that a change in sample volume could be used to adjust the position and response of a concentration-based calibration curve for this assay. For instance, an increase in sample volume from 10 µL to 20 µL would double the moles of analyte that were applied with each injection, thus shifting a concentration-based calibration curve for the assay to a two-fold lower range in concentration (Note: a calibration curve based on the moles of injected analyte would not be affected by the same change in sample volume). In this report, a sample volume of 20 µL was used in the final method to match the usable calibration range with the concentrations of free phenytoin that would be expected in serum at therapeutic levels.
Another item that was considered was the effect of HSA on the RDIA method. This was of interest because HSA has a high concentration in serum and is known to bind to phenytoin and structurally-related compounds.3–7,20 It was found that a sample containing 550 µM HSA (i.e., a representative serum concentration)19,21 did give a reproducible displacement peak in the RDIA method; however, this peak was smaller (i.e., 25–30% or less) than the displacement peaks seen for free phenytoin in spiked serum or representative phenytoin/HSA mixtures. The NIR fluorescent dye used in this report as a label is known to have little or no change in its fluorescence in the presence of serum or HSA9 and has no appreciable binding to HSA. Thus, the peak observed during the injections of HSA in the RDIA method was probably due to the displacement of label as this protein bound to the drug analog column. In this study, a correction for this effect was made for all drug/HSA mixtures and serum samples by subtracting the results for an HSA standard or serum sample that did not contain any phenytoin. This approach was satisfactory for this current report, but alternative approaches for avoiding or minimizing this effect are currently being examined.
Performance of RDIA method
Under the conditions employed in Figure 4, the lower limit of detection for the RDIA method was estimated to be 27–29 pmol (i.e., 1.3–1.4 µM for a 20 µL sample) at a signal-to-noise ratio of three when using either labeled antibodies or labeled Fab fragments. The upper limit of detection extended to at least 160–200 pmol (i.e., 8–10 µM for a 20 µL sample). This dynamic range was a good match with the levels expected for free phenytoin in clinical samples.9,19 The fact that the lower and upper limits of detection were larger than the amount of retained label in the drug analog column indicated that only a fraction of the analyte was required to displace this label for detection. This feature was attractive for the measurement of free drug fractions, where minimal perturbation of the sample is desired. In addition, this feature suggested that longer drug analog columns and slower flow rates could be used in other applications of the RDIA method to provide lower limits of detection by allowing for more efficient displacement of the label by the analyte.
As shown in Figure 3, displacement peaks were obtained within 20–30 s of sample injection. The total time per analysis, when using the injection of label between samples and no pH step for elution, was less than 5–10 min per sample once two more applications of label had been made onto the drug analog column. The method was also found to be robust. For instance, the two sets of curves in Figure 4 were obtained using different batches of the drug analog support and labeled agent, yet these curves still gave a consistent response and calibration range. In addition, the amount of drug analog support that was required per column was only a few milligrams and each drug analog column could be used for several months and hundreds of cycles with no significant signs of degradation.
The accuracy of this method was tested by using it to measure the free concentration of phenytoin in both mixtures of this drug with physiological concentrations of HSA and in serum spiked with phenytoin, with the latter being used as a representative clinical sample (Note: additional clinical samples will be considered in future studies). The results are summarized in Table 1. Identical samples were examined by using ultrafiltration as a reference method to measure the free drug fractions.9 The RDIA results differed from those obtained by ultrafiltration by only 4–11% and all overlapped within ±1 standard error of the mean for the reference values. As has been observed for the UFIDA method, the overall assay precision in the RDIA technique was mainly determined by factors that affected the amount of label that was present for each injection (e.g., the amount of injected label, the timing of the label and assay injections, and the flow rate).4,9,10 The precision of the RDIA method for the tested samples was 7–23% (average coefficient of variation, 13%), which was comparable to or slightly higher than the 4–13% (average, 7%) seen when using ultrafiltration. However, the RDIA method was significantly faster than the ultrafiltration method, which had a total analysis time of at least 1 hour per sample.9 The RDIA method also required much smaller amounts of sample than ultrafiltration (i.e., 20 µL versus 1.0 mL).
Table 1.
Determination of free phenytoin concentrations by ultrafiltration and RDIA
| Measured Free Phenytoin Conc. (µM)a | ||
|---|---|---|
| Sample | Ultrafiltration | RDIA |
| 550 µM HSA + 10 µM Phenytoin | 2.4 (±0.3) | 2.6 (±0.6) |
| 550 µM HSA + 16 µM Phenytoin | 3.7 (±0.2) | 4.1 (±0.3) |
| Serum + 12 µM Phenytoin | 2.5 (±0.1) | 2.4 (±0.2) |
All measurements were made in duplicate or triplicate at 37°C and in pH 7.4, 0.067 M potassium phosphate buffer. The values in parentheses represent ±1 standard error of the mean. The data for the drug/protein mixtures were obtained using labeled antibodies and the serum results were obtained using labeled Fab fragments.
CONCLUSIONS
This report described the development of an RDIA method as a new format for flow-based immunoassays. This approach was illustrated by using the detection of free drug fractions of phenytoin as a model. Several factors were considered in the creation and optimization of this assay, such as the sample application conditions, the design of the affinity support, the amount and type of label that was applied, and approaches for regenerating the column between sample injections. The final assay was found to be a robust method that produced a signal within 20–30 s of sample injection. This technique gave good correlation with ultrafiltration for samples containing clinically-relevant levels of free phenytoin; however, the RDIA method required much less time and sample to perform these measurements.
