Abstract
The availability of multiple teleost (bony fish) genomes is providing unprecedented opportunities to understand the diversity and function of gene duplication events using comparative genomics. Here we examine multiple paralogous genes of γ-glutamyl transferase (GGT) in several distantly related teleost species including medaka, stickleback, green spotted pufferfish, fugu and zebrafish. Through mining genome databases, we have identified multiple GGT orthologs. Duplicate (paralogous) GGT sequences for GGT1 (GGT1 a and b), GGTL1 (GGTL1 a and b) and GGTL3 (GGTL3 a and b) were identified for each species. Phylogenetic analysis suggests that GGTs are ancient proteins conserved across most metazoan phyla and those paralogous GGTs in teleosts likely arose from the serial 3R genome duplication events. A third GGTL1 gene (GGTL1c) was found in green spotted pufferfish; however this gene is not present in medaka, stickleback or fugu. Similarly, one or both paralogs of GGTL3 appear to have been lost in green spotted pufferfish, fugu and zebrafish. Syntenic relationships were highly maintained between duplicated teleost chromosomes, among teleosts and across ray-finned (Actinopterygii) and lobe-finned (Sarcopterygii) species. To assess subfunction partitioning, six medaka GGT genes were cloned and assessed for developmental and tissue specific expression. Based upon these data, we propose a modification of the “duplication-degeneration-complementation” (DDC) model of subfunction partitioning where quantitative differences rather than absolute differences in gene expression are observed between gene paralogs. Our results demonstrate that multiple GGT genes have been retained within teleost genomes. Questions remain however regarding the functional roles of multiple GGTs in these species.
BACKGROUND
Glutathione (GSH) is an abundant intracellular thiol which plays an important role in protecting cells against toxic insult and reactive oxygen species. Depletion in GSH generates a cellular sensitivity to oxidants resulting in an induced antioxidant response mediated by an induction of phase II enzymes including γ-glutamyl transferases (GGTs). GGTs are transmembrane enzymes consisting of a heavy chain and a light chain. They are the only known group of enzymes that cleave γ-glutamyl amide bonds and facilitate glutathione metabolism and turnover providing cells with local cysteine supply.
GGT gene/protein sequences are found in most species examined including archaebacteria, eubacteria, protocysts, yeast, insects, and vertebrates suggesting a basic conserved function for these enzymes throughout evolution (Suzuki et al., 89′; Chikhi et al., 99′; Park et al., 2005). GGT initiates the glutathione degradation in extracellular matrix by cleaving the glutamyl bond of glutathione or glutathione conjugates, producing cysteinyl-glycine. The dipeptide is further hydrolyzed by dipeptidase, producing cysteine, which is the limiting amino acid in glutathione synthesis. Since the intact glutathione molecule is resistant to digestion by any peptidases, GGT is considered an essential component of glutathione catabolism (Zhang and Forman, 2009). Intracellular GSH levels are depleted when GSH conjugates are continuously excreted from cells during detoxification. GGT in turns plays pivotal role in replenishment of the intracellular GSH to support cellular detoxification mechanism(s) (Dickinson and Forman, 2002).
In human and rat, splice variants and alternate promoter usage are common mechanisms for diversification of GGT expression. For instance, analysis of rat GGT1 mRNA reveals seven unique GGT1 transcripts from a single GGT1 gene ranging in size from 2.2 to 2.6 Kb (Taniguchi and Ikeda, 98′). All seven transcripts share a common GGT1 open reading frame but differ in their 5′ untranslated regions (UTR’s) (Chikhi et al., 99′). Genomic mapping of 5′UTRs leads to the discovery of five unique promoters (P1–P5) driving GGT1 expression. Each promoter was then found to be uniquely responsive to cellular stressors, such as hyperoxia, hypoxia, and exogenous chemicals (Zhang and Forman, 2009).
Multiple GGT genes are present in mammals with up to 13 proteins predicted in human (Heisterkamp et al., 2008). Multiple copies of GGT are likely due to gene and/or genome duplication events during vertebrate evolution (Taylor et al., 2001; Venkatesh and Yap, 2005). One proposed mechanism for duplications is the serial “2R” genome duplication hypothesis, which states that the entire vertebrate genome is a result of two rapid and successive rounds of genome duplication around the time of the divergence of jawless and jawed vertebrates, approximately 500 million years ago (Mya) (Taylor et al., 2001). Additionally, in a stem lineage of ray-finned fish (Actinopterygii), a third and fish-specific genome duplication (the “FSGD” or 3R hypothesis) has occurred prior to the radiation of the teleostean fishes but after this lineage diverged from tetrapods (Hedges and Kumar, 2002). Observations supporting the 3R hypothesis include the facts that (1) many paralogous genes in teleosts appear to have originated at the same time; (2) ray-finned fish share many of the gene duplicates; and (3) paralogous regions on different chromosomes maintain conserved synteny (Volff, 2005).
Ray-finned fish comprise ~24,000 extant species and are among the most diverse and successful group of vertebrates (Venkatesh, 2003). These organisms represent a large diversity of phenotypic characteristics and maintain considerable genetic diversity. It appears that much of the complexity of the teleost genome is a result of successive rounds of gene and/or genome duplication (Meyer and Van de Peer, 2005, Innan and Kondrashov, 2010). Because larger genomes might facilitate functional diversification and extend gene families, the presence of multiple gene copies is believed to have had a large impact on evolution of vertebrates in general (Crow and Wagner, 2006).
To date, GGT or GGT orthologs have not been thoroughly described in any fish species. In this study we have conducted an exhaustive search for GGT sequences in teleost genomes including medaka, stickleback, green spotted pufferfish, fugu and zebrafish. In each species examined we have identified multiple copies of GGT sequences. The identification of duplicate GGT genes in these species provides an opportunity to compare species-specific retention of these genes among distantly related teleosts and to evaluate putative mechanisms of gene conservation and subfunction partitioning (Postlethwait, 2007).
METHODS
Chemicals
Tert-butyl hydroquinone (tBHQ) was purchased from Alfa Aesfar, USA. Chemicals were prepared fresh in high performance liquid chromatography (HPLC)-grade dimethyl sulfoxide (DMSO) before use.
Test animals
Medaka (Oryzias latipes) are small (3–5 cm adult length) oviparous freshwater fish native to rice paddies of Japan, Korea, and eastern China. Male and female fish were collected from an orange-red line and maintained under standard recirculating aquaculture conditions. Water was maintained at a constant temperature of 25°C and photoperiod was kept at a constant light-dark cycle of 16hr:8hr. Fish husbandry and all experimental procedures with animals were carried out according to the NCSU Institutional Animal Care and Use Committees (IACUC) animal guidlines. All exposures were conducted on medaka larvae at 1 day post-hatch (dph) in six-well culture plates containing 5 ml medaka rearing solution (ERM; 5.1 mM NaCl, 120 μM KCl, 198 μM MgSO4 and 81 μM CaCl2; pH7.2). tBHQ prepared in DMSO was spiked into ERM at a final concentration of 100 μM. Vehicle concentration did not exceed 0.1% of total volume. Exposure times ranged from 15 minutes to 6 hrs. At each sampling time point larvae were removed from solution, rinsed with fresh ERM and snap-frozen for RNA isolation. All exposures were conducted with three biological replicates containing 3 pooled fish per replicate. All exposures were replicated three times each.
