SUMMARY
Dyslipidemia and atherosclerosis are associated with reduced insulin sensitivity and diabetes, but the mechanism is unclear. Gain-of-function of the gene encoding deacetylase SirT1 improves insulin sensitivity, and could be expected to protect against lipid abnormalities. Surprisingly, when transgenic mice overexpressing SirT1 (SirBACO) are placed on atherogenic diet, they maintain better glucose homeostasis, but develop worse lipid profiles and larger atherosclerotic lesions than controls. We show that transcription factor cAMP response element binding protein (Creb) is deacetylated in SirBACO mice. We identify Lys136 is a substrate for SirT1-dependent deacetylation that affects Creb activity by preventing its cAMP-dependent phosphorylation, leading to reduced expression of glucogenic genes, and promoting hepatic lipid accumulation and secretion. Expression of constitutively acetylated Creb (K136Q) in SirBACO mice mimics Creb activation and abolishes the dyslipidemic and insulin-sensitizing effects of SirT1 gain-of-function. We propose that SirT1-dependent Creb deacetylation regulates the balance between glucose and lipid metabolism, integrating fasting signals.
INTRODUCTION
Patients with type 2 diabetes develop more severe and more extensive atherosclerosis that contributes to their increased risk of cardiovascular disease (CVD) and related mortality (National Institute of Diabetes and Digestive and Kidney Diseases, 2005). Thus, it is important to understand the mechanism linking diabetes and atherosclerosis. Insulin resistance is a prominent feature of type 2 diabetes and an independent risk factor for atherosclerosis (Howard et al., 1996). The mechanism linking dyslipidemia with insulin action remains unclear (Haeusler and Accili, 2008), but alterations of hepatic insulin sensitivity are sufficient to bring about changes of lipid metabolism reminiscent of diabetic dyslipidemia (Biddinger et al., 2008; Han et al., 2009).
We and others have reported that genetic gain-of-function or pharmacologic activation of the NAD+-dependent protein deacetylase SirT1 improve insulin sensitivity in rodents (Banks et al., 2008; Baur et al., 2006; Pfluger et al., 2008). Moreover, SirT1 overexpression in endothelial cells increases endothelial nitric oxide synthase (eNOS) function (Chen et al., 2008; Li et al., 2007; Zhang et al., 2008), and sirtuins reduce inflammation in the vessel wall, and improve hepatic and macrophage cholesterol metabolism (Chen et al., 2008; Li et al., 2007). These and germane findings (Schwer and Verdin, 2008) raise the question of whether the insulin-sensitizing effects of sirtuins can prevent atherosclerosis.
To answer this question, we placed transgenic mice carrying an extra copy of the SirT1 gene (Banks et al., 2008) on a cholesterol-rich (Western-type) diet (WTD), and determined their susceptibility to dyslipidemia and atherosclerosis. Surprisingly, we show that SirT1 gain-of-function has detrimental effects on lipid metabolism, despite its beneficial effects on glucose metabolism. We show that these effects are associated with deacetylation-dependent inhibition of the cAMP response element binding protein (Creb). Creb promotes hepatic gluconeogenesis (Chrivia et al., 1993) and inhibits lipid synthesis (Herzig et al., 2003). Its activity is regulated by several cofactors, two of which–Torc2 and Cbp–are also deacetylated by SirT1 (Liu et al., 2008). However, it’s unknown whether Creb itself is a SirT1 substrate and how this might affect the cAMP response. We report that SirT1 directly deacetylates Creb and identify Lys136 as a site of SirT1-dependent Creb deacetylation that modulates its protein kinase A (PKA)– dependent phosphorylation. We demonstrate that a constitutively acetylated Creb mutant (K136Q) reverses the effects of SirT1 on hepatic lipid synthesis and deposition, as well as glucose homeostasis, indicating that Creb deacetylation plays a central role in the paradoxical dissociation between glucose and lipid metabolic effects observed in SirT1 transgenics.
RESULTS
Increased dyslipidemia and atherosclerosis in SirBACO:Ldlr–/ mice
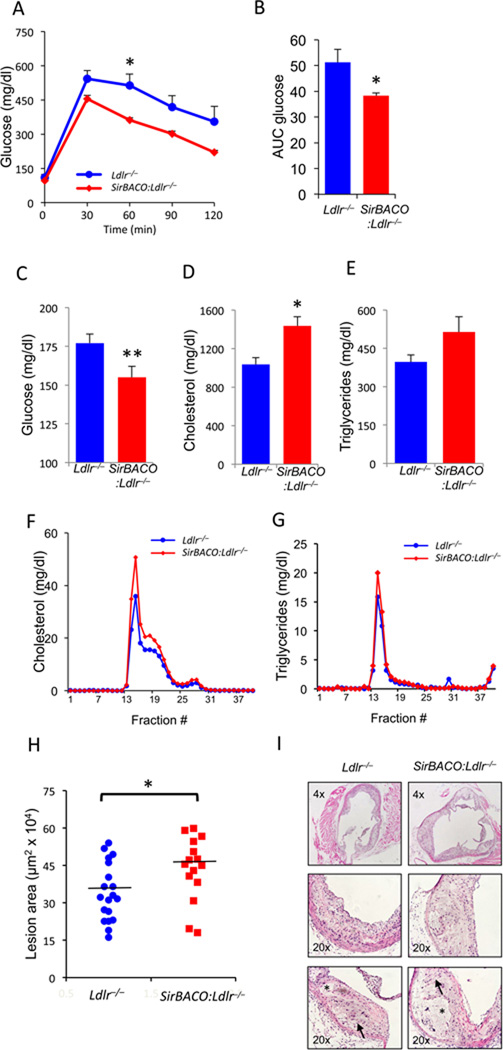
To test the effects of SirT1 gain-of-function on lipid metabolism and atherosclerosis, we intercrossed SirT1-transgenic mice (SirT1 Bacterial Artificial Chromosome Overexpressor, SirBACO) (Banks et al., 2008) and Ldlr−/− mice, subjected double mutant mice to WTD and analyzed the resulting phenotypes. SirBACO:Ldlr−/− mice displayed better glucose tolerance (Figure 1A,B) and lower fasting glucose than Ldlr−/− controls (Figure 1C). Strikingly, the improvement of glucose metabolism was associated with a worsening lipid profile, characterized by increased total cholesterol (Figure 1D), a trend toward increased triglycerides (TG) (Figure 1E) and elevated VLDL- and LDL-cholesterol and VLDL-TG (Figure 1F, G). These changes were not present in mice fed standard chow (Figure S1A–D), and were independent of changes in insulin levels (Figure S1E–H).
Figure 1. Metabolic characterizations of WTD-fed SirBACO:Ldlr−/− mice.
(A–B) IPGTT time courses (A) and areas under the curve (B) (*=P<0.05, n=5 each).
(C–E) Measurements of fasting plasma glucose (C), total cholesterol (D) and TG (E) (**=P<0.01, *=P<0.05, n=18–24).
