Abstract
Regulation of gene expression using small interfering RNA (siRNA) is a promising strategy for research and treatment of numerous diseases. In this study, we develop and characterize a delivery system for siRNA composed of polyethylenimine (PEI), polyethylene glycol (PEG), and mannose (Man). Cationic PEI complexes and compacts siRNA, PEG forms a hydrophilic layer outside of the polyplex for steric stabilization, and mannose serves as a cell binding ligand for macrophages. The PEI-PEG-mannose delivery system was constructed in two different ways. In the first approach, mannose and PEG chains are directly conjugated to the PEI backbone. In the second approach, mannose is conjugated to one end of the PEG chain and the other end of the PEG chain is conjugated to the PEI backbone. The PEI-PEG-mannose delivery systems were synthesized with 3.45 – 13.3 PEG chains and 4.7 – 3.0 mannose molecules per PEI. The PEI-PEG-Man-siRNA polyplexes displayed a coarse surface in Scanning Electron Microscopy (SEM) images. Polyplex sizes were found to range from 169nm to 357nm. Gel retardation assays showed that the PEI-PEG-mannose polymers are able to efficiently complex with siRNA at low N/P ratios. Confocal microscope images showed that the PEI-PEG-Man-siRNA polyplexes could enter cells and localized in the lysosomes at 2 hours post-incubation. Pegylation of the PEI reduced toxicity without any adverse reduction in knockdown efficiency relative to PEI alone. Mannosylation of the PEI-PEG could be carried out without any significant reduction in knockdown efficiency relative to PEI alone. Conjugating mannose to PEI via the PEG spacer generated superior toxicity and gene knockdown activity relative to conjugating mannose and PEG directly onto the PEI backbone.
Keywords: HRPT, luciferase, polyethylenimine, mannose, polyethylene glycol, PEI, PEG, nanoplexes, nanoparticles, polyplexes, siRNA, RNA interference, non-viral, gene medicine
INTRODUCTION
Gene therapy using RNAi (RNA interference) has significant potential for treatment of a variety of diseases as well as a tool for biomedical research (Behlke, 2006). RNAi is a natural cellular mechanism by which a specific mRNA is targeted for degradation through inhibition of the synthesis of the encoded protein (Akhtar and Benter, 2007). Selectively silencing post-transcriptional mRNA by RNAi represents a promising new approach for the inhibition of gene expression in vitro and in vivo (Fire et al., 1998). SiRNA, 21–27 base pair double-stranded RNA, plays a vital role in initiating the RNAi mechanism (Caplen et al., 2001). However, siRNA is susceptible to enzymatic degradation and has a relatively large molecular weight with a net negative charge that lowers its capacity to penetrate the cell membrane (Abe et al., 2009).
To overcome this challenge, both viral and non-viral vectors have been developed to make siRNA delivery more efficient (Aigner, 2007; Kim and Kim, 2009). Viral vectors such as adenoviruses and retroviruses are very effective delivery systems (Zhang and Godbey, 2006). However, concerns about their immunogenic nature and inadvertent gene expression changes following random integration into the host genome still exist (Abbas et al., 2008; Martin and Caplen, 2007). Polyethylenimine (PEI) is a synthetic cationic polymer that has been widely used to deliver oligonucleotides, siRNA and plasmid DNA in vitro and in vivo (Abbas et al., 2008; Aigner et al., 2002; Boussif et al., 1995; Hobel and Aigner, 2010; Intra and Salem, 2008; Nimesh and Chandra, 2009; Pearce et al., 2008; Petersen et al., 2002; Zhang et al., 2008; Zintchenko et al., 2008b). PEI electrostatically condenses nucleic acids and forms stable nanoparticles or polyplexes (Intra and Salem, 2008). Branched PEI with higher molecular weights (<25kDa) is more efficient for in vitro transfection because it condenses nucleic acids more effectively than linear PEI (Godbey et al., 1999). Every third atom of PEI is a protonatable nitrogen atom, which enables the “proton sponge” effect over a wide range of pH. PEI will buffer the acidification within the endosome after endocytosis resulting in endosomal chloride accumulation. This results in osmotic swelling and rupture allowing for endosomal escape of the PEI/siRNA polyplexes (Boussif et al., 1995). Although cationic PEI has promising potential as a gene delivery vehicle, it is also associated with high toxicity relative to other non-viral vectors. (Fischer et al., 2003; Intra and Salem, 2008; Nimesh and Chandra, 2009; Swami et al., 2007). PEI can be modified to reduce toxicity and its free amine groups can be used to conjugate cell binding or targeting ligands (Beyerle et al., 2010; Biswal et al., 2010; Bonsted et al., 2008; Breunig et al., 2008; Diebold et al., 1999a; Kang et al., 2010; Kim et al., 2008; Merkel et al., 2009b; Moore et al., 2008; Ogris M, 1999; Patnaik et al., 2010; Petersen et al., 2002; Zintchenko et al., 2008a).
Various modifications have been applied to PEI to reduce toxicity and increase target specificity (Kawakami and Hashida, 2007). They include coupling to other macromolecules like polyethylene glycol (PEG), either alone (Katayose and Kataoka, 1997; Mao et al., 2006; Merkel et al., 2009a; Petersen et al., 2002; Sagara and Kim, 2002) or in combination with ligands for tissue specific targeting (Biswal et al., 2010; Hobel and Aigner, 2010; Kang et al., 2010; Kunath et al., 2003b; Ogris M, 1999; Schiffelers et al., 2004; Sun et al., 2008). Conjugation of PEG to the polyplexes or nanoparticles creates a hydrophilic layer around the nanoparticles that provides steric stabilization thereby reducing aggregation of the particles. The PEG layer increases the circulatory half-life of the nanoparticles or polyplexes and reduces toxicity (Owens and Peppas, 2006; Peracchia et al., 1999). A surface PEG chain larger than 2kDa is more efficient at steric stabilization and increasing the circulation half-life. This minimum MW is due to the inflexibility of shorter PEG chains (Gref et al., 1994; Leroux et al., 1995; Peracchia et al., 1997). PEG can be cleared by renal, fecal or hepato-biliary routes depending on their molecular weight or modification (Yamaoka et al., 1994, 1995). The majority of PEG 6kDa is excreted in the urine 12 hours after intravenous administration in humans. It is approved by the FDA for internal use due to its nontoxic and non-immunogenic characteristics (Fishburn, 2008; Pang and Fiume, 2001; Webster et al., 2007).