Genome analysis
Mining of GGT sequences in individual genomes (medaka, stickleback, green spotted pufferfish, fugu, zebrafish, Xenopus, chicken, mouse and human) was performed using public databases: Ensembl genome browser (http://www.ensembl.org), National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) and Joint Genome Institute (JGI, http://genome.jgi-psf.org/ for fugu only). With each species a generalized BLAST search was conducted using human GGT1 as the BLAST query. Complete open reading frames (ORFs) were determined by identifying the start and stop codons for each gene. Predicted GGT sequences were then BLASTed back to the Entrez NR protein database to ensure identification of the GGT Pfam domain and sequence similarity with the homologous GGTs.
Synteny analysis
Using medaka GGTs as anchor sites, comparison of gene neighbors for each GGT paralog and/or ortholog was conducted in medaka, stickleback, green spotted pufferfish, fugu, zebrafish, frog (Xenopus), chicken, mouse and human using the BioMart v0.5 program in Ensembl (http://www.ensembl.org/biomart). Comparisons of paralogous genes within species were conducted on presumed duplicate chromosomes. All comparisons were performed using flanking loci in genomes of each species examined (see Supplementary file 1 for the genome versions). Analysis of syntenic relationships was conducted either manually or through the BioMart program (Kasprzyk et al., 2004). The Ensembl Gene IDs of GGTs examined are listed in Supplementary file 2.
Phylogenetic analyses
A dataset of GGT sequences was constructed for phylogenetic analysis by downloading a large number of amino acid sequences from GENBANK using PSI Blast (Altschul et al., 97′; Altschul and Koonin, 98′). These were augmented with with GGT sequences identified through lab work. The combined sequence set was aligned using the multiple sequence alignment software FSA version 1.15 (Bradley et al., 2009), and trimmed to remove columns that fell outside the GGT Pfam domain (See Supplementary file 5 for detailed methods). The resulting alignment was 915 columns long and contained 277 sequences. Individual sequence lengths ranged from 343 to 542 amino acids, with a median length of 518 amino acids. The phylogeny was estimated in a Bayesian framework under the C20+ 4 model (Quang Le et al., 2008). The inference was conducted using the software package PhyloBayes 3.1g (Lartillot et al., 2009). We report the posterior probability of each split in order to indicate support.
RNA Isolation and cloning of GGT cDNAs in medaka
Total RNA was isolated from medaka embryos, larvae or adult tissues using the RNA BEE reagent (Tel-test, USA) according to the manufacturer’s instructions. Reverse transcription was performed with 1 μg total RNA using Superscript III RNase H− reverse transcriptase (Invitrogen) and oligo-dT12–18. Primer pairs (GGT1a-F1 and GGT1a-R1, L1a-F1 and L1a-R1, and L3a-F1 and L3a-R1) targeting three putative medaka GGT cDNAs (GGT1, GGTL1 and GGTL3, respectively) were designed and used for amplification. PCR was performed in a 50-μl mixture consisting of 10 ηg of first strand cDNA, 1 × PCR buffer (20 mM Tris/HCl pH 8.4, 50 mM KCl), 1 μM of each primer, 0.2 mM dNTPs, 1.5 mM MgCl2 and 5 U of Advantage 2 DNA polymerase (BD Clontech). The PCR program consisted of initial denaturation at 94°C for 1 min, followed by 35 cycles of amplification (denaturation at 94°C for 30 sec, combined annealing and extension at 68°C for 3 min) and a final extension at 68°C for 5 min in a thermocycler (MJ Research). To confirm the 5′- and 3′-ends of the three GGT cDNAs, 5′- and 3′-RACE PCR was conducted using the Marathon RACE cDNA Amplification kit (BD Clontech) according to the manufacturer’s recommendations. Briefly, a mixture of poly (A)+ RNA was purified from total RNA extracted from brain, intestine, kidney and liver of medaka using the PolyATract System kit (Promega, USA) and used as a source of template in RACE PCR. 5′-RACE PCR was performed using gene-specific nested primers (GGT1a-5′GSP1 and GGT1a-5′GSP2 for GGT1a, L1a-5′GSP1 and L1a-5′GSP2 for GGTL1a, and L3a-5′GSP1 and L3a-5′GSP2 for GGTL3a). 3′-RACE PCR was performed using gene-specific nested primers (GGT1a-3′GSP1 and GGT1a-3′GSP2 for GGT1a, L1a-3′GSP1 and L1a-3′GSP2 for GGTL1a, and L3a-3′GSP1 and L3a-3′GSP2 for GGTL3a). Full-length cDNAs were obtained by PCR amplification using a pair of primers targeting at 5′-and 3′-ends of each of the GGT cDNAs (GGT1a-F2 and GGT1a-R2 for GGT1a, L1a-F2 and L1a-R2 for GGTL1a and L3a-F2 and L3a-R2 for GGTL3a). PCR products were cloned into pCR2.1 TA vector (Invitrogen, USA) for DNA sequencing.
Complete ORFs for medaka GGT1b, GGTL1b and a partial ORF for GGTL3b were amplified with the PCR primer pairs GGT1b-F1 and GGT1b-R1, L1b-F1 and L1b-R1, and L3b-F1 and L3b-R1, respectively. 5′ and 3′ RACE for GGT1b, GGTL1b and GGT3 was conducted as described above with the following primer sets (GGT1b-5′GSP1 and GGT1b-5′GSP2 for GGT1b, L1ba-5′GSP1 and L1b-5′GSP2 for GGTL1b, and L3b-5′GSP1 and L3b-5′GSP2 for GGTL3b) and (GGT1b-3′GSP1 and GGT1b-3′GSP2 for GGT1b, L1b-3′GSP1 and L1ba-3′GSP2 for GGTL1ba, and L3b-3′GSP1 and L3b-3′GSP2 for GGTL3b). Full-length PCR products were cloned into pCR2.1 TA vector (Invitrogen, USA) for DNA sequencing. All GGT cDNA clones were sequenced in both directions to ensure maximum coverage, and verify correct ORFs.
Quantitative PCR
Quantitative real-time PCR (qPCR) was performed using first-strand cDNA as template with SYBR Green PCR Master Mix (Applied Biosystems, USA) in the ABI 7300 system (Applied Biosystems) according to the manufacturer’s instructions. First-strand cDNA was diluted to 1/50 and 5 μl was used for each real-time PCR reaction. The following primer pairs were used for GGT amplification and tested for efficiency and target specificity: GGT1a (GGT1a-F1 and GGT1a-R3), GGT1b (GGT1b-F1 and GGT1b-R2), GGTL1a (L1a-F3 and L1a-R3), GGTL1b (L1b-F3 and L1b-R3), GGTL3a (L3a-F3 and L3a-R3), and GGTL3b (L3b-F3 and L3b-R3). Medaka 18S rRNA was amplified with 18S-F and 18S-R using 5 μl first-strand cDNA diluted to 1/500 as template. The PCR profile consisted of a first step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. A dissociation curve which detects any non-specific amplification, including formation of primer-dimers, was run by an additional program of 95°C for 5 sec, 60°C for 10 min and 95°C for 5 sec at the end of the PCR profile. Relative gene expression levels were normalized to the 18S rRNA levels in the respective samples. To analyze the results, Ct value (the cycle number at which the fluorescence signal in a PCR reaction reaches a threshold) was calculated using the 7300 System SDS software (Applied Biosystems, USA). qPCR experiment for developmental expression analysis was conducted using RNA isolated from three biological replicates each consisting of 10-pooled embryos/larvae (30 total) for each developmental stage examined. For tissue-specific expression analysis, qPCR was carried out with RNA samples from 5 individual medaka adults for each sex (n=5). To study the larval response to oxidative stress, RNA was sampled from 5 biological replicates each containing 10-pooled larvae (50 total) for each time point investigated (n=5). Each transcript was PCR-amplified in duplicate on 96-well plates and all data were tested for significant differences within either treatment or developmental period using the Prism4 software package (GraphPad Software Inc., San Diego, CA). Data were logarithmically transformed as needed to improve equality of variances (ANOVA, p value < 0.05), followed by Newman-Keuls Multiple Comparison test and are represented as the mean relative mRNA level ± SEM. Principal Component Analysis (PCA) was performed on the developmental, spatial and inductive GGT gene expression data using the R package (version 2.12.1, http://www.r-project.org). The analysis was carried out with R function “prcomp” with option “retx” set to true. Each PCA 2-D plot was drawn with the top two principal components as X and Y axes (Raychaudhuri et al., 2000).