(F–G) FPLC fractionation of lipoprotein cholesterol (F) and TG (G) in pooled sera (6 mice each). All measurements were carried out after a 5-hr fast.
(H) Measurements of aortic root lesion areas following 14 weeks on WTD (*=P <0.05, n=15–19 each). A horizontal line indicates mean area in each group.
(I) H&E staining of representative aortic root lesions, with arrows indicating cholesterol clefts, and asterisks indicating necrotic cores. Data are expressed as means ± SEM.
Consistent with the plasma lipid values, we observed a 28% increase of aortic root atherosclerotic lesion area (P<0.05) (Figure 1H, I). There was no difference in necrotic core area (Figure S1I). The phenotype was present in both genders and was independent of macrophage function, as it failed to be transferred by transplanting bone marrow from SirBACO:Ldlr−/− into Ldlr−/− mice (data not shown).
SirT1 increases hepatic lipid content and secretion in WTD-fed mice
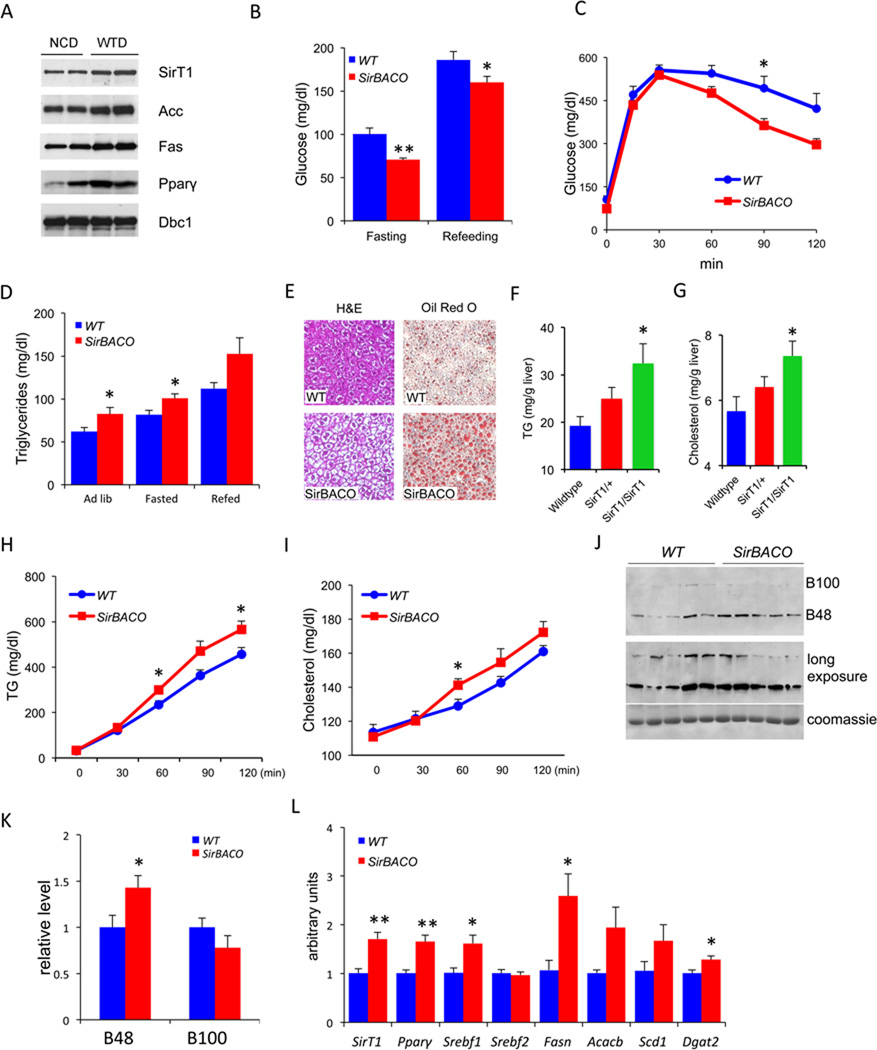
To determine the role of SirT1 in the observed phenotype of euglycemia with dyslipidemia, we first analyzed the effect of WTD on hepatic SirT1 expression in wild-type C57BL6 mice. SirT1 levels rose ~twofold following 2 weeks on WTD, as did Acc, Fas, and Pparγ levels (Figure 2A). Thus, the transgenic SirT1 gain-of-function can be viewed as mimicking a pathophysiological response to WTD. Conversely, SirT1−/− mice show decreased levels of Fas and Acc1 in basal conditions (Figure S2A). Due to their poor health, a more detailed characterization of these mice was not possible.
Figure 2. Transgenic overexpression of SirT1 increases hepatic lipid content and secretion upon WTD feeding.
(A) Western blots analysis of liver proteins from male C57BL/6J mice after 4 weeks WTD feeding. Mice were fasted for 7 hours. Dbc1 is used as a loading control.
(B–K) Metabolic analyses of SirBACO mice and control littermates (WT) after 2 weeks of WTD, including glucose (B) (n=10–11 each), IPGTT (C) (n=5–6 each), plasma TG (D) (n=10–11 each), hepatic H&E and Oil red-O staining (E), hepatic TG (F) and cholesterol content (G) following an 8-hr fast (n=6–10).
(H–I) Triton assays to measure TG (H) and cholesterol secretion (I) (n=6–7 each).
(J) Representative ApoB western blot following Triton injection.
(K) Quantification of multiple Triton secretion experiments (n=5 each).
(L) Q-PCR analysis of hepatic gene expression (n=5 each). In all experiments, **=P<0.01, *=P<0.05. Data are expressed as means ± SEM.
Next, we examined the regulation of glucose and lipid metabolism in SirBACO mice independently of the Ldlr deletion. On a normal diet, these mice have normal metabolic parameters (Figure S2B, C). Strikingly, 2 weeks of WTD recapitulated aspects of the SirBACO:Ldlr−/− mouse phenotype. Thus, glucose levels were significantly lower and glucose tolerance greater in WTD-fed SirBACO compared to control mice (Figure 2B, C) with normal insulin levels (Figure S2D), while TG rose in both ad lib and fasted conditions (Figure 2D and S2E, F). We observed similar trends with longer duration of the diet (data not shown).
To examine the mechanism of lipid abnormalities, we measured hepatic lipid synthesis, content, and secretion. Hepatic de novo lipogenesis demonstrated a trend toward increased levels (Fig. S2G); TG and cholesterol content increased (Figure 2E–G) in a SirT1 copy number-dependent fashion in SirBACO hemizygous and homozygous mice. Secretion of TG (Figure 2H), cholesterol (Figure 2I), and the main VLDL apoprotein, ApoB48 (Figure 2J, K) (Ginsberg et al., 2005) also rose.