The RDIA method is not limited to phenytoin or the measurement of free drug fractions but could be applied to any analyte for which an appropriate label and immobilized analyte analog (or structurally related agent)10–12 is available or can be developed. The use of an immobilized analog of the analyte should provide a stable support that can be utilized with a variety of elution conditions, as has been noted for other methods that use similar materials (e.g., a one-site immunometric assay).10–13 The fact that the label, rather than a sample component, is bound to the immobilized analog and used for signal generation is another attractive feature. As an example, it was shown in this report that this feature makes it possible to regenerate a phenytoin analog column by simply applying more label between sample injections. The fact that the RDIA method eliminates the need for any pre-incubation of the sample with a labeled binding agent or the need for a separate elution buffer should lead to a significant reduction in analysis time for this approach when compared to other flow-based immunoassays (e.g., the UFIDA method, one-site immunometric assay, and traditional displacement assays).4,9,10–13 These advantages make the RDIA method attractive for future work requiring the high-throughput analysis of drugs or biological agents in pharmaceutical and clinical samples.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Basiaga and M. Shortridge for their assistance in characterizing the silica supports or Fab fragments. J.E. Schiel was supported, in part, by the Society of Analytical Chemists of Pittsburgh through an ACS Division of Analytical Chemistry Graduate Fellowship. This work was supported by the National Institutes of Health (NIH) under grant R01 GM044931 and the University of Nebraska Research Council was carried out in facilities renovated with support under NIH grant RR015468-01.
REFERENCES
- 1.Herve F, Urien S, Albengres E, Duche J-C, Tillement JP. Clin. Pharmacokin. 1994;26:44–58. doi: 10.2165/00003088-199426010-00004. [DOI] [PubMed] [Google Scholar]
- 2.Smith QR, Fisher C, Allen DD. In: Blood-Brain Barrier: Drug Delivery and Brain Pathology. Kobiler D, Lustig S, Shapira S, editors. New York: Kluwer Academic/Plenum Publishers; 2000. pp. 311–321. [Google Scholar]
- 3.Thummel KE, Shen DD. In: Goodman and Gilman's The Pharmacological Basis of Therapeutics. 10th ed. Hardman JG, Limbird LE, editors. New York: McGraw-Hill; 2001. pp. 1924–2023. [Google Scholar]
- 4.Clarke W, Schiel JE, Moser A, Hage DS. Anal. Chem. 2005;77:1859–1866. doi: 10.1021/ac040127x. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Ohnmacht CM, Hage DS. J. Chromatogr. B. 2004;809:137–145. doi: 10.1016/j.jchromb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Roberts WL, Annesley TM, De BK, Moulton L, Juenke JM, Moyer T. Therapeut. Drug. Monit. 2001;23:148–154. doi: 10.1097/00007691-200104000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Warner A, Privitera M, Bates D. Clin. Chem. 1998;44:1085–1095. [PubMed] [Google Scholar]
- 8.Miller TD, Pinkerton TC. Anal. Chim. Acta. 1985;170:295–300. [Google Scholar]
- 9.Ohnmacht CM, Schiel JE, Hage DS. Anal. Chem. 2006;78:7547–7556. doi: 10.1021/ac061215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moser AC, Hage DS. Chap. 29. In: Hage DS, editor. Handbook of Affinity Chromatography. New York: Taylor and Francis; 2006. [Google Scholar]
- 11.Freytag JW, Dickinson JC, Tseng SY. Clin. Chem. 1984;30:417–420. [PubMed] [Google Scholar]
- 12.Guanratna PC, Wilson GS. Anal. Chem. 1993;65:1152–1157. doi: 10.1021/ac00057a009. [DOI] [PubMed] [Google Scholar]
- 13.Hage DS, Nelson MA. Anal. Chem. 2001;73:198A–205A. [PubMed] [Google Scholar]
- 14.Kim HS, Hage DS. Chap. 3. In: Hage DS, editor. Handbook of Affinity Chromatography. New York: Taylor and Francis; 2006. [Google Scholar]
- 15.Larsson PO. Methods Enzymol. 1984;104:212–223. doi: 10.1016/s0076-6879(84)04091-x. [DOI] [PubMed] [Google Scholar]
- 16.Middendorf L, Amen J, Bruce R, Draney D, DeGraff D, Gewecke J, Grone D, Humphrey P, Little G, Lugade A, Narayanan N, Oommen A, Osterman H, Peterson R, Rada J, Raghavachari R, Roemer Sn. In: Near-Infrared Dyes for High Technology Applications. Daehne S, editor. Netherlands: Kluwer Academic; 1998. pp. 21–54. [Google Scholar]
- 17.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal. Biochem. 1985;150:76. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 18.Clarke W, Chowdhuri AR, Hage DS. Anal. Chem. 2001;73:2157–2164. doi: 10.1021/ac0009752. [DOI] [PubMed] [Google Scholar]
- 19.Tietz NW, editor. Textbook of Clinical Chemistry. Philadelphia: Saunders; 1986. [Google Scholar]
- 20.Ohnmacht CM, Chen S, Tong Z. J. Chromatogr. B. 2006;836:83–91. doi: 10.1016/j.jchromb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 21.Peters T Jr, editor. All About Albumin: Biochemistry, Genetics and Medical Applications. San Diego: Academic Press; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.