RESULTS
Characterization of teleost GGTs
Through screening the medaka genome database (v.200406, http://dolphin.lab.nig.ac.jp/medaka/) we identified multiple candidate GGT sequences exhibiting a high degree of sequence similarity with human GGT1 [GenBank: NM_053840] used as the TBLASTX query. Analyses of gene arrangement and structure within the medaka genome demonstrated that each GGT sequence represents a unique gene with a defined genomic locus, intron-exon boundaries and 5′- and 3′-UTRs. Three distinct GGT and GGT-like (GGTL) genes were initially identified and complete open reading frames (ORFs) for putative medaka GGT1a (Human homolog GGT1), GGTL1a (human homolog GGT5) and GGTL3a (human homolog GGT7) (Heisterkamp et al., 2008) sequences were mapped to chromosome 12 (nucleotide 66,948 to 73,643 on sense strand), chromosome 12 (nucleotide 78,894 to 94,420 on antisense strand) and chromosome 7 (nucleotide 16,938 to 26,716 on antisense strand), respectively. Primer pairs targeting 5′- and 3′-ends of the ORF for each of the three GGTs were used in RT-PCR with medaka liver RNA as template. Full-length cDNA for each gene was cloned to verify sequence and identify potential pseudogenes. Medaka GGT1a consists of a 1719-bp ORF which encodes a 572-amino acid protein. The full-length cDNA of medaka GGTL1a contains an ORF of 1677 bp and encodes a predicted protein of 558 amino acids while the full-length cDNA of medaka GGTL3a contains an ORF of 2040 bp encoding for a predicted protein of 679 amino acids (Table 1). Alignment of putative protein sequence for each medaka GGT is shown in Figure 1.
Table 1.
Characteristics of the medaka GGT cDNAs.
| GGT1a | GGT1b | GGTL1a | GGTL1b | GGTL3a | GGTL3b | |
|---|---|---|---|---|---|---|
| GenBank accession number | HQ213987 | HQ213988 | HQ213989 | HQ213990 | HQ213991 | HQ213992 |
| Length of cDNA cloned (bp) | 2394 | 1818 | 4586 | 1620 | 3875 | 17831 |
| Length of ORF (bp) | 1719 | 1818 | 1677 | 1620 | 2040 | 17831 |
| Putative protein length (amino acids) | 572 | 605 | 558 | 539 | 679 | 5941 |
| Predicted protein weight (kDa) | 62 | 66 | 60 | 58 | 73 | ND2 |
| Predicted isoelectric point | 6.29 | 6.58 | 8.82 | 8.39 | 5.06 | ND2 |
cloning based upon predicted ORF, 5′RACE for this paralog was inconclusive
ND - Not Determined base on partial sequence information
Figure 1. Multiple alignment of the putative medaka GGT proteins.
Peptide sequences of medaka GGTs were aligned by ClustalW 2.0. The number at the beginning of each row indicates the amino acid position. Identical residues were highlighted in black while similar residues were highlighted in grey by BOXSHADE. Pfam γ-glutamyl transferase domain (ID 01019) is indicated on the top of the alignment. Asterisk above the amino acid denotes the N-terminus of the putative light chain.
In a secondary search, additional medaka GGT sequences were identified using a TBLASTN search in the Ensembl database (http://www.ensembl.org). In each instance a duplicate set of GGT genes were found within the medaka genome, namely medaka GGT1b, GGTL1b and GGTL3b. GGT1b was mapped to chromosome 9 (nucleotide 21,062,230 to 21,071,861 on antisense strand), GGTL1b was mapped to chromosome 9 (nucleotide 21,107,117 to 21,112,976 on antisense strand) and GGTl3b was mapped to chromosome 5 (nucleotide 26,874,946 to 26,876,226 on sense strand). As with the first set of GGTs, RT-PCR amplification of the ORF for each gene was conducted with medaka liver and/or whole medaka hatchling RNA template to verify gene sequence and identify possible pseudogenes. The complete ORF for medaka GGT1b consists of 1818 bp and encodes a 605-amino acid protein. Medaka GGTL1b contains an ORF of 1620 bp and encodes a predicted protein of 539 amino acids while the medaka GGTL3b cDNA contains a partial ORF of 1783 bp encoding a predicted protein of 594 amino acids (Table 1). All PCR primers for GGT gene cloning and quantification are listed in Supplementary file 3.
Individual GGT sequences (both nucleic acid and protein) were BLASTed back to the NCBI NR database to cross check and validate homology to previously reported GGT sequences. On the basis of similarity to the human sequence, medaka sequences were subsequently annotated as GGT1a/b, GGTL1a/b and GGTL3a/b. To further validate the sensitivity of the annotation, analysis was augmented by examining relation to γ-glutamyl transferase Pfam domain (ID 01019) as illustrated in Figure 1. The results of these complementary approaches are candidate GGTs which authenticity can be supported by evidence from BLAST reciprocity.
To determine the presence of multiple paralogous GGT genes in other ray-finned fish, we mined the genomes of stickleback, green spotted pufferfish, fugu, and zebrafish. In each instance, we found multiple GGT sequences (Fig. 2). As with medaka, each GGT sequence was confirmed by assessing similarity with previously reported GGTs and alignment with the GGT Pfam domain. As demonstrated in Figure 2, six GGT sequences were found in each fish species with the exception of fugu where only five GGT sequences were identified. GGT1 is present in all species tested as duplicate alpha and beta paralogs (GGT1a and GGT1b). Duplicates of GGTL1 were also identified in each fish species examined except green spotted pufferfish, which has three copies (designated GGTL1a, L1b and L1c). Medaka and stickleback have duplicate copies of GGTL3 whereas green spotted pufferfish and fugu both have single copy of this gene. No GGTL3 ortholog was found in zebrafish rather, four novel GGT genes were identified (Fig. 2). Non-teleosts species including frog (Xenopus tropicalis), chicken, rat, mouse and human each have a single copy of GGT1, GGTL1 and GGL3. In the human genome (Ensembl genome version GRCh37), nine additional GGT sequences (GGT2, GGT3P, GGT4P, GGT8P, GGTLC1, GGTLC2, GGTLC3, GGTLC4P and GGTLC5P) have been identified and annotated unique to humans (Heisterkamp et al., 2008). Among the nine sequences, five labeled with “P” are pseudogenes, and thus they are not listed in Figure 2. GGT6 orthologs are present in all the genomes investigated (Fig. 2), but sequence analyses demonstrated that GGT6s have significant dissimilarity to other GGT paralogs thus these sequences were not included in further analyses.
Figure 2. Symbolic diagram of GGT paralogs and orthologs in selected species.