Analyses of gene expression showed increased levels of mRNAs encoding lipogenic enzymes and their transcriptional regulators, including peroxisome proliferator-activated receptor gamma (Pparγ), Srebp-1c (Srebf1), fatty acid synthase (Fasn), acetyl-CoA carboxylase 2 (Acacb), diacylglycerol O-acyltransferase 2 (Dgat2) and stearoyl-coenzyme A desaturase 1 (Scd1), whereas the cholesterol synthesis regulator Srebp2 (Srebf2) (Figure 2L), and its target genes Ldlr and Pcsk9 (Figure S2H), were unchanged. We also observed increases of fatty acid β-oxidative genes Pparα and peroxisomal acyl-CoA oxidase (Acox) (Figure S2G). Thus, metabolic and gene expression data support the conclusion that SirT1 gain-of-function promotes hepatic lipid accumulation and secretion in response to WTD.
SirT1 gain-of-function decreases Creb acetylation
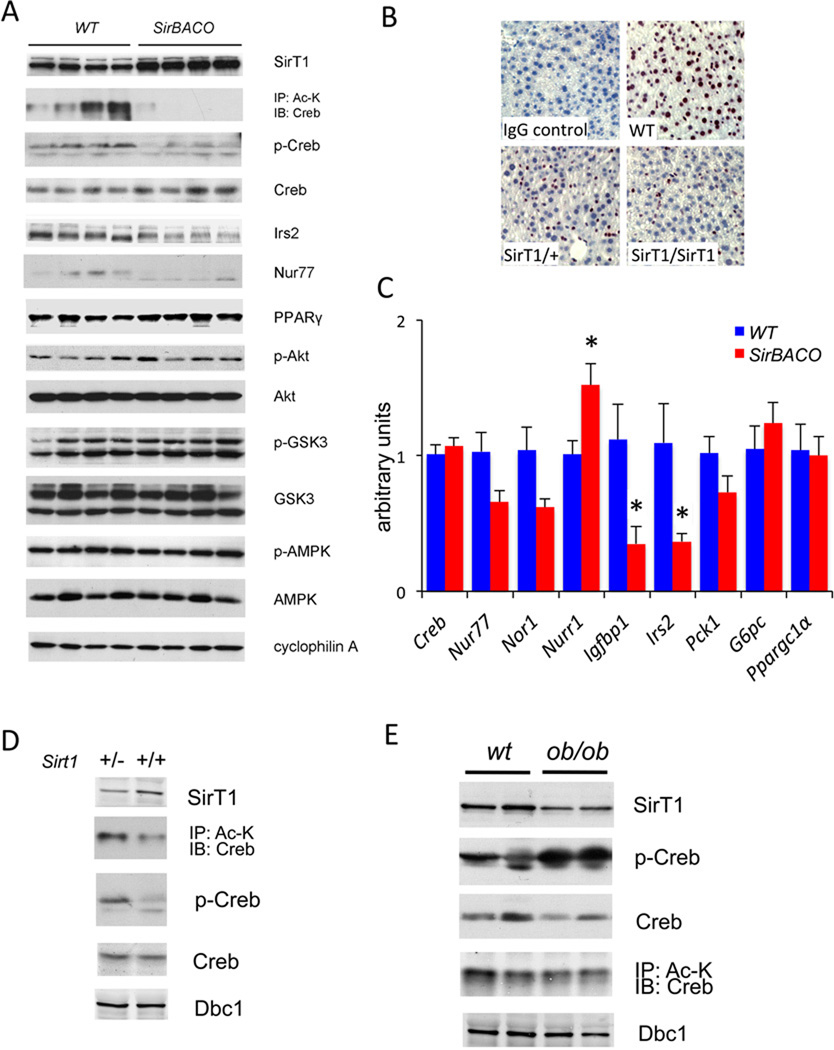
To understand the mechanism of decreased plasma glucose and increased liver fat content and secretion, we examined Akt activation, but failed to detect statistically significant differences in phospho-Akt or its substrate phosphor-Gsk3β between SirBACO and control mice (Figure 3A and S3A). These data are consistent with normal insulin signaling in SirBACO:Ldlr−/− mice (Figure S1J), and with our previous data in SirBACO mice under different experimental conditions (Banks et al., 2008).
Figure 3. SirT1 modulates Creb activity in liver.
(A) Western blots of liver proteins from WTD-fed mice (Quantification in Figure S3A).
(B) Immunohistochemical staining of phospho-Ser133 Creb in WTD-fed mice. 8-wk-old mice were fasted for 8-hr prior to isolating livers for analysis.
(C) Q-PCR analysis of hepatic gene expression in mice on WTD for 2 wk. Mice were fasted for 24-hr and then re-fed for 16-hr for RNA extraction (*= P<0.05, n=5 each). Data are expressed as means ± SEM.
(D) Western blots analysis of liver proteins from 8-wk-old SirT1 heterozygous knockouts (SirT1+/−) and control littermates (SirT1+/+).
(E) Western blots of liver proteins from 12-wk-old ob/ob mice and control littermates. Mice were fasted overnight prior to analysis. Dbc1 is used as a loading control.
Creb activates gluconeogenesis (Herzig et al., 2001) and inhibits lipid synthesis (Herzig et al., 2003). Hence, decreased Creb activity could explain the phenotype of WTD-fed SirBACO mice. We hypothesized that SirT1 gain-of-function could inhibit Creb function through deacetylation, resulting in decreased gluconeogenesis and elevated lipogenesis. Indeed, Creb acetylation was nearly absent in SirBACO mice, and phosphorylation of Ser133–the site required for Creb activation (Chrivia et al., 1993)– was substantially decreased when assessed by either western blotting (Figure 3A) or immunohistochemistry. The latter change occurred in a SirT1 transgene copy number-dependent manner (Figure 3B).
Expression of most Creb target genes was lower, consistent with decreased Creb activity. Thus, levels of Insulin receptor substrate-2 (Irs2) and Nur77, a gene that promotes gluconeogenesis and inhibits lipogenesis (Chao et al., 2007; Pols et al., 2008), decreased, as did mRNA levels encoding Nur77 (encoded by Nr4a1), Irs2, Igfbp1, and neuron-derived orphan receptor-1 Nor1 (encoded by Nr4a3). Pparγ, a target of Creb-dependent suppression (Herzig et al., 2003) showed increased levels, consistent with decreased Creb activity (Figure 3A). The only Creb target gene that was discordant with this pattern was Nurr1 (Nr4a2), whose levels rose by ~50% (Figure 3C). These data indicate that Creb deacetylation is accompanied by decreased expression of most–but not all–of Creb target genes.
Both SirT1 and Creb are active in fasted conditions (Mayr and Montminy, 2001; Schwer and Verdin, 2008). Accordingly, we did not detect differences in Creb phosphorylation, acetylation, Creb-dependent gene expression, and liver fat accumulation in re-fed mice (Figure S3B–E).