Specific ligands can bind to target cell surface receptors, which in turn can trigger receptor-mediated endocytosis (Park et al., 2008b). Mannose is often used as a ligand that binds mannose receptors on cells to induce receptor-mediated endocytosis, which increases the delivery efficacy (Diebold et al., 1999a). Its receptor is expressed on the surface of antigen presenting cells (APCs; dendritic cell, macrophages) and liver endothelial cells (Diebold et al., 2002), etc. A number of studies have shown that mannosylated nanoparticles enter Raw264.7 cells via receptor-mediated endocytosis (Jiang et al., 2009b; Kim et al., 2006; Park et al., 2008a; Zhou et al., 2007b).
In this study, we designed and synthesized a siRNA delivery system that incorporates PEI, PEG, and mannose. The mannosylated pegylated PEI delivery systems were designed in two different constructs. In our first approach, mannose and PEG chains were directly conjugated to the PEI backbone. In our second approach, mannose was conjugated to PEI via a PEG chain spacer (Table 1). The PEI-PEG-mannose constructs were then characterized for capacity to complex with siRNA, surface morphology and shape, gene and mRNA knockdown efficiency and toxicity.
Table 1.
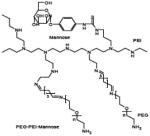
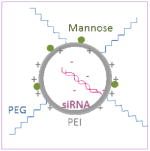
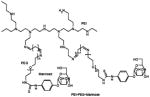
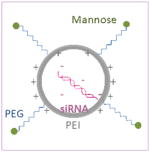
Schematical diagram showing chemical structures of two different mannosylated pegylated polyethylenimine constructs are depicted on the left column and schematical diagrams of the constructs after complexation with siRNA are shown on the right column. Mannose and PEG are directly conjugated to the PEI backbone in construct #1, whilst mannose is conjugated to the PEI via a PEG spacer in construct #2.
| Structure | Schematic diagram | |
|---|---|---|
| Construct #1 Mannose-PEI-PEG |
|
|
| Construct #2 PEI-PEG-mannose |
|
|
MATERIALS AND METHODS
Materials
Branched PEI 25kDa and α-D-mannopyranosylphenyl isothiocyanate (MPITC) were purchased from Sigma-Aldrich (St. Louis, MO). PEG 2kDa with one amine terminus was purchased from Creative PEGWorks (Winston Salem, NC). DMSO, glacial acetic acid and EDTA were purchased from Fisher (Pittsburgh, PA). Bio-Gel P-2 Gel is from Bio-Rad (Hercules, CA). Maleic anhydride, toluene, glutaraldehyde 50%, methanol and sulfuric acid were purchased from Sigma-Aldrich (St. Louis, MO). All the siRNAs (DS scrambled negative control, RLuc-S1 DS positive control, Cy-3™, NC1, and HPRT) and pDNA (psiCHECK™-2, Promega, Madison, WI) were kindly provided by Integrated DNA Technologies (Coralville, IA).
Synthesis
PEI-PEG
500mg PEG was dissolved in 10ml toluene and heated up to 100°C. Maleic anhydride solution (33.1mg in 40ml toluene) was added into the PEG solution in small increments. After the introduction of the maleic anhydride, the temperature was increased to 110°C and then the mixture was refluxed for 12 hours. Excess toluene was removed using rotary evaporation and the recovered product was dissolved in 2.5ml deionized water which was then purified using a P-2 column. The elution was lyophilized and redissolved in 10ml methanol. Then, 1ml maleic anhydride-PEG was slowly added into 50% w/v glutaraldehyde solution containing 50μl glutaraldehyde and 300μl methanol. The reaction was stopped by 2 hours and the mixture loaded into a P-2 column. The elution was immediately added to a 2ml PEI solution (~150mg PEI) for overnight reaction. PEI-PEG was recovered after dialysis (MWCO 10,000, Pierce Biotechnology Inc., Rockford, IL) for 3 days and lyophilized (Labconco FreeZone4.5, Kansas City, MO).
Mannose-PEI-PEG
After the PEI was PEGylated, as described above, 80μl of MPITC solution (dissolved at a concentration of 0.125mg/μl in DMSO) was added to PEI-PEG for overnight mannosylation. The conjugate was recovered after dialysis and lyophilization.
PEI-PEG-Mannose
80 μl MPITC solution was added to 2ml PEG solution (dissolved using NaHCO3 buffer, 8.516mg/ml, pH 8.4) containing 50mg PEG for overnight reaction, followed by the addition of 50μl of 50% w/v glutaraldehyde solution and 300μl methanol. The reaction was stopped in 2 hours using a P-2 column separation. The collection was added to PEI solution for overnight reaction. PEI-PEG-mannose was recovered after dialysis and lyophilization.
1H NMR
Confirmation of the presence of mannose, PEI and PEG in the conjugate was achieved by dissolving the products in 1.0% w/v DCl/D2O (Cambridge Isotope Lab, Andover, MA) at ~40mg/ml and characterizing using proton nuclear magnetic resonance (1H NMR, Bruker Avance 300)
Resorcinol assay
Resorcinol (Riedel-de Haen, Seelze, Germany) was dissolved at 6mg/ml. D-(+)-mannose predried at 110°C overnight was used as a standard. Mannose was dissolved in 1% w/v HAc at a concentration ranging from 9.0854 to 908.54μg/ml. PEI-PEG-mannose and mannose-PEI-PEG samples were dissolved using 1% w/v HAc at appropriate concentrations. Each test mixture, consisted of 20μl mannose/sample solution, 20μl resorcinol, 50μl pristane (Acros, Fair Lawn, NJ), and 100μl 75% w/v sulfuric acid, was subjected to vortex for 30 seconds, heating at 93°C for 30 minutes and cooling down to room temperature for 30 minutes in a 96-well plate. The absorbance was recorded at 480nm.