Each square represents a GGT gene identified in BLAST search of genomes in Ensembl or GenBank. See Additional file 2 for Ensembl Gene IDs and/or GenBank accession numbers. Alphabets in squares denote the duplicate or replicate copies of the GGT genes. Four additional GGTs were found in zebrafish genome while human also has multiple unique GGT sequences which have been annotated in Ensembl. The human GGT pseudogenes described previously are not listed. Asterisks indicate the two sets of species-specific GGTs are not replicate copies of a single gene.
Estimates of GGT gene trees
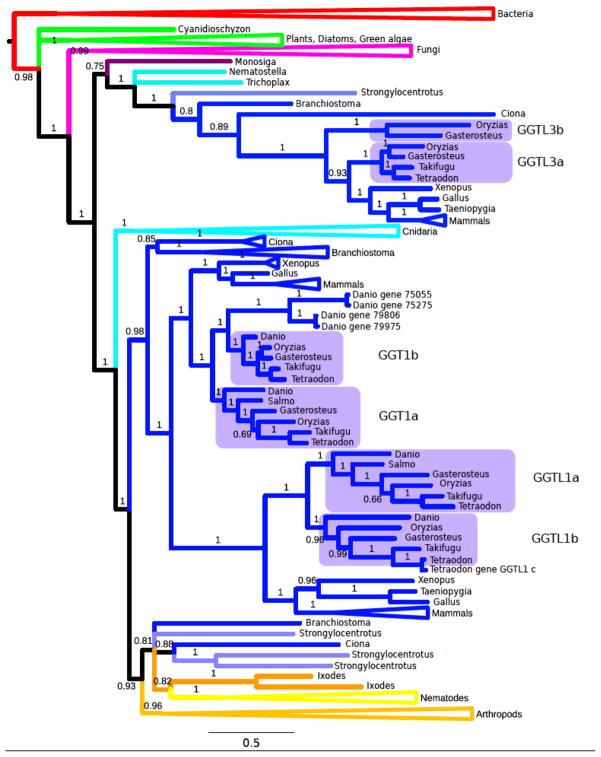
We estimated the phylogeny of the GGT amino acid sequences from across the tree of life (Fig. 3). The phylogeny estimate is broadly consistent with recent estimates of the phylogeny of chordates and other animals (Delsuc et al. 2008). Vertebrates are found to be more closely related to the Tunicates than to the Cephalochordates (lancelets). The Cnidaria (e.g. Nematostella and Hydra) are found to be placed more basally than any other Animal group except for the Choanoflagellates (e.g. Monosiga).
Figure 3. Bayesian phylogenetic inference of GGT genes.
Triangles indicated clades that have been collapsed to save space. Colors indicate the taxonomic group of sequences. Red indicates Bacteria, green indicates Archaeplastidae (red algae, green algae, and plants), dark purple indicates basal animals (e.g. Monosiga), cyan indicates Cnidaria, orange indicates Arthropoda, yellow indicates Nematoda, light blue indicates non-vertebrate Deuterostomes, and blue indicates Chordates. Vertebrate GGTs divide into GGTL3 (upper blue clade) and GGT1/GGTL1 (lower blue clade). The GGT1/GGTL1 clade further divides into the GGT1 clade and the GGTL1 clade. In teleosts the GGT1, GGTL1, and possibly the GGTL3 clade then each divide into alpha and beta forms. Each of the branch labels represents the corresponding posterior probabilities.
The phylogeny estimate allowed identification of a related but distinct clade of paralogous GGT-like protein sequences (Supplementary Figure Sf2). This clade contains several sequences from Bacteria and Archaea, as well as Eukaryotic sequences from a variety of groups including Fungi, Plants, and Animals. In addition, several sequences in this clade were annotated as having Cephalosporin acylase activity in addition to GGT activity. Therefore, this clade may have resulted from a duplication of an ancestral GGT gene that occurred before the divergence of Bacteria and from Eukaryotes. Alternatively, these sequences may be the result of horizontal gene transfer from Bacteria to Eukaryotes.
The phylogeny estimate supports the division of Vertebrate GGTs into the three families GGT1, GGTL1, and GGTL3. The phylogeny estimate additionally supports the hypothesis that the GGT1 and GGTL1 families arose by gene duplication in the vertebrate lineage after divergence from Branchiostoma but before the divergence of amphibians and teleost fish. Non-vertebrate animals, including the non-chordate Deuterostomes (such as Stronglyocentratus) and Protostomes (including Nematodes and Arthropods) appear not to have undergone the GGT1 versus GGTL1 duplication.
Within the teleost fish, we see that the GGT1 and GGTL1 families have been duplicated into GGT1a/GGT1b and GGTL1a/GGTL1b before the divergence of zebrafish from other teleost fish. In contrast, the duplication separating the GGTL3a and GGTL3b genes appears to fall with moderate support, not on the fish lineage, but on the lineage leading up to the divergence of fish and amphibians.
Within the GGTL1 family, Tetraodon has three sequences including GGTL1a, GGTL1b and GGTL1c. While the Tetraodon sequence GGTL1a is within the GGTL1a family, the Tetraodon sequences GGTL1b and GGTL1c both cluster within the GGTL1b sub-family. Since the Tetraodon GGTL1b and GGTL1c are much closer to each other than to any other sequences, it is most likely that they are the result of a single gene duplication within the Tetraodon clade. The GGTL3 clade is represented by the smallest number of teleost sequences, with two sequences from medaka, two from stickleback, and one from each of Tetraodon and Takifugu. No GGTL3 sequences were identified from zebrafish.
Synteny analysis
Comparisons of gene synteny in genomic regions flanking each GGT gene reveal that gene organization is well conserved among duplicated teleost chromosomes, between teleost species, and across vertebrates (Figs. 4a and 4b). Medaka GGT1a and GGTL1a were found in a head-to-tail arrangement on chromosome 12. Medaka GGTL3a was identified on chromosome 7. Duplicate (paralogous) copies of medaka GGT1 (GGT1b), and GGTL1 (GGTL1b) were identified on chromosome 9 in the same head-to-tail arrangement. Medaka GGTL3b was identified on chromosome 5. The gene neighborhood flanking GGT1a-L1a on chromosome 12 and GGT1b-L1b on chromosome 9 reveal over 34 pairs of gene duplicates (Figs. 4a and 4b). Twenty-four pairs of duplicated genes were found in the gene neighborhood flanking medaka GGTL3a on chromosome 5 and GGTL3b on chromosome 7. Order and arrangement of genes between duplicated chromosomes was often maintained but in several instances, inversions and changes in gene order were observed..
Figure 4. Gene synteny of GGT neighborhoods.

Chromosomes/scaffolds harboring the duplicate GGT genes were compared and duplicated neighboring genes were aligned. GGT paralogs within species and across species are connected by solid lines and dotted lines respectively. Approximate chromosomal/scaffold position of the genes are indicated. (A) Syntenic gene neighborhood of GGT1-GGTL1. (B) Syntenic gene neighborhood of GGTL3. “?” denotes GGTL3b was expected but not found in green spotted pufferfish. (C) Unique GGT genes identified in zebrafish genome. Four additional GGT family members were located in chromosome 1 of zebrafish genome (Zv8; Ensembl database) spanning from nucleotide 58.17 Mb to 58.40 Mb. Curved arrows indicate that the pair of genes share 98–100% sequence identity.