SirT1 loss-of-function activates Creb
Next, we assessed Creb acetylation and phosphorylation in animals with reduced SirT1 levels. SirT1 haploinsufficient mice showed increased acetyl-Creb and phospho-Creb (Figure 3D). In the liver of ob/ob mice, a genetic model of insulin resistance and obesity, total Creb levels were decreased, but the ratio of acetyl-Creb to total Creb increased due to reduced hepatic SirT1 expression (Figure 3E). These data indicate that SirT1 regulation of Creb is not limited to the SirBACO model, but is mirrored in models of insulin-resistant diabetes.
SirT1 regulates Creb activity through deacetylation
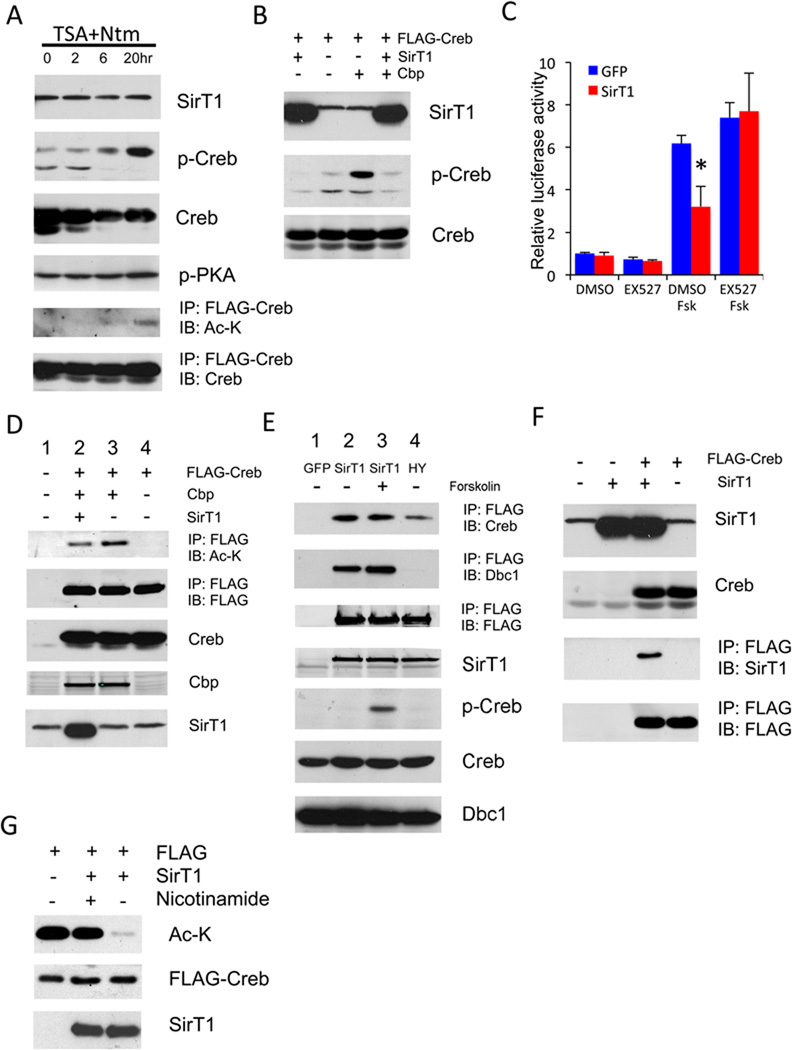
We asked whether acetylation affects Creb activity. Concurrent inhibition of class I/II histone deacetylases (HDACs) by trichostatin A (TSA) and class III HDACs by nicotinamide increased Creb Ser133 phosphorylation and acetylation without affecting phosphorylation levels of the Creb kinase PKA (Figure 4A), suggesting that acetylation modulates Creb phosphorylation independent of PKA. Indeed, overexpression of the Creb acetylase Cbp (Lu et al., 2003) was sufficient to promote Creb phosphorylation, while SirT1 overexpression blocked it (Figure 4B). In the presence of the SirT1 inhibitors, nicotinamide or EX527 (Napper et al., 2005), forskolin-induced Creb phosphorylation increased in magnitude and duration (Figure S4A), further supporting the notion that deacetylation of Creb by SirT1 negatively regulates its activity.
Figure 4. SirT1 regulates Creb acetylation and phosphorylation.
(A) Western blots of anti-FLAG immunoprecipitates obtained from 293 cells overexpressing FLAG-tagged Creb following treatment with 1 µM trichostatin A (TSA) and 5 mM nicotinamide (Ntm) for the indicated times.
(B) Western blots of extracts from 293 cells transfected with FLAG-tagged Creb in combination with Cbp or SirT1.
(C) CRE-luciferase assays from McA-RH7777 cells transduced with GFP or SirT1 adenovirus. (*= P<0.05, n=3). Data are expressed as means ± SEM.
(D) Immunoprecipitation of FLAG-Creb from 293 cells cotransfected with Cbp or SirT1.
(E) Coimmunoprecipitation of endogenous Creb by FLAG-tagged WT or H363Y SirT1 mutant in 293 cells following 30 min treatment with forskolin (5 µM).
(F) Co-immunoprecipitation of SirT1 by FLAG-tagged Creb in 293 cells.
(G) In vitro Creb deacetylation assay with purified Creb and SirT1.
We tested whether pharmacological SirT1 inhibition affected Creb activity. Consistent with the in vivo gene expression data, inhibition of SirT1 deacetylase activity resulted in higher levels of Creb targets Nur77, Nor1, Irs2, Igfbp1 and glucose-6-phosphatase (encoded by G6pc) in hepatoma cells (Figure S4B). Conversely, SirT1 overexpression repressed forskolin-induced Creb-luciferase activity by 50%, and EX527 treatment preempted Creb inhibition by SirT1 (Figure 4C). Overexpression of SirT1 in hepatoma cells had similar effects, increasing expression of Ppar γ, Dgat2, Acaca and Acacb, repressing Nur77, Tnf-α, Il-1α and Il-6 (Pfluger et al., 2008); the only discordant change was Nurr1 (Figure S4C). These data suggest that SirT1 affects Creb function via deacetylation.
Creb is a SirT1 substrate
We sought to determine whether Creb is a SirT1 substrate. We transfected 293 cells with FLAG-tagged Creb, SirT1, and Cbp in various combinations. Creb acetylation was enhanced by Cbp (Figure 4D, top panel, lane 3), and decreased by SirT1 (Figure 4D, top panel, lane 2). The phosphorylation-deficient Creb mutant M1 (S133A) was similarly deacetylated by SirT1, suggesting that deacetylation doesn’t require Ser133 phosphorylation (Figure S4D). We then asked whether SirT1 physically interacts with Creb. We detected endogenous Creb by immunoblotting, following immunoprecipitation of FLAG-SirT1 transfected in 293 cells (Figure 4E, lanes 2 and 3), demonstrating that SirT1 binds to endogenous Creb and that this interaction is unaffected by Creb phosphorylation. Conversely, we detected SirT1 by immunoblotting of Creb immunoprecipitates obtained from cells co-expressing SirT1 and FLAG-tagged Creb (Figure 4F). Notably, unlike the SirT1-interacting protein Dbc1 (Zhao et al., 2008), Creb bound the mutant SirT1 H355Y, albeit with reduced efficiency (Figure 4E, lane 4).