Cell Culture
Raw264.7 cells (ATCC, Manassas, VA), a murine macrophage cell line that is known to express mannose receptors and that is typically hard to transfect, was selected for in vitro experiments because macrophages are a potential target for this delivery system. The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Hyclone), penicillin-streptomycin (100units penicillin; 100ug streptomycin/ml, Gibco). The cells were maintained at 37 C in a humidified, 5% carbon dioxide atmosphere.
Amplification and purification of pDNA
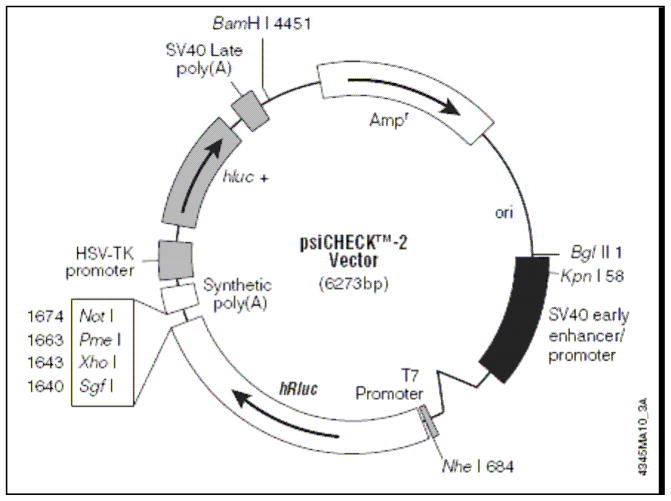
The psiCHECK™-2 (Promega, Madison, WI) is a 6.3 kb pDNA designed to monitor a quantitative measurement of RNAi (Figure 2). It has genes encoding for firefly (hluc+) luciferase and Renilla (hRluc) luciferase with each luciferase having a HSV-TK or SV40 promoter respectively. The firefly reporter gene has been constructed to serve as an intraplasmid standard so that the Renilla luciferase signal can be normalized to the firefly luciferase signal. The pDNA was transformed in E. coli DH5α (Invitrogen) and amplified in LB Broth media at 37 °C overnight on a plate shaker set at 250 rpm. The pDNA was extracted with Wizard® Plus Maxipreps DNA Purification System (Promega) followed by removal of bacterial endotoxin contamination with Endotoxin Removal Kit (MiraCLEAN®) according to the manufacturers’ protocols. Purified pDNA was dissolved in Tris–EDTA buffer and its purity and concentration were determined by UV absorbance at 260 and 280 nm.
Figure 2.
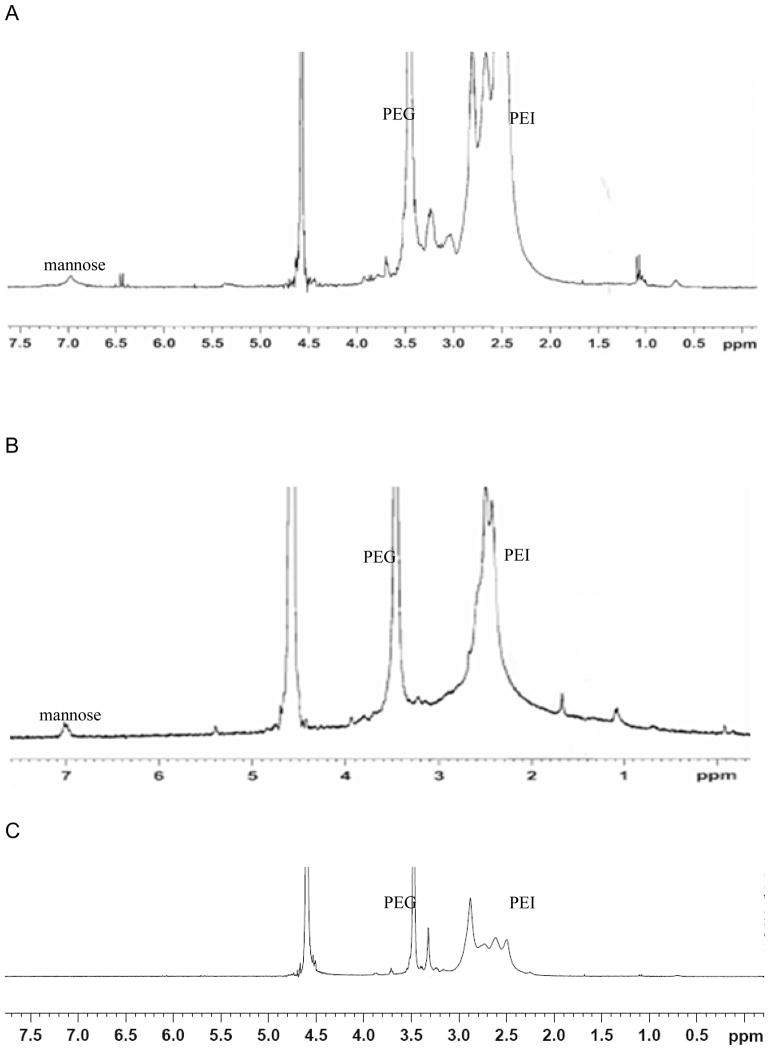
1H NMR spectras of mannose-PEI-PEG (A), PEI-PEG-mannose (B) and PEG-PEI (C). Mannose-PEI-PEG showed peaks of PEG (PEG-H) at 2.6 ppm, PEI (PEI-H) at 3.5 ppm and mannose (phenyl-H) at 7ppm. PEI-PEG-mannose also showed each component at the same peaks.
Preparation of PEI-siRNA polyplexes
PEI/siRNA polyplexes were formed at desired N/P (nitrogen in cationic polymer per phosphate in nucleic acid) ratios with predetermined amounts of siRNA and polymer solutions. A mass per phosphate of 325Da for RNA and mass per charge of 43 for PEI were used to calculate N/P ratio. The polymer solution was dropped into siRNA solution, and then the mixture was vortexed for 20 seconds followed by 30 minutes incubation at room temperature.