In comparison to medaka, similar arrangements of GGT a and b paralogs were observed on chromosomes, linkage groups or scaffolds of additional teleosts examined. Gene order is well conserved between medaka, stickleback, green spotted pufferfish and fugu (see Figs. 4a and 4b with reference to medaka GGT1a-L1a and L3a). Zebrafish exhibits little syntenic similarity to the other teleosts examined. Zebrafish GGT1a was identified on chromosome 10 however the locus for GGTL1a has not yet been assigned in Ensembl. Zebrafish GGT1b and L1b are located on chromosome 8 in a similar orientation as observed in other teleosts however gene synteny exhibits little conservation (Fig. 4a). A third GGTL1 gene, (GGTL1c) was identified in green spotted pufferfish located adjacent to GGTL1b on chromosome 12 (Fig. 4a).
Comparison of the GGT1a-L1a regions between teleosts and non-teleost species additionally reveals a high degree of conserved synteny. The head-to-tail arrangement of these two genes is retained among fish, amphibian, bird and mammals. Note however that only a single ortholog for GGT1, GGTL1 and GGTL3 was identified in frog (Xenopus), chicken, mouse and human genomes. Comparison of local gene neighborhoods of medaka and mouse GGT1a-L1a demonstrates up to 17 common genes occurring within a single homologous chromosomal region. A similar pattern of gene organization is observed in humans; however, genes are distributed between chromosomes h12 and h22. For GGTL3a, approximately 20 genes were found in common between teleosts and mammals (within a 40 MB region) distributed on a single contiguous chromosome (Fig. 4B).
In the zebrafish genome, four additional unique GGT genes were identified and annotated with the last 5 digits of the respective Ensembl Gene IDs (Fig. 2). Each gene was mapped to a defined locus on chromosome 1 and together form a gene cluster. All five of these GGT sequences are missing the N-terminal ends in the predicted protein sequences (data not show). In gene similarity analysis two pairs are formed among the four (Supplementary file 4) and exhibit 98–100% amino acid sequence similarities within each group.
Developmental expression of medaka GGTs
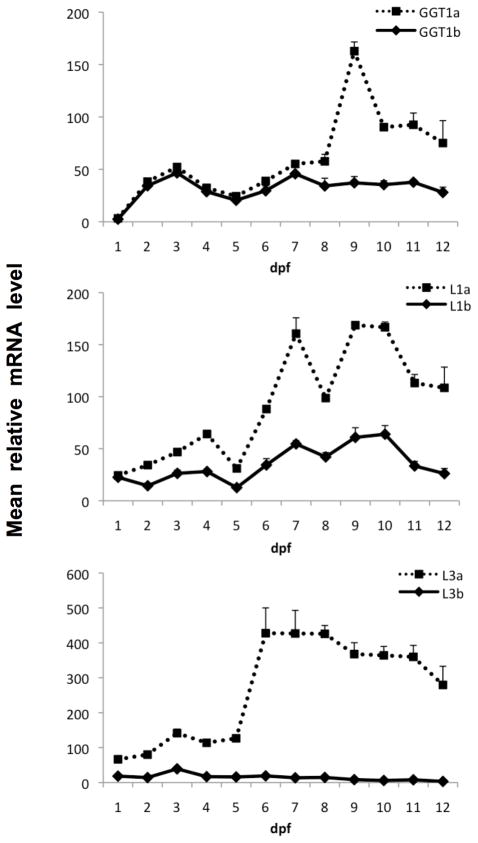
Quantitative PCR was used to gain further insight into the ontogenesis of each GGT sequence during medaka embryonic development. Gene-specific primers were designed for each of the six medaka GGT genes. qPCR analysis was performed using total RNA isolated from embryonic and larval stages between 1–12 dpf. A standard curve showing the relationship of the concentration of the PCR template and the Ct value was plotted for each of the gene detections (data not shown). All GGT primer pairs exhibited efficiencies >98% (R2 = 0.999). Figure 5 illustrates the relative quantification of medaka GGT expression. mRNA transcripts of all six GGTs were detectable 1 dpf. A steady increase in expression was observed for each GGT except GGTL3b which remained consistently low over the 12-day examination period. Expression levels for GGT1a and GGT1b were similar until 9 dpf where GGT1a rapidly increased up to 3 fold concurrent with hatching. GGT1a expression subsequently decreased at 10 dpf but remained significantly higher than GGT1b throughout the remainder of examination. Expression of GGTL1a and GGTL1b was similar up to 6 dpf where the GGTL1a mRNA increased by 3 fold. A slight increase in GGTL1b expression occurred between 6–10 dpf. Expression of both GGTL1a and GGTL1b dropped subsequent to hatching. GGTL3a was expressed over the course of development and exhibited a sudden increase in expression at 6 dpf. GGTL3b expression however remained low throughout the duration of embryonic development. Of the six GGT genes GGTL3a maintained the highest level at 6 dpf.
Figure 5. Developmental expression of medaka GGT genes.
Relative mRNA levels of medaka GGT genes within medaka embryos and larvae (1 to 12 dpf) as measured by qPCR. Data were normalized to 18S rRNA levels and are represented as the mean relative mRNA level ± SEM (n=5 pooled samples).
Principal Component Analysis (PCA) was used to visualize similarities and differences in developmental expression of the six medaka GGTs. In our analysis PC1 refers to the principal component exhibiting the most variation among the GGTs while PC2 refers to the principal component exhibiting the second most variation. Illustrated in Figure 8A, PC1 explains 63% of the variation in the entire data set while PC2 explains 20%. GGT1a and GGT1b exhibit a clear distinction in developmental expression with defined separation on PC2 in later (d8-12) stages of development. Developmental expression patterns of GGTL3a and GGTL3b are significantly divergent with distinct separation occurring within both principal components PC1 and PC2. There appears to be little separation between developmental expression of GGTL1a and GGTL1b both of which cluster with expression of GGTL3a.
Figure 8. PCA plots of GGT expression in (A) developmental profile, (B) tissue distribution and (C) response to tBHQ exposure.
Arrows indicate the positions of GGT genes analysed with R program on the two PCs which explain the largest amount of variation in the mRNA expression. (A) d, day post fertilization. (B) M, male; F, female; b, brain; gi, gill; gu, gut; h, heart; k, kidney; l, liver; m, muscle; o, ovary; s, spleen; t, testis. (C) Samples plotted are duration of exposure in (minutes) with 100 μM tBHQ.
Tissue distribution of medaka GGTs
Tissue specific expression for each GGT gene was conducted in 10 selected adult medaka tissues including brain, heart, gill, gut, kidney, liver, muscle, spleen, testis and ovary. GGT transcripts were expressed across a wide variety of tissues with differential patterns occurring in most tissues (Fig. 6). In brain and ovary, expression of GGTL3a and b was dominant while GGT1a and b was weak. Conversely, kidney and gut both expressed abundant levels of GGT1a and b with low levels of GGTL3a and b. Gill, heart, and spleen each expressed abundant levels of GGTL1a and b transcripts. Between paralogus GGTs, kidney, liver and gut exhibited abundant GGT1a transcripts with little expression of GGT1b. In brain and kidney GGTL1a was abundantly expressed while GGTL1b was expressed at lower level. In gonads, the converse is observed with GGTL3a being highly expressed and GGTL3b having a moderate expression in testis. In ovary GGTL3b transcripts were highly abundant and exhibited maximal expression compared to all other GGTs in all tissues. While expression of GGTL3a was also abundant, GGTL3b was more than 10 fold higher than GGTL3a. Several examples of sexual dimorphic expression were additionally evident for these analyses. In brain, gut, muscle and kidney, females expressed higher mRNA levels of GGTL3b compared to males.