To demonstrate that SirT1 deacetylates Creb directly, and not through interactions with Cbp or Torc2 (Liu et al., 2008), we performed in vitro deacetylation assays with purified FLAG-Creb and SirT1. Addition of SirT1 to the reaction mix resulted in complete Creb deacetylation that was entirely reversed by the SirT1 inhibitor, nicotinamide (Figure 4G). Next, we asked whether repression of Creb by SirT1 requires the latter’s deacetylase activity. In 293 cells, a catalytically inactive SirT1 mutant (H355Y) (Luo et al., 2001) failed to repress Creb acetylation and phosphorylation, while repression by wild type SirT1 was blunted by EX527 (Figure S4E), indicating that Creb inhibition is dependent on the deacetylase activity of SirT1. We also saw inhibition of Creb by SirT1 in adipocytes (Figure S4F), suggesting that it’s a general event.
SirT1 deacetylates Creb on Lys136
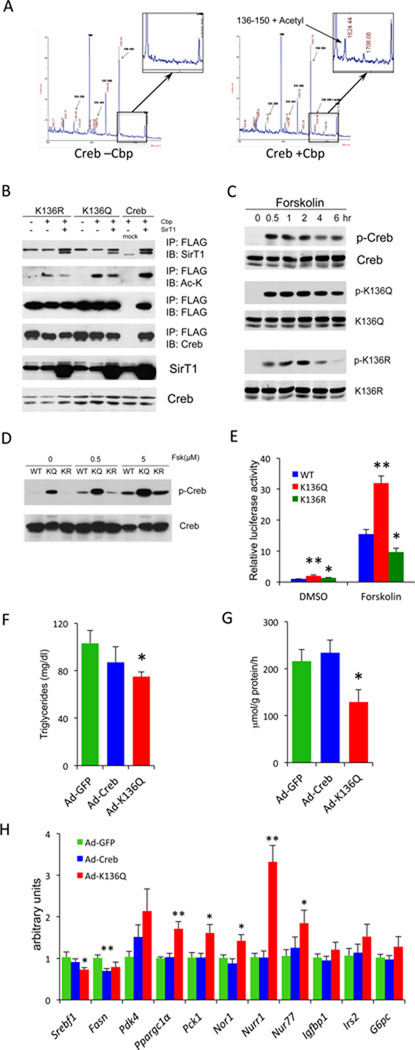
To identify SirT1 deacetylation sites, we immunprecipitated Creb from cells expressing Cbp or control DNA, purified the immunoprecipitated protein from polyacrylamide gels, and subjected it to MALDI-MS and LC-MS/MS. Out of fifteen lysine residues present in Creb, ten are clustered in the bZIP domain, and two (Lys123 and Lys136) are adjacent to the cAMP-dependent phosphorylation site, Ser133 (Figure S5A). We detected acetyl- Lys136, but not acetyl-Lys123 (Figure 5A). Interestingly, Lys136 is the only lysine residue in this region conserved across Creb homologs in different species (Figure S5A).
Figure 5. Creb Lys136 is a SirT1 substrate.
(A) Mass spectra obtained from FLAG-Creb isolated from 293 cells cotransfected with Cbp (+Cbp) or control DNA (–Cbp).
(B) Immunoprecipitation of FLAG-tagged WT, K136Q or K136R Creb from 293 cells cotransfected with Cpb or SirT1.
(C) Western blots of protein extracts from 293 cells transfected with WT, K136Q or K136R Creb after 5 uM forskolin treatment for the indicated times.
(D) Western blot of 293 cells expressing wild type Creb (WT), K136Q (KQ) or K136R (KR) mutant. Cells were harvested after 30 min forskolin treatment at indicated doses.
(E) CRE-luciferase assays from McA-RH7777 cells transfected with WT, K136Q or K136R Creb (**=P<0.01, *=P<0.05, n=4).
(F–H) Effects of in vivo transduction of C57BL/6J mice with wild-type (Ad-Creb) or K136Q (Ad-K136Q) Creb adenoviruses after 1 wk on WTD. Mice were fasted overnight and then refed for 4 hours.
(F) Plasma TG levels (*=P<0.05 vs. GFP control, n=6–7).
(G) Hepatic de novo lipogenesis (*=P<0.05 vs. GFP control, n=6–7).
(H) Q-PCR analysis of hepatic gene expression (**=P<0.01, *=P<0.05 vs. GFP control, n=6). Data are expressed as means ± SEM.
To evaluate the role of Lys136 in Creb function, we mutated it to glutamine (K136Q) as acetylation mimetic, or to arginine (K136R) as deacetylation mimetic. Neither mutation affected Creb binding to SirT1, but both decreased Cbp-dependent Creb acetylation (Figure 5B), consistent with the notion that this is a site of Creb acetylation in intact cells (Lu et al., 2003). Moreover, both mutations preempted the ability of SirT1 inhibitors to stimulate Creb phosphorylation (Figure S5B), indicating that Lys136 is the acetylation site involved in regulating Creb activation.
Time-course analyses of forskolin induction indicated that phosphorylation of the K136Q mutant was more rapid and long lasting, while that of the K136R mutant was more slow and short-lived than WT (Figure 5C). The K136Q mutant mimicked the effect of SirT1 inhibition (Figure S5B). Dose-response studies showed that the K136Q mutation increased Creb sensitivity to forskolin-induced phosphorylation, whereas the K136R mutation restored it to normal (Figure 5D). Moreover, in CRE transactivation assays, the K136Q mutant increased Creb reporter gene activity, while the K136R mutant decreased compared to WT Creb, indicating that the acetylation state of K136 directly affects Creb activity (Figure 5E). To sum up, these data show that Lys136 acetylation increases Creb sensitivity to PKA-dependent phosphorylation.
Gene expression studies in hepatoma cells further support these conclusions. Expression of K136Q resulted in greater basal or forskolin-induced levels of phosphoenolpyruvate-carboxykinase (Pck1), Igfbp1, G6pc, Nur77, Nor1 and Nurr1, and suppressed Fasn and Acaca (Figure S5C), mirroring the effects seen in SirBACO mice and supporting the idea that acetylation activates Creb. In addition, the ability of insulin to suppress forskolin-induced expression of Pck1, Igfbp1, and G6pc was impaired. The lipogenic genes do not respond to insulin induction in this cell type, so this aspect of the phenotype could not be properly evaluated.