Size distribution and surface morphology analysis
SiRNA/polymer polyplexes were formed as described above. Then the polyplex solutions were sprayed using All-Glass Nebulizer (PELCO, Redding, CA) onto silicon wafers. The silicon wafers were stained with 4% osmium tetroxide vapor under the hood overnight and mounted on aluminum stubs using liquid colloid silver adhesives followed by overnight drying at room temperature. The specimens were observed using scanning electron microscopy (SEM, Hitachi S-4800). Polyplex size and zeta potential measurements were conducted using the Zetasizer Nano ZS (Malvern, Southborough, MA). Briefly, the polyplexes were prepared as described above in nuclease-free deionized water at an N/P ratio of 7. The size measurements were performed at 25°C at a 173° scattering angle. The mean hydrodynamic diameter was determined by cumulative analysis.
Gel Retardation Assay
SiRNA/PEI polyplexes were loaded in a 2% agarose gel with 0.5μg/ml Ethidium Bromide and run at 60V in TAE buffer for 45 minutes. The gels were visualized with a UV transilluminator (Spectroline, Westbury, NY) and photographed by Panasonic DMC-FX30 digital camera.
Intracellular Trafficking
RAW264.7 cells were plated in 0.01% (w/v) poly L-lysine coated 8-well chamber slides (Lab-Tak) at 1.2 × 104/well concentration and incubated overnight. For lysosomal staining, the cells were incubated with 75nM Lysotracker® Green DND-26 (Molecular Probes®) containing opti-MEM media (Gibco) for 30 minutes then transfected with polymer/Cy3-labeled siRNA polyplexes in fresh opti-MEM. At pre-determined time points, the cells were washed with PBS and fixed with 4% paraformaldehyde and mounted using DAPI-containing Vectashield mounting medium for nuclear staining (VECTOR Laboratories, UK). Then the slides were covered with coverslips followed by 4°C storage in the dark before visualization under the multiphoton/confocal microscope (Bio-Rad Radience 2100MP and Zeiss Confocal 710).
Assays to evaluate knockdown of gene and mRNA expression
Dual Luciferase Assay
RAW264.7 cells were seeded on a T25 flask (6.8 × 106 cells) on day 0 and incubated overnight. PsiCHECK™-2 was transfected using PEI in opti-MEM media on day 1 for 4 hours, then trypsinized to be replated onto 24-well plates (3 × 105 cells/well). Then RLuc-S1 DS positive control siRNA, which encodes a complementary sequence for Renilla luciferase gene in psiCHECK™-2 vector, was transfected using each different polymer in opti-MEM media on day 2 and the cells were incubated for 4 hours followed by overnight incubation in complete media. On day 3, luciferase gene expression was analyzed using the Dual Luciferase Reporter System (Promega, Madison, WI) according to the product manual and using a Lumat LB 9507 (Berthold Technologies, Bad Wildbad, Germany). Plasmid DNA/PEI complexes or siRNA/polymer complexes were prepared as described above at precalculated N/P ratios. DS scrambled neg. siRNA, non-targeting sequences in the human, mouse, or rat transcriptome, served as a negative control. The Renilla luciferase gene expression was normalized to firefly luciferase as internal control expression and expressed as relative gene expression (100% to DS scrambled neg. transfected with PEI). The data was reported as mean ± standard deviation for triplicate samples using Microsoft Excel and Prism® software (GraphPad Software, Inc). Every experiment was repeated at least twice.
Real-Time PCR
Cells were seeded onto 48-well plate (8 × 104 cells/well) for endogenous gene knockdown on day 0. Then NC1 (negative control) and HPRT siRNAs were transfected using various polymers in opti-MEM media on day 1 and incubated for 24 hours. Total RNA was extracted (Promega, SV96 Total RNA Isolation System) on day 2 followed by cDNA synthesis and real-time PCR. 150ng of total RNA was used for reverse transcription using Superscript II reverse transcriptase (Invitrogen, San Diego, CA). cDNA equivalent to 40 ng total RNA was analyzed by real-time PCR in triplicate using Immolase polymerase (Bioline, Randolph, MA) on AB7900HT (Applied Biosystems). The data was reported as mean ± standard deviation from triplicate RT-PCR reactions of each triplicate sample using Microsoft Excel and Prism® software (GraphPad Software, Inc). Every experiment was repeated at least twice.
Cytotoxicity assay
Cytotoxicity was determined using the MTS assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega, Madison, WI) using the protocol provided by the manufacturer. In brief, the Raw264.7 cells were seeded in 96-well plates (5 × 105cells/well) and incubated overnight. The polymer containing DMEM media were prepared at different concentrations and added to cells for incubation. 20μl MTS solution per 100μl media was added to each well for further 1 to 3 hours incubation at 37 C in a humidified, 5% CO2 atmosphere. The plate was then measured at 490nm absorbance (SpectraMax Plus384, Molecular Device) and the relative cell viability calculated using standard curves. Experiments were repeated in triplicate.
Statistical Analysis
Group data are reported as mean ± SD. Differences between groups were analyzed by one-way ANOVA analysis with Tukey’s post-test. Levels of significance were accepted at P<0.05. Statistical analyses were performed using Prism software (Graphpad Software, Inc., San Diego, CA.).
RESULTs
PEI, PEG and mannose are detected in final product 1H NMR spectra
PEI-PEG-mannose and mannose-PEI-PEG showed peaks at δ 7.0 (MPITC phenyl-H), δ 3.5 (PEG-H), and δ 2.6 (PEI-H) indicating successful incorporation of PEI, PEG and mannose in the final product (Figure 2). The signal at 7ppm was relatively weak but indicated the existence of mannose (MPITC) which was further verified and quantified using the resorcinol assay. When comparing these spectra to the control spectra of PEI-PEG (Figure 2C), the 3.5ppm peak matched to PEG and the 2.6ppm corresponded to the PEI hydrocarbon chain. The conjugation/substitution ratio was calculated based on the peak areas ratio between 3.5ppm PEG and 2.6ppm PEI (Table 2). The molar ratios of PEI to PEG were 3.704 in PEI-PEG, 0.962 in mannose-PEI-PEG and 1.23 in PEI-PEG-mannose. There was no significant difference between the 1H NMR spectra of mannose-PEG-PEI and PEG-PEI-mannose.
