Abstract
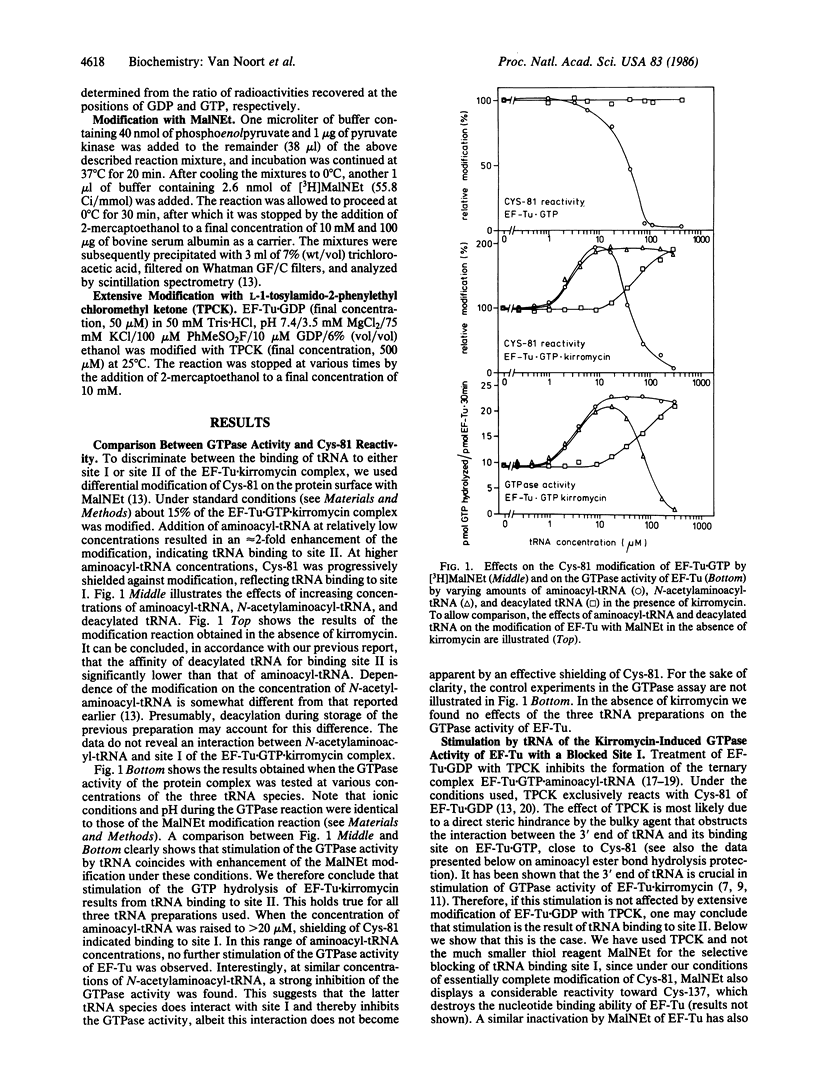
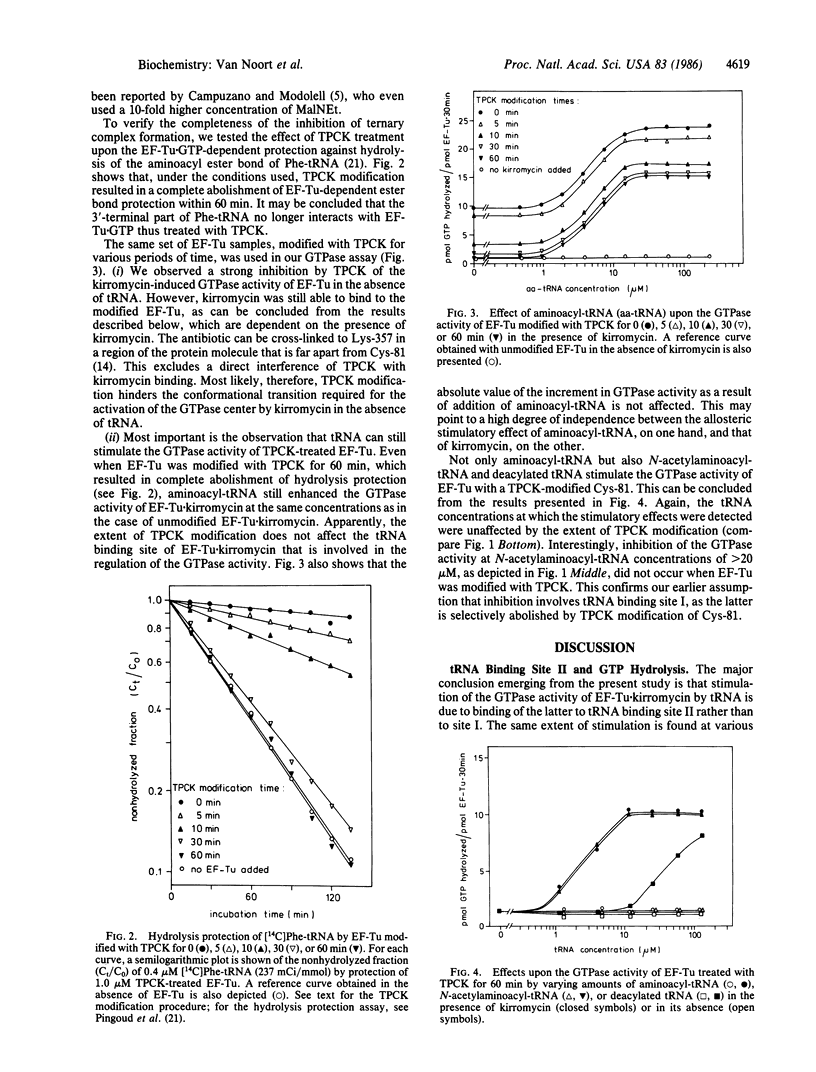
Interaction of the elongation factor EF-Tu with the antibiotic kirromycin results in activation of the GTPase center of the factor and in induction of an additional tRNA binding site (tRNA binding site II to distinguish it from the classical tRNA binding site I). Activation of the GTPase center under these conditions is stimulated by addition of tRNA. Two-fold evidence is presented that this stimulation is due to tRNA binding to site II rather than to site I. First, a strong correlation is observed between stimulation of the GTPase activity and enhancement of the reactivity of Cys-81 of EF-Tu toward N-ethylmaleimide at various concentrations of aminoacyl-tRNA, deacylated tRNA, and N-acetylaminoacyl-tRNA. The latter effects signal tRNA binding to site II. Stimulation of the kirromycin-induced GTPase activity by tRNA binding to the factor also occurs when binding to site I is completely abolished. Such an abolishment was achieved by treating EF-Tu extensively with the thiol reagent L-1-tosylamido-2-phenylethyl chloromethyl ketone. EF-Tu X GTP thus treated has lost its ability to protect the ester bond of aminoacyl-tRNA. The relevance of these data for the sequence of events during protein synthesis and for control of translational fidelity is discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K., Kawakita M., Nakamura S., Ishikawa K., Kaziro Y. Studies on the polypeptide elongation factors form E. coli. VI. Characterization of sulfhydryl groups in EF-Tu and EF-Ts. J Biochem. 1974 Sep;76(3):523–534. doi: 10.1093/oxfordjournals.jbchem.a130596. [DOI] [PubMed] [Google Scholar]
- Bhuta P., Chládek S. Effect of thiostrepton and 3'-terminal fragments of aminoacyl-tRNA on EF-Tu and ribosome-dependent GTP hydrolysis. Biochim Biophys Acta. 1982 Aug 30;698(2):167–172. doi: 10.1016/0167-4781(82)90132-4. [DOI] [PubMed] [Google Scholar]
- Bhuta P., Chládek S. Stimulation of Escherichia coli elongation factor Tu-dependent GTP hydrolysis by aminoacyl oligonucleotides in the presence of aurodox. FEBS Lett. 1980 Dec 15;122(1):113–116. doi: 10.1016/0014-5793(80)80414-5. [DOI] [PubMed] [Google Scholar]
- Bhuta P., Kumar G., Chládek S. Elongation factor Tu.ribosome dependent guanosine 5'-triphosphate hydrolysis: elucidation of the role of the aminoacyl transfer ribonucleic acid 3' terminus and site(s) involved in the inducing of the guanosinetriphosphatase reaction. Biochemistry. 1982 Mar 2;21(5):899–905. doi: 10.1021/bi00534a014. [DOI] [PubMed] [Google Scholar]