K136Q mutant Creb reverses the phenotype of WTD-fed SirBACO mice
To study the metabolic consequences of the K136Q mutation in vivo, we transduced C57BL mice with adenoviruses encoding WT or K136Q mutant Creb and subjected them to WTD. The adenoviruses affected neither glucose levels nor glucose tolerance (Figure S5D–G), but the K136Q mutant did decrease plasma TG and lowered hepatic de novo lipogenesis, as assessed by tracer methods, by nearly 50% (Figure 5F,G). In addition, expression of lipogenic genes (Srebf1, Fasn) was decreased, while that of glucogenic genes (Pgc1α, Pck1, Nur77) was increased, consistent with the notion that Creb acetylation mimics Creb activation by cAMP (Figure 5H).
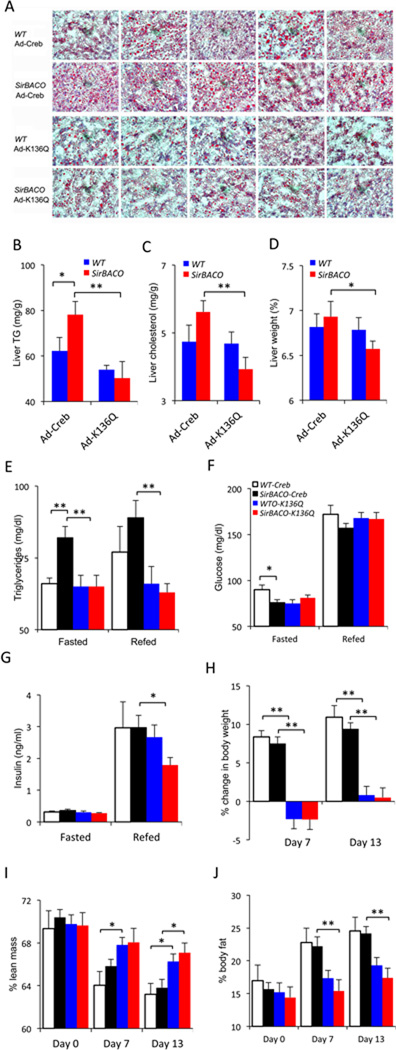
Next, we evaluated the ability of the K136Q mutant Creb to prevent WTD-induced dyslipidemia and steatosis in SirBACO mice. We administered adenoviruses encoding WT or K136Q mutant Creb to SirBACO mice and control littermates and placed them on WTD. After 2 weeks on the diet, SirBACO mice treated with WT Creb still demonstrated the same abnormalities seen in untreated mice: increased hepatic lipid, as well as plasma TG and cholesterol, lower blood glucose, and unchanged insulin levels compared to control littermates (Figure 6A–G). In contrast, SirBACO mice treated with Ad-K136Q showed similar glucose, TG and cholesterol levels to wildtype controls in the fasted or refed states (Figure 6E–G, S6A–B). Administration of Ad-K136Q also prevented diet-induced weight gain and reduced body fat content in both control and SirBACO mice (Figure 6H–J, S6C). These data show that preventing Creb deacetylation reverses the metabolic features of WTD-fed SirBACO mice.
Figure 6. Creb K136Q prevents lipid abnormalities in WTD-fed SirBACO mice.
(A–C) Hepatic lipid content characterized by Oil Red-O staining (A), TG (B) and cholesterol measurements (C) (n=7–9 each).
(D) Liver weight (n=7–9).
(E–G) Plasma TG (D), glucose (E) and insulin (F) after 24-hour fasting (Fasted) followed by 4-hour re-feeding (Refed) (n=7–9).
(H–J) Body weight change (H) and composition, with lean (I) and fat mass (J) n=7–9) during WTD feeding. In all experiments **=P<0.01, *=P<0.05. Open bar: Ad-Creb in WT mice; black bar: Ad-Creb in SirBACO mice; blue bar: Ad-K136Q in WT mice; red bar: Ad-K136Q in SirBACO mice. Data are expressed as means ± SEM.
DISCUSSION
The question addressed in this work was whether SirT1, by virtue of its ability to prevent diabetes (Banks et al., 2008; Pfluger et al., 2008) would also protect against atherosclerosis. Surprisingly, our investigations reveal an apparent proatherogenic phenotype in mice carrying an extra copy of SirT1.
Are sirtuins good or bad for atherosclerosis?
The link between SirT1 and lipid metabolism has been the subject of intense investigation. Sirt1−/− mice display reduced HDL cholesterol and TG and a blunted response to LXR agonists that has been linked to reduced SirT1-dependent activation of LXR through deacetylation (Li et al., 2007). The predisposition of SirBACO mice to WTD-induced hepatosteatosis and the rise of plasma TG are consistent with these findings; however, we found no evidence of either increased HDL cholesterol, as would be expected based on the Sirt1−/− phenotype, nor did we see changes in cholesterol efflux and foam cell formation from SirBACO macrophages (data not shown). Potential explanations for this discrepancy include variations in genetic background and SirT1 expression levels, or source of macrophages (Li et al., 2007).
The phenotype of liver-specific SirT1 ablation is also consistent with that of SirBACO mice: decreased liver fat accumulation in response to WTD, lower Srebf1 and Fasn levels (Chen et al., 2008), as well as decreased Pparγ-dependent gene expression, increased inflammation and ER stress (Purushotham et al., 2009). In our study we observed decreased inflammation and ER stress (Figure S3F), as was seen in another model of SirT1 gain-of-function (Pfluger et al., 2008). Unlike the latter study, we saw increased hepatosteatosis in WTD-fed SirBACO mice. Although it’s possible that this apparent discrepancy is due to the different diets used in the two studies (cholesterol-rich vs. high-fat), we should point out that in our previous work we didn't see protection from hepatosteatosis in high fat-fed SirBACO mice that were protected from diabetes, as documented by glucose clamps and other metabolic data (Banks et al., 2008). The explanation for the difference between the two transgenic lines may not be unrelated to unusual features of the mice studied by Pfluger et al.: (i) extreme insulin sensitivity on the normal diet (low body weight, near-undetectable fasting insulin levels, near-flat glucose excursions during GTT); (ii) uncharacteristic response to the diet, in which glucose levels remained unchanged compared to the normal diet, insulin levels remained low, and yet glucose and insulin tolerance became altered in wild-type mice (Pfluger et al., 2008).
Previous work demonstrated anti-atherogenic effects of SirT1 in macrophages and vascular endothelial cells (Zhang et al., 2008). The design of our study allowed us to parse the integrated contribution of SirT1 gain-of-function in different tissues to atherosclerosis. Our data indicate that the proatherogenic effects of SirT1 on hepatic lipid metabolism trump its “peripheral” benefits on macrophages and vasculature. In fact, SirBACO macrophages appear to have largely normal functions in cholesterol uptake and inflammation, and failed to affect atherosclerosis development in bone marrow transfer experiments (data not shown).
We conclude that the effects of SirT1 are pleiotropic, and that in type 2 diabetes, sirtuin agonists might have undesired effects on lipid metabolism. However, such effects seemingly arise from increased insulin sensitivity, not from insulin resistance, adding to a growing body of evidence that suggests that excessive insulin action, as opposed to insulin resistance, predisposes to the characteristic plasma lipid profile of type 2 diabetes (Han et al., 2009; Shimomura et al., 2000).