Table 2.
PEI/PEG molar ratios and mannose content in PEI-PEG-mannose and mannose-PEI-PEG constructs. The PEI to PEG molar ratio was calculated based on the area of 1H NMR peaks. Resorcinol measurements were used to determine the content of mannose (μmol mannose per 1mg polymer) in each construct.
| Formulation | PEI/PEG molar ratio | Mannose content (μmol/mg) |
|---|---|---|
| Mannose-PEI-PEG | 0.962 | 0.18679 |
| PEI-PEG-Mannose | 1.23 | 0.12122 |
| PEI-PEG | 3.704 | N/A |
Mannose quantity is determined in final constructs using the resorcinol assay
The resorcinol assay is a commonly used method to determine the content of monosaccharides in polymeric structures (Diebold et al., 1999a). The mannose content in our products was determined using this technique (Table 2). PEI-PEG-mannose had 0.12μmol/mg of mannose and mannose-PEI-PEG had 0.19 μmol/mg mannose.
Pegylated polyplexes are spherical and coarse in appearance
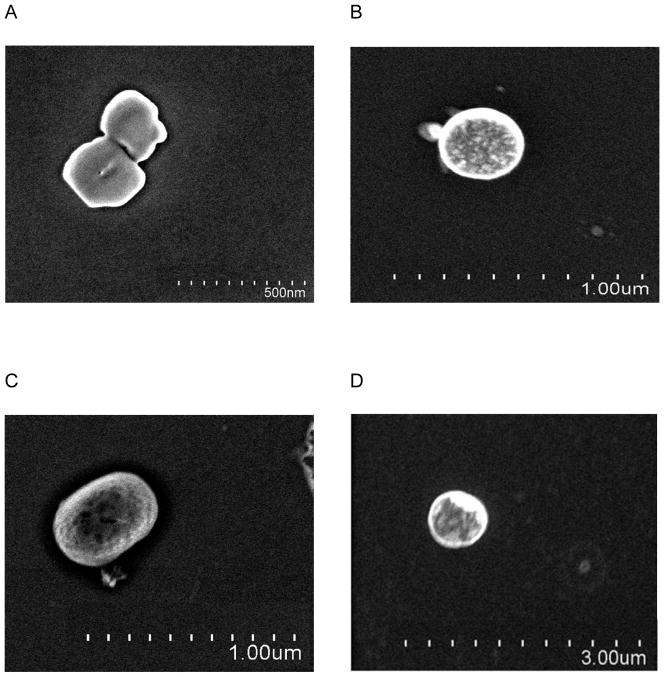
SEM images were used to observe surface morphology and shape of the polymer/siRNA polyplexes. The polymers/siRNA polyplexes without pegylation were spherically or semi-spherically shaped with porous but otherwise smooth surfaces (Fig. 3A). In contrast, the PEGylated polyplexes had more coarse features on the surface (Fig. 3B-D). The particle sizes of various PEI polymers/siRNA polyplexes are shown in Table 3. PEI/siRNA polyplexes and PEI-PEG/siRNA polyplexes had 214.57nm and 201.80nm average diameters respectively. Man-PEI-PEG formed polyplexes with siRNA with an average size of 169.10nm. PEI-PEG-mannose/siRNA polyplexes had an average size of 357.33nm. The pegylated PEI/siRNA polyplexes and PEI-PEG-Man/siRNA polyplexes displayed zeta potentials (24.37mV and 21.63mV respectively) that were similar to unmodified PEI (21.94mV). The zeta potential of polyplexes prepared using Man-PEI-PEG had a lower value of 10.46mV.
Figure 3.
SEM images of mannosylated pegylated PEI/siRNA polyplexes. (A) PEI/siRNA polyplexes, (B) PEI-PEG/siRNA polyplexes, (C) Mannose-PEI-PEG/siRNA polyplexes, (D) PEI-PEG-mannose/siRNA polyplexes. All the polyplexes were prepared with 1μM siRNA at N/P ratio 5.
Table 3.
Hydrodynamic size and zeta potential of the polymer/siRNA polyplexes. All the polyplexes were prepared with 1μM siRNA at N/P ratio 7. All data were represented as mean ± SD (n = 3).
| Zeta Potential (mV) | Size (nm) | |
|---|---|---|
| PEI | 21.94 ± 0.50 | 214.57 ± 29.00 |
| PEI-PEG | 24.37 ± 1.63 | 201.80 ± 4.95 |
| Man-PEI-PEG | 10.46 ± 0.91 | 169.10 ± 9.54 |
| PEI-PEG-Man | 21.63 ± 6.31 | 357.33 ± 92.90 |
PEI-PEG-mannose and mannose-PEI-PEG efficiently complex siRNA and retard siRNA migration in gel electrophoresis assays
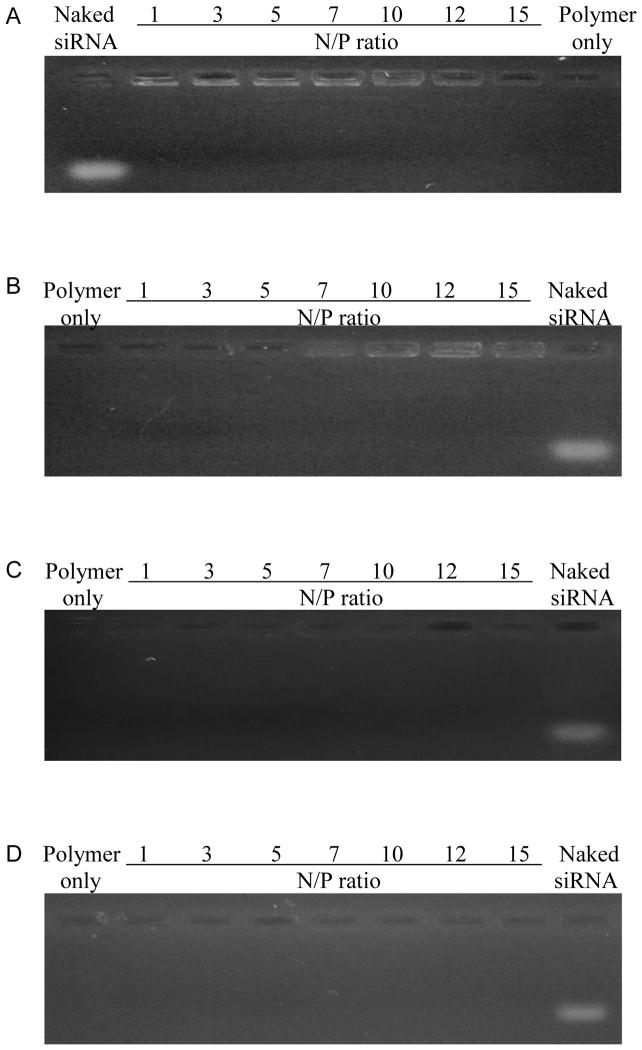
For the mannosylated and pegylated PEI products we generated to be capable of delivering siRNA, they must be able to complex and condense the siRNA as efficiently as PEI alone. Cationic polymers form complexes with anionic siRNAs and compact them by ionic/electrostatic interactions. This complexation capacity was analyzed using a gel electrophoresis assay. The migration of siRNA on the agarose gel was retarded with the use of all the polymer constructs. All the constructs including PEI (Fig. 4A) and PEI-PEG (Fig. 4B) showed excellent complexation with siRNA even at low N/P ratios of 1 that prevented siRNA migration in the gel retardation assays. Both mannose-PEI-PEG and PEI-PEG-mannose showed complete exclusion of EtBr from N/P ratios 1 to 15 indicating a broad and strong complexation capacity with siRNA (Fig. 4C and D). Based on this result, the following studies were carried out using polyplexes prepared within this range of N/P ratios.