Figure 6. Tissue specific expression of medaka GGT genes.
Quantitative, real-time PCR data showing expression of medaka GGTs in tissues of 6-month-old Orange-Red medaka males (white bars) and females (gray bars). Relative mRNA levels were measured in brain, gill, gut, heart, kidney, liver, skeletal muscle, spleen, and gonads (testis and ovary). Data were normalized to 18S rRNA levels and are represented as the mean relative mRNA level ± SEM (n=5 individual samples).
To determine spatial relationships and patterns of GGT expression we conducted PCA with expression data from all six GGT genes in male and female tissues (Fig. 8B). A prominent separation is observed among GGT1, GGTL1 and GGTL3 genes in PC1 and PC2. Defined clusters include GGT1b-GGTL1a-GGTL1b driven by expression of these genes in medaka gill, heart and spleen; GGTL3a-GGTL3b driven by expression in medaka brain and ovary, and GGT1a and b driven predominantly by expression in medaka gut and kidney. Noticeable through this analysis is a distinct differentiation between expression patterns of GGT1a and GGT1b paralogs. Both the GGTL1 group and GGTL3 group exhibit a similar distance from GGT1a. The eigenvalues of PC1 and PC2 are 32% and 26%, respectively.
Induction of medaka GGT genes
Given the role of GGTs in the antioxidant defense pathway, we determined whether medaka GGTs were inducible following treatment with pro-oxidants. In these experiments medaka larvae were treated with 100 μM tBHQ, a model oxidant (Xu et al., 2005) and assessed for alterations in GGT expression between 15 min and 6 hrs. Using qPCR, we demonstrated that several of the six medaka GGT genes exhibited significant gene induction with this treatment (Fig. 7). Within 15 minutes GGT1b mRNA exhibited a steady increase in mRNA abundance and peaked with a 3.5 fold induction after 6 hrs. Maximal induction of GGT1a occurred after 1 hr followed by a sharp decrease in expression compared to control levels. Comparatively, GGTL1a and GGT1b levels initially decreased followed by a transient 2-fold induction at 1 hr. By 6 hrs induction of GGTL1a and GGT1b had subsided. Expression of GGTL3 duplicates demonstrated considerable differences following induction with tBHQ. While GGTL3b exhibited up to a 3 fold induction within 1 hr of exposure, GGTL3a levels remained constant with little increase during the course of the exposure.
Figure 7. Exposure of medaka larvae to tBHQ.
Medaka larvae (1 dpf) were exposed to tBHQ in six-well plates containing 5 ml medaka rearing solution (5.1 mM NaCl, 120 μM KCl, 198 μM MgSO4 and 81 μM CaCl2; pH7.2) and 100 μM tBHQ in DMSO. Vehicle concentration did not exceed 0.1% of total volume. Exposure times ranged from 15 minutes to 6 hrs. At each sampling time point larvae were removed from solution, rinsed with fresh medaka rearing medium and snap-frozen for RNA isolation and qPCR analysis with GGT specific primer pairs. Data were normalized to 18S rRNA levels and are represented as the mean relative mRNA level ± SEM (n=5 individual samples).
PCA analysis of larval tBHQ exposures (Fig. 8C) are similar to those observed for the developmental profile (Fig. 8A). GGTL1a and GGTL1b are closely related while GGT1a and GGTL3a appear distantly separated from their respective duplicates. GGT1a clusters with GGTL3a on the PCA plot due to an inductive response occurring at the earlier time points. Separation of GGTL3b and GGT1b appears to be due to inductive effects occurring in later time points. PC1 represents 59% of the variation in the entire data set while PC2 represents 25%.
DISCUSSION
Combining comparative genomics and conventional molecular approaches, our laboratory has identified multiple GGT transcripts in teleost fish. Our comparative approach encompasses the use of several established genomic databases for teleosts including medaka, green spotted pufferfish, fugu, stickleback and zebrafish. Through reciprocal BLAST analysis, assessment of Pfam domains, phylogenetics and gene synteny, we established that teleost GGTs are co-orthologs of GGTs from lobe-finned descendants. Additionally, for each GGT1, GGTL1 and GGTL3 observed in these teleosts, gene duplicates (paralogs) were identified. Comparative, surveys in genome databases of higher vertebrates suggest the presence of only a single copy for each of GGT1, GGTL1 and GGTL3.
We estimated the phylogeny of GGT sequences from a wide range of taxa across the tree of life. It was necessary to conduct such a broad phylogenetic survey of all GGT sequences in order to identify the evolutionary relationship of GGT sequences to each other, and thus avoid confusing orthologous and paralogous sequences. This is because interpretations of a phylogenetic analysis can be undermined when paralogous sequences are mistakenly taken to be orthologous sequences. The phylogeny of GGT sequences shows that vertebrate GGTs are the result of at least three sequential duplication events. The first duplication event occurred prior to the divergence of extant animals and lead to the creation of the GGTL3 family and the ancestor of the GGT1 and GGTL1 families. The second duplication event created the GGT1 and GGTL1 families from their ancestor, and appears to have occurred early on the vertebrate lineage before the divergence of ray-finned and lobe-finned fishes.
Most notably, pairs of GGT families were identified among a subset of teleost genomes which are divided into alpha and beta subfamilies by a third duplication. This distinction permits the resolution of alpha and beta GGTs from the list of candidate GGTs previously described. Chromosomal segregation and topology of teleost GGT1a/GGT1b suggest that the these two copies present in the fish genome are more closely related to each other than to any tetrapod GGT1; the same is true for GGTL1a/L1b.
It thus appears that alpha and beta versions of GGTs in the GGT1, GGTL1 and possibly GGTL3 families arose from a duplication event in the ray-finned fish lineage after the divergence of the tetrapods but prior to the teleost radiation. This is consistent with the fish-specific whole genome duplication event and further supports the notion that teleost GGTs are co-orthologs of mammalian GGTs (Volff, 2005).
To further assess the relationship among these duplicates we examined neighboring gene arrangements of GGTs both within and among species. Conserved syntenic regions defined by closely linked orthologous genes on a single chromosome or a chromosomal fragment in each of two or more different species provides critical information concerning how genes and genomes evolve (Postlethwait, 2007). Assessment of gene synteny demonstrated that gene content among GGT containing chromosomes in medaka, stickleback, green spotted pufferfish and fugu are highly conserved. Gene order and arrangement varies both between GGT a and b chromosome pairs within species (paralogs) and among species (orthologs), suggesting significant shuffling of gene order has occurred with divergence of these teleost lineages. Syntenic relationships between closely related species such as medaka and stickleback, and green spotted pufferfish and fugu, were highly maintained providing further support for the phylogenetic relatedness of these species as previously suggested (Mitani et al., 2006). Zebrafish exhibited little synteny with any of the fish genomes examined likely due to extensive intra-chromosomal rearrangement (Kasahara et al., 2007).