- Bocchini V., Parlato G., De Vendittis E., Sander G., Parmeggiani A. Energetic aspects of the EF-Tu-dependent GTPase activity. A study using the antibiotic kirromycin. Eur J Biochem. 1980 Dec;113(1):53–60. doi: 10.1111/j.1432-1033.1980.tb06138.x. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Modolell J. Effects of antibiotics, N-acetylaminoacyl-tRNA and other agents on the elongation-factor-Tu dependent and ribosome-dependent GTP hydrolysis promoted by 2'(3')-O-L-phenylalanyladenosine. Eur J Biochem. 1981 Jun;117(1):27–31. doi: 10.1111/j.1432-1033.1981.tb06298.x. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Modolell J. Hydrolysis of GTP on elongation factor Tu.ribosome complexes promoted by 2'(3')-O-L-phenylalanyladenosine. Proc Natl Acad Sci U S A. 1980 Feb;77(2):905–909. doi: 10.1073/pnas.77.2.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinali G., Wolf H., Parmeggiani A. Effect of kirromycin on elongation factor Tu. Location of the catalytic center for ribosome-elongation-factor-Tu GTPase activity on the elongation factor. Eur J Biochem. 1977 May 2;75(1):55–65. doi: 10.1111/j.1432-1033.1977.tb11503.x. [DOI] [PubMed] [Google Scholar]
- Fasano O., Bruns W., Crechet J. B., Sander G., Parmeggiani A. Modification of elongation-factor-Tu . guanine-nucleotide interaction by kirromycin. A comparison with the effect of aminoacyl-tRNA and elongation factor Ts. Eur J Biochem. 1978 Sep 1;89(2):557–565. doi: 10.1111/j.1432-1033.1978.tb12560.x. [DOI] [PubMed] [Google Scholar]
- Guesnet J., Parlato G., Parmeggiani A. Interaction between the different domains of aminoacyl-tRNA and the elongation-factor-Tu x kirromycin complex. Eur J Biochem. 1983 Jul 1;133(3):499–507. doi: 10.1111/j.1432-1033.1983.tb07492.x. [DOI] [PubMed] [Google Scholar]
- Helk B., Sprinzl M. Interaction of unfolded tRNA with the 3'-terminal region of E. coli 16S ribosomal RNA. Nucleic Acids Res. 1985 Sep 11;13(17):6283–6298. doi: 10.1093/nar/13.17.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högenauer G. The stability of a codon transfer RNA complex. Eur J Biochem. 1970 Feb;12(3):527–532. doi: 10.1111/j.1432-1033.1970.tb00882.x. [DOI] [PubMed] [Google Scholar]
- Ivell R., Sander G., Parmeggiani A. Modulation by monovalent and divalent cations of the guanosine-5'-triphosphatase activity dependent on elongation factor Tu. Biochemistry. 1981 Nov 24;20(24):6852–6859. doi: 10.1021/bi00527a017. [DOI] [PubMed] [Google Scholar]
- Jonák J., Petersen T. E., Clark B. F., Rychlík I. N-Tosyl-L-phenylalanylchloromethane reacts with cysteine 81 in the molecule of elongation factor Tu from Escherichia coli. FEBS Lett. 1982 Dec 27;150(2):485–488. doi: 10.1016/0014-5793(82)80795-3. [DOI] [PubMed] [Google Scholar]
- Jonák J., Sedlácek J., Rychlík I. Mode of action of N-tosyl-L-phenylalanylchloromethane on the elongation protein-synthesizing S 3 factor from Bacillus stearothermophilus. Biochim Biophys Acta. 1973 Jan 19;294(2):322–328. doi: 10.1016/0005-2787(73)90304-3. [DOI] [PubMed] [Google Scholar]