Figure 4.
Gel retardation assay results of various polyplex formulations. (A) PEI/siRNA polyplexes, (B) PEI-PEG/siRNA polyplexes, (C) Mannose-PEI-PEG/siRNA polyplexes, (D) PEI-PEG-mannose/siRNA polyplexes. All the polyplexes were prepared with 1μM siRNA from N/P ratio 1 to 15.
Mannosylated pegylated PEI/siRNA polyplexes are efficiently endocytosed by RAW264.7 cells
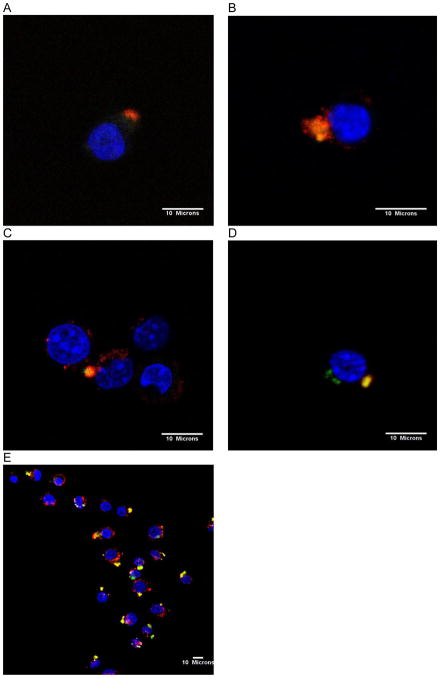
To track the cellular uptake and distribution of mannosylated pegylated PEI/siRNA complexes (N:P ratio of 7), siRNA and endosomes were labeled with red and green fluorophores, respectively. At 2 hours post-transfection, most of the polymers/Cy3-siRNA complexes (shown in red) were successfully internalized in the RAW264.7 cells. PEI/siRNA and PEI-PEG/siRNA polyplexes were taken up by cells and localized in vesicular structures as seen by yellow fluorescence due to the green staining of lysosome and red siRNA signal in close proximity (Fig. 5A and B). Mannose-PEI-PEG/siRNA complexes were also found in the endosomes (Fig. 5C). PEI-PEG-mannose/siRNA polyplexes showed the most wide-spread endocytosis with relatively even distribution in a large group of cells (Fig. 5D and E). Several images including Figure 5C and 5E showed polyplexes in the cytoplasm at the perinuclear region as observed by the separation of the green and red signal (nuclei stained with blue) suggesting release of the polyplexes from the endosomes.
Figure 5.
Confocal microscopy images showing intracellular trafficking of polyplexes. Raw264.7 cells were stained with Lysotracker Green (green), incubated with polyplexes formed using Cy-3 labelled siRNA (red), and then mounted with DAPI containing mounting solution after fixation. Co-localization of polyplexes and lysosomes are shown as a yellow signal. (A) PEI/siRNA polyplexes, (B) PEI-PEG/siRNA polyplexes, (C) Mannose-PEI-PEG/siRNA polyplexes, (D) PEI-PEG-mannose/siRNA polyplexes, (E) PEI-PEG-mannose/siRNA polyplexes in lower magnification. Images were taken at 2 hours after transfection and all the polyplexes were prepared with 1μM siRNA at N/P ratio 7.