Sirtuins and polyphenols
Polyphenols like resveratrol have been suggested to promote sirtuin activity (Howitz et al., 2003). But their effect on hepatic lipid metabolism stands in apparent contrast to the phenotype of SirBACO mice (Baur et al., 2006; Hou et al., 2008). In light of the demonstration that polyphenols activate AMPK (Canto et al., 2009; Lagouge et al., 2006), we suggest that their metabolic effects are independent of SirT1. Alternatively, it’s possible that their effect exceeds that of a single extra copy of SirT1 in SirBACO mice; however, in view of the fact that key SirT1 substrates are hypoacetylated in SirBACO mice (Banks et al., 2008; Pfluger et al., 2008), this explanation seems unlikely.
Two fasting signals
The identification of Creb as a bona fide SirT1 substrate reveals a biochemical and genetic pathway linking two critical “fasting” signals: cAMP and NAD+. It also allows us to integrate these two proteins with another regulator of the fasting response, FoxO1 (Accili and Arden, 2004). Interestingly, acetylation plays a permissive role in FoxO1 (Qiang et al., 2010) and Creb phosphorylation (this report). But phosphorylation inactivates FoxO1, while activating Creb. It can thus be envisioned that SirT1 activation in response to falling nutrient levels promotes a shift from Creb- to FoxO1-dependent metabolism (Liu et al., 2008), leading to increased lipid utilization. Our findings can also help reconcile conflicting reports on Creb and lipid metabolism. While dominant-negative Creb (A-Creb) or Creb RNAi cause liver steatosis in mice (Herzig et al., 2003), Creb inhibition by antisense oligonucleotide results in lower plasma and liver cholesterol and TG in diabetic rats but not in ob/ob mice (Erion et al., 2009). The dissociation between Creb levels and its phosphorylation/acetylation in ob/ob mice indicates that changes in acetylation can offset the effects of decreased protein levels.
Two recent findings in human studies support our conclusions that Creb regulates lipid metabolism: clinical trials of glucagon receptor antagonists demonstrate an improvement of glucose levels in patients with diabetes associated with a rise in LDL-cholesterol (Engel et al., 2011). And genetic mutations of the Creb family gene CrebH are associated with alterations of TG clearance leading to hypertriglyceridemia (Lee et al., 2011). Thus, we tentatively propose that the mechanism identified in our study is relevant to human disease pathophysiology.
In summary, we demonstrate a dyslipidemic effect of SirT1 gain-of-function, mediated through Creb deacetylation. This effect might limit the therapeutic value of SirT1 agonists in diabetes treatment. Further studies of the pathway linking SirT1 and Creb will provide alternative approaches to the modulation of lipid metabolism by insulin sensitizers.
EXPERIMENTAL PROCEDURES
Reagents
We cultured McA-RH7777 and HEK-293T cells (ATCC, Manassas, VA) in DMEM with 10% fetal bovine serum (Mediatech, Inc., Manassas, VA). We used Hygromycin B (Sigma-Aldrich) to select for stable clones expressing CRE-luciferase. We used the following antibodies: monoclonal and polyclonal anti-acetylated-Lysine, p-Akt (T308 or S473), p-Creb, total Creb, p-AMPK (Cell Signaling), Fas, PPARγ, cyclophillin A, p-PKA, α-tubulin, RXRα, FXR (Santa Cruz); SirT1, Irs2, p-Acc (Millipore-Upstate); mouse Nur77 (eBioscience); Dbc1 (Bethyl Laboratories); FLAG M2 (Sigma-Aldrich); LXRα (R&D systems); apoB and HA (Abcam). Expression vectors for FLAG-Creb (# 22968) and phosphorylation-defective Creb-M1 (# 22969) were from Addgene Inc. (Cambridge, MA). Creb-K136Q and Creb-K136R mutants were made by site-direct mutagenesis (GenScript USA Inc., Piscataway, NJ). Creb-K136Q adenovirus were made and purified by Welgen Inc. (Worcester, MA). FLAG-SirT1 and SirT1-H363Y were cloned into pEGFP-N1 for transient expression. pGL4.29[luc2P/CRE/Hygro] was from Promega (Madison, WI), and TransIT-LT1 transfection reagent from Mirus Bio LLC (Madison, WI).
Mice
SirBACO mice on C57BL/6J background (Banks et al., 2008) were mated with Ldlr−/− mice to generate SirBACO:Ldlr−/− mice. SirT1+/− mice have been described (Li et al., 2007), and C57BL/6J and Lepob/+ mice were from Jackson Laboratories (Bar Harbor, ME). The western diet used in these experiments contains 42% milk fat, 0.15% cholesterol (TD88137, Harlan Teklad). Atherosclerotic lesions were analyzed as described (Han et al., 2006). All adenovirus administrations were at a dose of 1×109 vp/g. The Columbia University Animal Care and Utilization Committee have approved all animal experiments.
Protein Studies
For native immunoprecipitation, we lysed cells in FLAG IP buffer (50mM Tris pH 7.4, 150mM NaCl, 0.2% Triton-X100, 10% glycerol) supplemented with 2µM Trichostatin A (TSA) (Sigma-Aldrich), 10mM nicotinamide (Sigma-Aldrich), protease and phosphatase inhibitors (Boston Bioproducts), and incubated 1 mg protein with Anti-FLAG M2 Affinity Gel (Sigma-Aldrich) overnight at 4°C. After 4 washes with FLAG IP buffer, proteins were eluted by 3x FLAG peptides in FLAG IP buffer. Acetylated-Lysine IP was carried out following Tansey WP’s protocol. Mouse tissue was lysed in TSD buffer (50mM Tris pH 7.4, 1% sodium dodecyl sulfate (SDS), 5mM DTT) with 2µM TSA and 10mM nicotinamide, boiled for 10 minutes, and centrifuged. Thereafter, we transferred 0.6 mg protein to 1.2mL TNN buffer (50mM Tris pH7.4, 250mM NaCl, 5mM EDTA and 0.5% NP-40) with inhibitors, pre-absorbed with 50µL Protein A/G agarose slurry (Santa Cruz), and immunoprecipitated using 5µL anti Acetylated-Lysine antibody (Cell Signaling). Immune complexes were bound to protein A/G agarose slurry, extensively washed and eluted by boiling in 0.1 mL sample buffer.
For western blotting, we lysed cells or tissues in buffer containing 20mM Tris, pH 7.4, 150mM NaCl, 10% glycerol, 2% NP-40, 1mM EDTA pH 8.0, 0.2% SDS, 0.5% NaDOC supplemented with protease and phosphatase inhibitors. Protein were denatured under reducing condition and separated on SDS-PAGE prior to western blotting using either ECL substrate kit (Thermo Scientific) or Odyssey Infrared Imagining System (LI-COR Biosciences).