- Labuda D., Pörschke D. Codon-induced transfer RNA association. A property of transfer RNA involved in its adaptor function? J Mol Biol. 1983 Jun 15;167(1):205–209. doi: 10.1016/s0022-2836(83)80042-4. [DOI] [PubMed] [Google Scholar]
- Labuda D., Pörschke D. Multistep mechanism of codon recognition by transfer ribonucleic acid. Biochemistry. 1980 Aug 5;19(16):3799–3805. doi: 10.1021/bi00557a023. [DOI] [PubMed] [Google Scholar]
- Labuda D., Striker G., Porschke D. Mechanism of codon recognition by transfer RNA and codon-induced tRNA association. J Mol Biol. 1984 Apr 25;174(4):587–604. doi: 10.1016/0022-2836(84)90085-8. [DOI] [PubMed] [Google Scholar]
- Leberman R., Antonsson B., Giovanelli R., Guariguata R., Schumann R., Wittinghofer A. A simplified procedure for the isolation of bacterial polypeptide elongation factor EF-Tu. Anal Biochem. 1980 May 1;104(1):29–36. doi: 10.1016/0003-2697(80)90272-9. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Rigler R., Laggner P. Structural variability of tRNA: small-angle x-ray scattering of the yeast tRNAphe-Escherichia coli tRNAGlu2 complex. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5891–5895. doi: 10.1073/pnas.79.19.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. A semi-quantitative treatment of missense and nonsense suppression in the strA and ram ribosomal mutants of Escherichia coli. Evaluation of some molecular parameters of translation in vivo. J Mol Biol. 1974 Apr 5;84(2):297–313. doi: 10.1016/0022-2836(74)90586-5. [DOI] [PubMed] [Google Scholar]
- Parlato G., Guesnet J., Crechet J. B., Parmeggiani A. The GTPase activity of elongation factor Tu and the 3'-terminal end of aminoacyl-tRNA. FEBS Lett. 1981 Mar 23;125(2):257–260. doi: 10.1016/0014-5793(81)80733-8. [DOI] [PubMed] [Google Scholar]
- Parlato G., Pizzano R., Picone D., Guesnet J., Fasano O., Parmeggiani A. Different regions of aminoacyl-tRNA regulate the function of elongation factor Tu. J Biol Chem. 1983 Jan 25;258(2):995–1000. [PubMed] [Google Scholar]
- Parmeggiani A., Sander G. Properties and regulation of the GTPase activities of elongation factors Tu and G, and of initiation factor 2. Mol Cell Biochem. 1981 Mar 27;35(3):129–158. doi: 10.1007/BF02357085. [DOI] [PubMed] [Google Scholar]
- Pestka S. Assay for nonenzymatic and enzymatic translocation with Escherichia coli ribosomes. Methods Enzymol. 1974;30:462–470. doi: 10.1016/0076-6879(74)30046-8. [DOI] [PubMed] [Google Scholar]
- Picone D., Parmeggiani A. Transfer ribonucleic acid deprived of the C-C-A 3'-extremity can interact with elongation factor Tu. Biochemistry. 1983 Sep 13;22(19):4400–4405. doi: 10.1021/bi00288a009. [DOI] [PubMed] [Google Scholar]