Pegylation and mannosylation of PEI does not reduce knockdown efficiency relative to PEI alone
Dual Luciferase Assay
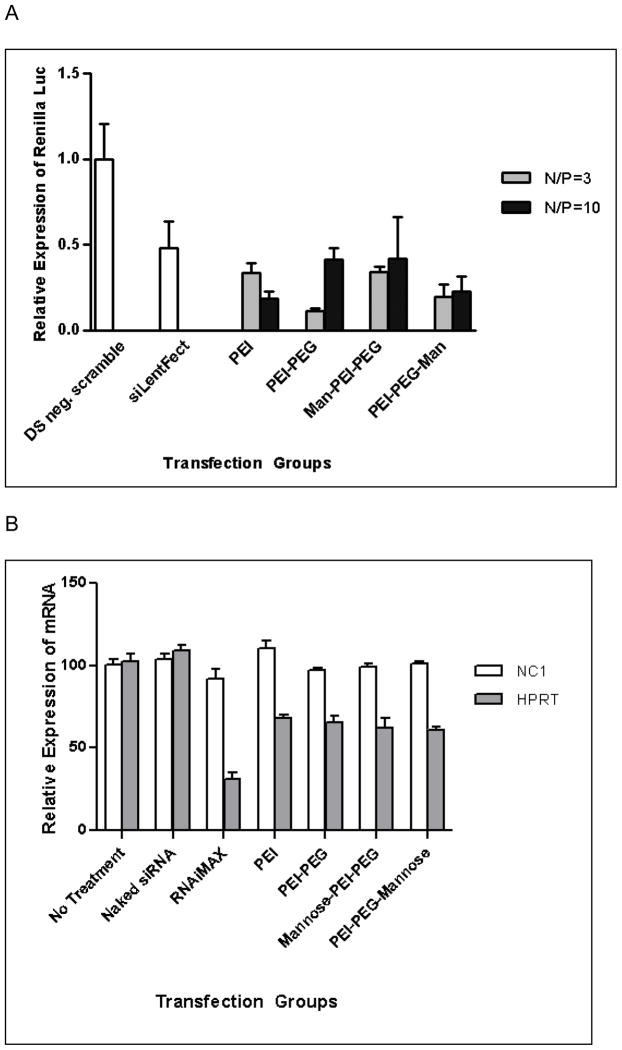
In vitro transfection was performed using the dual-luciferase reporter system to evaluate the siRNA delivery potential of mannosylated pegylated PEI delivery systems. Relative gene expression levels were analyzed after transfecting siRNA only targeting Renilla luciferase mRNA and leaving firefly gene expression as an internal control. All the polymers successfully delivered siRNA and reduced target gene expression (Figure 6A). PEI/siRNA polyplexes inhibited Renilla gene expression to 33.6% at an N/P ratio of 3 and 18.8% at an N/P ratio 10. At an N/P ratio of 3, PEI-PEG showed the strongest knockdown efficiency of 88.7%, which is significantly higher than the other transfection groups including a commercial transfection reagent, siLentFect (51.8%). However, increasing the N/P ratio of PEI-PEG to 10 reduced the knockdown efficiency to 58.6%. There was no statistically significant difference in gene silencing efficiency of PEI-PEG between the N/P ratio of 3 and 10 (P>0.05). Mannose-PEI-PEG/siRNA polyplexes generated 34.2% and 42.0% gene expression when used at N/P ratios 3 and 10, respectively. PEI-PEG-mannose decreased gene expression down to 19.9% at N/P ratio 3 and 22.9% at N/P ratio 10. No significant difference was found between the N/P ratios of 3 and 10 in both mannosylated pegylated PEI’s (P>0.05). Furthermore, the modified polymers showed no significant difference in gene silencing efficiency in comparison to unmodified PEI (P>0.05). Gene silencing efficiency in cells treated with polyplexes prepared at N/P ratios of 5 and 7 was not significantly different to cells treated with polyplexes prepared at N/P ratios of 3 and 10 (data not shown).
Figure 6. Knockdown of luciferase expression and HPRT mRNA expression.
A. Relative gene expression of Renilla luciferase is presented with firefly luciferase used an internal control. From the left, DS scrambled neg with PEI, siLentFect/hRluc siRNA PEI/hRluc siRNA, PEI-PEG/hRluc siRNA, mannose-PEI-PEG/hRluc siRNA, and PEI-PEG-mannose/hRluc siRNA with N/P ratios at 3 and 10. B. Relative expression of HPRT mRNA levels with NCI used as a control. From the left, no treatment, Naked siRNA, RNAiMax/HPRT siRNA, PEI/HPRT siRNA, PEI-PEG/HPRT siRNA, mannose-PEI-PEG/HPRT siRNA, PEI-PEG-mannose/HPRT siRNA.
Real-Time PCR
Endogenous gene knockdown was carried out using HPRT (Hypoxanthine-Guanine Phosphoribosyl Transferase) siRNA transfection. HPRT is a ubiquitously expressed enzyme that is commonly used as a positive control for endogenous gene knockdown experiments. PEI/siRNA polyplexes resulted in a 68.31% gene expression at N/P ratio 10 and PEI-PEG/siRNA resulted in a 65.80% remaining gene expression. The tricomponent polymers, mannose-PEI-PEG and PEI-PEG-mannose, further enhanced gene delivery showing 62.15% and 61.19%, respectively. Whilst pegylation and mannosylation marginally improved knockdown at the mRNA level, none of these differences were significant.
Pegylation of PEI reduces toxicity relative to PEI alone
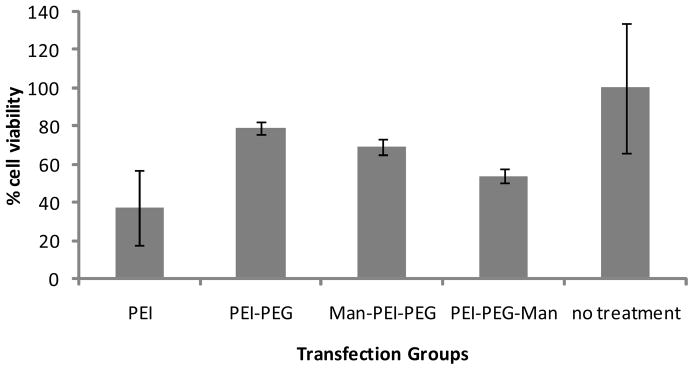
Raw264.7 cells were treated with various groups of polyplexes to evaluate toxicity. At a working concentration of 0.0078mg/ml, PEI showed the highest toxicity resulting in 37.5% cell viability (Figure 7). PEI-PEG resulted in higher cell viability (79.1%) when compared to PEI alone. Mannose-PEI-PEG (68.9%) and PEI-PEG-mannose (53.9%) also resulted in higher cell viabilities than PEI alone. The modified PEI polymers demonstrated lower cytotoxicity relative to unmodified PEI. Cell viabilities were decreased by increasing concentrations of polymers for all the groups tested. However, the reduction in cytotoxicity when using modified polymers in comparison to unmodified PEI increased as the concentration of the polymers incubated with the cells increased (data not shown).
Figure 7. Cytotoxicity.