Assessment of teleost genomes suggests that chromosomal organization in most teleosts consist of paired chromosomes which are likely derived from a single common protochromosome prior to a whole genome duplication event (Naruse et al., 2004). In medaka, evidence supports paralogy for medaka chromosomes 12/9, and 7/5, and green spotted pufferfish chromosomes 4/12 and 9/11. Our analysis suggests that large syntenic regions are well conserved between these chromosome pairs. This is consistent with the organization found for groupings of GGT1a/L1a: GGT1b/L1b and L3a/L3b providing further support for these pairings in each species except zebrafish. Zebrafish zfGGT1a was identified on chromosome 10, however location of GGTL1a was limited to a scaffold designation. As such, no assessment can be made in regards to the “head-to-tail” arrangement of these two genes as observed with the remaining species examined. Additionally there is no supporting evidence for orthology between medaka chromosome 12 and zebrafish chromosome 10 (Naruse et al., 2004) suggesting some disparity between the origin of these two chromosomes. Conversely, zebrafish GGT1b and GGTL1b were found in a head-to-tail arrangement on zebrafish chromosome 8 which is consistent with the pairing of GGT1b and GGTL1b in other teleost species. Zebrafish chromosome 8 is also thought to be orthologous to medaka chromosome 9 and green spotted pufferfish chromosome 12 each derived from the same protochromosome. Four additional GGT genes were found on zebrafish chromosome 1. To date, no paralogous chromosome in other species has been identified for zebrafish chromosome 1. This is likely due to either the deletion of an entire chromosome in an ancestor of zebrafish or that the paralogous chromosome has been redistributed to other chromosomes by translocation. As such there is no evidence for gene duplicates for any of these four GGT genes (Kasahara et al., 2007).
Following the whole genome duplication event the last common ancestor to medaka, green spotted pufferfish and zebrafish maintained 24 chromosomes and had undergone eight major inter-chromosomal rearrangements (Kasahara et al., 2007). Based upon our findings we concur that medaka likely has preserved the ancestral genomic structure without undergoing major inter-chromosomal rearrangements while green spotted pufferfish has undergone several fusion events (Kasahara et al., 2007). In comparison, the zebrafish genome has incurred multiple inter-chromosomal rearrangements through extensive translocation. This is likely why we are unable to demonstrate a paired relationship between zebrafish chromosomes 10 and 8.
Across vertebrate groups, gene synenty surrounding GGT1/GGTL1 and GGTL3 is significantly retained. Within a 40-Mb region of the mouse genome up to 17 genes were found on both teleosts and mouse chromosomes. Comparison to human however suggests that these same 17 genes are split between two chromosomes h12 and h22. This is not surprising given that comparative gene mapping demonstrates extensive gene shuffling since the divergence of medaka and mouse lineages (Naruse et al., 2004).
While there is still significant debate whether increased copy number is due to whole genome duplications or reflects multiple independent local duplication events, the FSGD hypothesis appears to be the most parsimonious (Postlethwait, 2007). Subsequent to a whole genome duplication (WGD) event gene duplicates have several possible fates. The classical model of gene duplication assumes redundancy in gene function(s) after duplication, with relaxed selection often resulting in deleterious mutations, pseudogene formation and eventual nonfunctionalization of one member of the pair (Force et al.,99′; Innan and Kondrashov, 2010). When nonfunctionalization does not occur, the classical model has gene duplicates maintained by mutation, fixation and positive selection resulting in neofunctionalization. In this scenario, one copy acquires a new protein activity while the second copy maintains the original function (Postlethwait et al., 2004, Innan and Kondrashov, 2010). In a third model, Force et al. (99′) proposed that gene duplicates are maintained by subfunction partitioning as a consequence of “duplication-degeneration-complementation”. In subfunction partitioning, deleterious mutations result in the simultaneous decay of specific regulatory regions or coding sequence of each gene copy. This decay means that the ancestral gene function(s) cannot be maintained unless both gene copies are retained. Subfunction partitioning Occurs rapidly and often gene pairs undergo subsequent independent evolutionary events resulting in eventual neofunctionalization, a process termed sub-neofunctionalization (SNF) (He and Zhang, 2005; Rastogi and Liberles, 2005). Subfunction partitioning is thought to be the dominant mechanism for maintenance of most gene duplicates in teleosts (Force et al., 99′; Postlethwait et al., 2004; Steinke et al., 2006, Innan and Kondrashov, 2010). The phylogenetic timing of the FSGD and the radiation of teleosts subsequent to this event provide suggestive evidence that subfunction partitioning and neofunctionalization may have contributed to the physiological plasticity, specification and evolutionary diversification of these organisms (Ohno, 99′; Taylor and Raes, 2004).
Our observation of clear expression differences between GGT1a (gut, kidney) and GGT1b (gill, heart) paralogs suggests that subfunction partitioning has likely occurred between several of the GGT paralogous pairs. Overall our mRNA expression data of for development, localization and induction show that (1) the variation between GGTL1a and GGTL1b is the least among the three paralogous pairs and is conserved in all three aspects (development, spatial, and induction); (2) the recent duplicates GGT1a and GGT1b appear most divergent in expression, compared to the other two a-b duplicate pairs (3) GGTL3a and GGTL3b only share similarity in tissue distribution, and are separated in developmental profiles and responses to oxidative stress; and (4) the inter-paralogous variation is condition dependent. It is noted however, that complete loss of gene expression was not observed in any one tissue or developmental stage, rather a differential degree of expression was observed between paralogs. While we recognize that our description does not follow the classical definition of subfunction partitioning, we hypothesize that a “quantitative subfunction partitioning” will manifest in differential abundance of each gene paralog in a specific tissue, developmental stage and/or gene induction. This interpretation of the “duplication-degeneration-complementation” (DDC) model does not require differential loss of subfunctions between the duplicates, but rather retention of the ancestral function through differential abundance in gene expression.
Expression of GGTs in response to tBHQ exposure suggests teleosts maintain an ability to respond to redox stress. Elevated expression of GGT may impart an ability to adapt to modifications in cellular environmental conditions. GGT expression is presumed to be coupled to intracellular glutathione concentrations (Zhang and Forman, 2009). GSH depletion generates a cellular sensitivity to oxidants resulting in an induced anti-oxidant response mediated by an induction of phase II enzymes including GGT. Increased liver GGT expression occurs in response to a range of oxidants, antioxidants and chemo-preventive compounds, including menadione, ethoxyquin, butylated hydroxytoluene and the naturally occurring plant constituent indol-3-carbinol (Hudson et al., 97′). GGT expression is additionally induced in alveolar type II cells in response to quinone toxicity (Kugelman et al., 94′) and in the epididymis in response to additional reactive oxygen species (ROS) (Markey et al., 98′). In each case, GGT transcripts are identified in cell types normally low in or devoid of GGT expression. Consensus elements for AP1, AP2, ARE and NFκB are present in mammalian GGT promoters, suggesting a redox sensitivity and potential responsiveness to various reactive oxygen species. Exact mechanisms of GGT induction following oxidative stress have not been elucidated. However, control of GSH levels by GGT expression is suggested to be a major component to the anti-oxidative stress response. Gene induction may be elicited by a direct alteration of redox sensitive transcription factors or result due to altered signaling cascades in response to GSH depletion (Wilhelm et al., 97′).
CONCLUSIONS
In summary we demonstrate the presence of multiple paralogous genes of γ-glutamyl transferase in several distantly related teleost species. It is likely that GGT paralogs arose from the serial 3R genome duplication event. There is some evidence that GGTL1 was further duplicated (three genes) in green spotted pufferfish or that this third duplicate was lost in medaka, stickleback, and fugu. Similarly one or both paralogs of GGTL3 was lost in green spotted pufferfish, fugu and zebrafish. Gene synteny is highly maintained both within species duplicates and among species including teleosts and lobe-finned descendants. Finally, we present a modification of the “duplication-degeneration-complementation” (DDC) model of subfunction partitioning where quantitative differences are observed in gene expression between gene paralogs. Questions remain however regarding the functional role of multiple GGT genes in teleosts, their role in the antioxidant defense process or their ability to impart plasticity for adaptation to novel aquatic environments.