Gene Expression Analysis
We isolated RNA with RNeasy kit (Qiagen) and DNase I (Qiagen) digestion, synthesized cDNA with High-capacity cDNA Reverse Transcription kit (Applied Biosystems), and performed quantitative real-time PCR (qPCR) with goTaq qPCR Master Mix (Promega) on Bio-Rad CFX96 Real-Time PCR system. Expression levels were calculated by ΔΔCt method using murine cyclophilin A or rat 36B4 as controls. Primer sequences are in Table S1.
Metabolic Measurements
TG secretion was measured following intravenous injection of Triton WR-1339 (0.5mg/g body weight) (Sigma-Aldrich) in 5-hr-fasted mice (Han et al., 2009). Blood samples were collected in EDTA capillaries. Hepatic lipids were extracted as described (Folch et al., 1957). Metabolites were measured with NEFA-HR(2), Cholesterol E, HDL-Cholesterol E (Wako Diagnostics), Infinity™ Triglyceride Reagent (Thermo Scientific), Mouse Insulin ELISA (Millipore), and OneTouch Glucometer (Lifescan, Inc.).
De novo Lipogenesis Assay
Hepatic de novo lipogenesis was measured as described (Zhang et al., 2006), using 1 mCi 3H2O (American Radiolabeled Chemicals) per mouse.
Histological Analysis
We used paraffin-embedded liver sections for hematoxylin and eosin, and frozen sections for Oil Red O staining. Immunohistochemical analysis of phospho-Creb in paraffin-embedded liver sections was carried out using phospho-Creb (Ser133) (87G3) antibody (Cell Signaling) at a dilution of 1:50 with dehydration.
Luciferase Assay
McA-RH7777 cells stably overexpressing CRE-driven luc2P were infected with SirT1 adenovirus at MOI=1, and pretreated with 10 µM EX527 overnight prior to 100µM forskolin for 5 hr. Luciferase activity was determined with Bright-Glo™ Luciferase Assay System (Promega).
Statistical analysis
We used unpaired 2-tail student’s t-test to evaluate differences and the customary value P<0.05 to declare statistical significance. All values are expressed as means ± standard error of means (SEM).
Highlights.
SirT1 gain-of-function predisposes to dyslipidemia and atherosclerosis
SirT1 deacetylates cAMP-response-element binding-protein (CREB) on K136
Constitutively acetylated CREB mutant K136Q lowers plasma lipids in SirT1 transgenics
CREB acetylation integrates cAMP-dependent with SirT1-dependent fasting responses
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Haeusler, K. Tsuchiya, A. Tall, I. Tabas, and G. Li for critical discussion of the data, W. Zhao, N. Kon, Y. Tang and M. Li for help with acetylation studies, J. Reusch and S. Pugazhenthi for Creb adenovirus, T. Kolar and A. Flete for technical support, and G. Heinrich for critical reading of the manuscript. This work was supported by NIH grants HL087123 and DK063608 (Columbia Diabetes & Endocrinology Research Center).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddinger SB, Hernandez-Ono A, Rask-Madsen C, Haas JT, Aleman JO, Suzuki R, Scapa EF, Agarwal C, Carey MC, Stephanopoulos G, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008;7:125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Zhang Z, Pei L, Saito T, Tontonoz P, Pilch PF. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol. 2007;21:2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Engel SS, Xu L, Andryuk PJ, Davies MJ, Amatruda J, Kaufman K, Goldstein BJ. Efficacy and Tolerability of MK-0893, a Glucagon Receptor Antagonist (GRA), in Patients with Type 2 Diabetes (T2DM) Diabetes. 2011;60(S1) 0309-OR. [Google Scholar]
- Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, et al. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci U S A. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method foRthe isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Ginsberg HN, Zhang YL, Hernandez-Ono A. Regulation of plasma triglycerides in insulin resistance and diabetes. Arch Med Res. 2005;36:232–240. doi: 10.1016/j.arcmed.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Accili D. The double life of Irs. Cell Metab. 2008;8:7–9. doi: 10.1016/j.cmet.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Liang CP, DeVries-Seimon T, Ranalletta M, Welch CL, Collins-Fletcher K, Accili D, Tabas I, Tall AR. Macrophage insulin receptor deficiency increases ER stress-induced apoptosis and necrotic core formation in advanced atherosclerotic lesions. Cell Metab. 2006;3:257–266. doi: 10.1016/j.cmet.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Han S, Liang CP, Westerterp M, Senokuchi T, Welch CL, Wang Q, Matsumoto M, Accili D, Tall AR. Hepatic insulin signaling regulates VLDL secretion and atherogenesis in mice. J Clin Invest. 2009;119:1029–1041. doi: 10.1172/JCI36523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Hedrick S, Morantte I, Koo SH, Galimi F, Montminy M. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-gamma. Nature. 2003;426:190–193. doi: 10.1038/nature02110. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G, O'Leary DH, Zaccaro D, Haffner S, Rewers M, Hamman R, Selby JV, Saad MF, Savage P, Bergman R. Insulin sensitivity and atherosclerosis. The Insulin Resistance Atherosclerosis Study (IRAS) Investigators. Circulation. 1996;93:1809–1817. doi: 10.1161/01.cir.93.10.1809. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lee JH, Giannikopoulos P, Duncan SA, Wang J, Johansen CT, Brown JD, Plutzky J, Hegele RA, Glimcher LH, Lee AH. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nature medicine. 2011;17:812–815. doi: 10.1038/nm.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Hutchins AE, Doyle CM, Lundblad JR, Kwok RP. Acetylation of cAMP-responsive element-binding protein (CREB) by CREB-binding protein enhances CREB-dependent transcription. J Biol Chem. 2003;278:15727–15734. doi: 10.1074/jbc.M300546200. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- National Institute of Diabetes and Digestive and Kidney Diseases. Bethesda, MD: U.S. Department of Health and Human Services, National Institute of Health; 2005. National Diabetes Statistics fact sheet: general information and national estimates on diabetes in the United States. [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols TW, Ottenhoff R, Vos M, Levels JH, Quax PH, Meijers JC, Pannekoek H, Groen AK, de Vries CJ. Nur77 modulates hepatic lipid metabolism through suppression of SREBP1c activity. Biochem Biophys Res Commun. 2008;366:910–916. doi: 10.1016/j.bbrc.2007.12.039. [DOI] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Banks AS, Accili D. Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. The Journal of biological chemistry. 2010;285:27396–27401. doi: 10.1074/jbc.M110.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, Simon MC, Diehl JA. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. The Journal of biological chemistry. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- Zhang QJ, Wang Z, Chen HZ, Zhou S, Zheng W, Liu G, Wei YS, Cai H, Liu DP, Liang CC. Endothelium-specific overexpression of class III deacetylase SIRT1 decreases atherosclerosis in apolipoprotein E-deficient mice. Cardiovasc Res. 2008;80:191–199. doi: 10.1093/cvr/cvn224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.