- Pingoud A., Urbanke C., Krauss G., Peters F., Maass G. Ternary complex formation between elongation factor Tu, GTP and aminoacyl-tRNA: an equilibrium study. Eur J Biochem. 1977 Sep;78(2):403–409. doi: 10.1111/j.1432-1033.1977.tb11752.x. [DOI] [PubMed] [Google Scholar]
- Pingoud A., Urbanke C., Wolf H., Maass G. The binding of kirromycin to elongation factor Tu. Structural alterations are responsible for the inhibitory action. Eur J Biochem. 1978 May;86(1):153–157. doi: 10.1111/j.1432-1033.1978.tb12294.x. [DOI] [PubMed] [Google Scholar]
- Richman N., Bodley J. W. Ribosomes cannot interact simultaneously with elongation factors EF Tu and EF G. Proc Natl Acad Sci U S A. 1972 Mar;69(3):686–689. doi: 10.1073/pnas.69.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Hearst J. E. Structure-function relations in E. coli 16S RNA. Cell. 1983 May;33(1):19–24. doi: 10.1016/0092-8674(83)90330-6. [DOI] [PubMed] [Google Scholar]
- Thompson R. C., Karim A. M. The accuracy of protein biosynthesis is limited by its speed: high fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTP[gamma S]. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4922–4926. doi: 10.1073/pnas.79.16.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Baller J., Doty P. Complementary oligonucleotide binding to the anticodon loop of fMet-transfer RNA. Nature. 1970 Feb 7;225(5232):508–510. doi: 10.1038/225508a0. [DOI] [PubMed] [Google Scholar]
- Van Noort J. M., Kraal B., Bosch L. A second tRNA binding site on elongation factor Tu is induced while the factor is bound to the ribosome. Proc Natl Acad Sci U S A. 1985 May;82(10):3212–3216. doi: 10.1073/pnas.82.10.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noort J. M., Kraal B., Bosch L., La Cour T. F., Nyborg J., Clark B. F. Cross-linking of tRNA at two different sites of the elongation factor Tu. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3969–3972. doi: 10.1073/pnas.81.13.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgenboom E., Vink T., Kraal B., Bosch L. Mutants of the elongation factor EF-Tu, a new class of nonsense suppressors. EMBO J. 1985 Apr;4(4):1049–1052. doi: 10.1002/j.1460-2075.1985.tb03737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. Interaction of siomycin with the acceptor site of Escherichia coli ribosomes. J Mol Biol. 1972 Jun 28;67(3):443–457. doi: 10.1016/0022-2836(72)90462-7. [DOI] [PubMed] [Google Scholar]
- Wolf H., Chinali G., Parmeggiani A. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4910–4914. doi: 10.1073/pnas.71.12.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort J. M., Duisterwinkel F. J., Jonák J., Sedlácek J., Kraal B., Bosch L. The elongation factor Tu.kirromycin complex has two binding sites for tRNA molecules. EMBO J. 1982;1(10):1199–1205. doi: 10.1002/j.1460-2075.1982.tb00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