Supplementary Material
Supplementary file 1 (table St1)- Versions of genome databases used in the current analyses.
Supplementary file 2 (table St2)- GenBank accessions and Ensembl Gene IDs of GGT genes in selected species.
Supplementary file 3 (table St3)- PCR primers for medaka GGT studies.
Supplementary file 4 (figure Sf1)- Dendrogram of the putative GGT genes in Danio rerio.
Supplementary file 5 – (text Sx1)- Phylogenetic methods
Supplementary file 6 (figure Sf2)- Un-collapsed phylogeny estimate.
Supplementary file 7 (text Sx2)-Figure sf2 legend
Acknowledgments
Supported by National Cancer Institute (R21CA105084-01A1 to SWK), and North Carolina Agricultural Research Grant (02225 to SWK). B.D.R. was supported by the National Evolutionary Synthesis Center (NSF EF-0905606).
This work was supported in part by National Cancer Institute (R21CA105084-01A1 to SWK), and North Carolina Agricultural Research Grant (02225 to SWK). B.D.R. was supported by the National Evolutionary Synthesis Center (NSF EF-0905606). We wish to thank Erin Kollitz, Erin Yost, Arnaud Van Wettere and Gwijun Kwon for medaka care, culture and maintenance. We also thank David Hinton for critical review of an early draft of this manuscript and Rudolf Wu, City University Hong Kong for medaka 18S rRNA normalization primer for real-time qPCR. We additionally thank Dr Peng Li, NHLBI, National Institutes of Health for assistance with the PCA analysis.
LIST OF ABBREVIATIONS
- GGT
gamma-glutamyl transferase
- GGTL
gamma-glutamyl transferase-like
- GSH
glutathione
References
- Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST--a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RK, Roberts A, Smoot M, Juvekar S, Do J, Dewey C, Holmes I, Pachter L. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi N, Holic N, Guellaen G, Laperche Y. Gamma-glutamyl transpeptidase gene organization and expression: a comparative analysis in rat, mouse, pig and human species. Comp Biochem Physiol B Biochem Mol Biol. 1999;122:367–380. doi: 10.1016/s0305-0491(99)00013-9. [DOI] [PubMed] [Google Scholar]
- Crow KD, Wagner GP. Proceedings of the SMBE Tri-National Young Investigators’ Workshop 2005. What is the role of genome duplication in the evolution of complexity and diversity? Mol Biol Evol. 2006;23:887–892. doi: 10.1093/molbev/msj083. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. doi: 10.1002/dvg.20450. [DOI] [PubMed] [Google Scholar]
- Dickinson DA, Forman HJ. Glutathione in defense and signaling: lessons from a small thiol. Ann N Y Acad Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. Genomics. Vertebrate genomes compared. Science. 2002;297:1283–1285. doi: 10.1126/science.1076231. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Groffen J, Warburton D, Sneddon TP. The human gamma-glutamyltransferase gene family. Hum Genet. 2008;123:321–332. doi: 10.1007/s00439-008-0487-7. [DOI] [PubMed] [Google Scholar]
- Hudson EA, Munks RJ, Manson MM. Characterization of transcriptional regulation of gamma-glutamyl transpeptidase in rat liver involving both positive and negative regulatory elements. Mol Carcinog. 1997;20:376–388. [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin IT, Takeda H, Morishita S, Kohara Y. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelman A, Choy HA, Liu R, Shi MM, Gozal E, Forman HJ. gamma-Glutamyl transpeptidase is increased by oxidative stress in rat alveolar L2 epithelial cells. Am J Respir Cell Mol Biol. 1994;11:586–592. doi: 10.1165/ajrcmb.11.5.7946387. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Markey CM, Rudolph DB, Labus JC, Hinton BT. Oxidative stress differentially regulates the expression of gamma-glutamyl transpeptidase mRNAs in the initial segment of the rat epididymis. J Androl. 1998;19:92–99. [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Mitani H, Kamei Y, Fukamachi S, Oda S, Sasaki T, Asakawa S, Todo T, Shimizu N. The medaka genome: why we need multiple fish models in vertebrate functional genomics. Genome Dyn. 2006;2:165–182. doi: 10.1159/000095103. [DOI] [PubMed] [Google Scholar]
- Naruse K, Tanaka M, Mita K, Shima A, Postlethwait J, Mitani H. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 2004;14:820–828. doi: 10.1101/gr.2004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin Cell Dev Biol. 1999;10:517–522. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- Park HJ, Moon JS, Kim HG, Kim IH, Kim K, Park EH, Lim CJ. Characterization of a second gene encoding gamma-glutamyl transpeptidase from Schizosaccharomyces pombe. Can J Microbiol. 2005;51:269–275. doi: 10.1139/w04-137. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Quang le S, Gascuel O, Lartillot N. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics. 2008;24:2317–2323. doi: 10.1093/bioinformatics/btn445. [DOI] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005;5:28. doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Stuart JM, Altman RB. Principal components analysis to summarize microarray experiments: application to sporulation time series. Proceedings of the Pacific Symposium on Biocomputing. 2000:455–466. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Braasch I, Meyer A. Many genes in fish have species-specific asymmetric rates of molecular evolution. BMC Genomics. 2006;7:20. doi: 10.1186/1471-2164-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kumagai H, Echigo T, Tochikura T. DNA sequence of the Escherichia coli K-12 gamma-glutamyltranspeptidase gene, ggt. J Bacteriol. 1989;171:5169–5172. doi: 10.1128/jb.171.9.5169-5172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi N, Ikeda Y. gamma-Glutamyl transpeptidase: catalytic mechanism and gene expression. Adv Enzymol Relat Areas Mol Biol. 1998;72:239–278. doi: 10.1002/9780470123188.ch7. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Meyer A. Genome duplication, divergent resolution and speciation. Trends Genet. 2001;17:299–301. doi: 10.1016/s0168-9525(01)02318-6. [DOI] [PubMed] [Google Scholar]
- Venkatesh B. Evolution and diversity of fish genomes. Curr Opin Genet Dev. 2003;13:588–592. doi: 10.1016/j.gde.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Yap WH. Comparative genomics using fugu: a tool for the identification of conserved vertebrate cis-regulatory elements. Bioessays. 2005;27:100–107. doi: 10.1002/bies.20134. [DOI] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Bender K, Knebel A, Angel P. The level of intracellular glutathione is a key regulator for the induction of stress-activated signal transduction pathways including Jun N-terminal protein kinases and p38 kinase by alkylating agents. Mol Cell Biol. 1997;17:4792–4800. doi: 10.1128/mcb.17.8.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forman HJ. Redox regulation of gamma-glutamyl transpeptidase. Am J Respir Cell Mol Biol. 2009;41:509–515. doi: 10.1165/rcmb.2009-0169TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1 (table St1)- Versions of genome databases used in the current analyses.
Supplementary file 2 (table St2)- GenBank accessions and Ensembl Gene IDs of GGT genes in selected species.
Supplementary file 3 (table St3)- PCR primers for medaka GGT studies.
Supplementary file 4 (figure Sf1)- Dendrogram of the putative GGT genes in Danio rerio.
Supplementary file 5 – (text Sx1)- Phylogenetic methods
Supplementary file 6 (figure Sf2)- Un-collapsed phylogeny estimate.
Supplementary file 7 (text Sx2)-Figure sf2 legend