Cytotoxicity of various polyplexes tested at the working concentration of 0.0078125 mg/ml. From the left; PEI, PEI-PEG, mannose-PEI-PEG, PEI-PEG-mannose, and no treatment group. The relative cell viability was calculated by normalizing to the non-treatment group.
DISCUSSION
In this study, we synthesized, characterized and tested a mannosylated pegylated PEI for potential delivery of siRNA. Pegylation has been previously shown to be of significant value in increasing the circulation time of nanoparticles and polyplexes (Brus et al., 2004; Owens and Peppas, 2006; Yamaoka et al., 1994). Mannose has previously been demonstrated to significantly increase binding of particles to cells that express the mannose receptor (Diebold et al., 1999a; Diebold et al., 1999b; Diebold et al., 2002; Hashimoto et al., 2006; Jiang et al., 2009a; Park et al., 2008b; Zhou et al., 2007a). To our knowledge, this is the first study to evaluate mannosylated pegylated PEI for siRNA delivery and it is the first study to characterize the effect of the location of the mannose ligand in mannosylated pegylated PEI constructs on knockdown efficiency.
PEG and mannose were either both directly conjugated onto the PEI backbone or mannose was conjugated to the PEI via a PEG spacer. 1H NMR spectra’s confirmed that both constructs had PEI, PEG and mannose present and the peaks strongly corresponded to previously reported values (Handwerger and Diamond, 2007; Sagara and Kim, 2002). The signal for mannose at 7 ppm was weak due to the relatively low proportion of mannose in the overall construct composition. For this reason, we used the resorcinol assay to quantify the amount of mannose present in each construct.
The surface chain density of PEG is a critical factor in improving stealth shielding of nanoparticles and polyplexes. The PEI to PEG ratio of PEI-PEG was 3.704, which suggests that every 25kDa PEI chain has 3.45 chains of 2kDa PEG. PEI-PEG-mannose had a 1.23 PEI/PEG ratio indicating 10.16 PEG chains per PEI. The 0.962 PEI/PEG ratio of mannose-PEI-PEG suggests that there are 13.3 PEG chains for each PEI. PEG chains have a larger range of motion at low surface coverage, that can lead to gaps in the PEG protective layer (Storm et al., 1995). For PEG chains to fully cover the surface of PEI/siRNA polyplexes, six short PEGs (5kDa) or one long PEG (20kDa) are needed (Brus et al., 2004; Petersen et al., 2002). Therefore, 3.45–13.3 chains of 2kDa PEG is expected to provide a satisfactory level of pegylation for steric stabilization. The 0.19μmol/mg of mannose in mannose-PEI-PEG and 0.12μmol/mg in PEI-PEG-mannose represent an average modification of 4.7 and 3.0 molecules of mannose per PEI, respectively. This quantity of mannose is expected to be sufficient for selective binding of mannose receptors on cells.
Polymer/siRNA polyplexes exhibited sizes in the range of 169.10nm and 357.33nm. This particle size range is suitable for efficient endocytosis by RAW264.7 cells. Larger particles outside of this range observed by SEM are likely clusters or aggregates of these smaller particles. The zeta potentials of PEI-PEG-Man/siRNA polyplexes showed positive values that were approximately 21.63mV and similar to unmodified PEI and pegylated PEI. This result suggested that PEI-PEG-Man could form stable polyplexes with siRNA. The zeta potential of Man-PEI-PEG/siRNA polyplexes was relatively low compared to PEI-PEG-Man/siRNA polyplexes. This could explain the lower cellular uptake of Man-PEI-PEG observed in our intracellular trafficking studies. The lower zeta potential could lead to weaker interactions with siRNA and the cell surface, which in turn could lead to decreased endocytosis of the polyplexes.
Branched PEI was selected as the backbone for our system because the complexation of branched PEI with siRNA has been reported to exceed that of linear PEI (Breunig et al., 2008). As a result, in gel retardation assays, no reduction in siRNA condensation properties was found with PEGylated PEIs. Overall, complete binding of siRNA was achieved with all the various constructs at N/P ratios of 3 and higher.
We selected RAW264.7 cells for evaluation of polyplex uptake, trafficking and knockdown because the RAW264.7 cells are a murine macrophage cell line that are known to express mannose receptors and that are considered to be typically hard to transfect. Macrophages are also a potential target for our pegylated mannosylated PEI delivery system (Diebold et al., 2002).
In confocal images, the cells endosomes/lysosomes were stained with Lysotracker Green™. Co-localization of the green signals with red signals associated with the Cy-3 labelled siRNA indicated that polyplexes were being internalized by endocytosis, which is consistent with previous reports on uptake by PEI/nucleic acid polyplexes (Lecocq et al., 2000). In addition, separate red signals seen in some cells were most likely due to siRNA that had released from the endosomal compartments. Thirty minutes after incubation, polyplexes had been internalized by cells and localized in cytoplasm. From another cellular uptake study using green fluorescent-labeled (Oregon Green 488) polymers and red fluorescent-labeled (Cy3) siRNA, we observed the two signals were separated in the cytoplasm (data not shown) 2 hours after transfection. This result indicated that siRNA complexes had been released from the lysosomal vesicles as well as from the polymers and were distributed in the cytosol. This release of siRNA is purported to be due to the proton sponge effect, which causes rupture of the endosomes because of the PEI’s strong buffering capacity (Boussif et al., 1995). Intracellular trafficking plays an important role in the fate of siRNA polyplexes because their spatial distribution does not correspond to simple diffusion (Jen and Gewirtz, 2000). Perinuclear localization of siRNA, as seen in Figure 5C and 5E, is required for successful gene silencing by interaction with RISC to induce RNAi. Interactions with RISC dictate siRNA localization even when siRNA is conjugated to cell-binding ligands such as the TAT peptide (Chiu et al., 2004). It suggests that the mannosylated pegylated PEI polymers developed in this study successfully protected siRNA during endocytosis and lysosomal escape in order to integrate siRNA into the RISC complex for correct RNAi processing.
Mannosylation and pegylation of PEI did not reduce gene knockdown efficiency relative to unmodified PEI showing no significant difference in relative gene expression. The advantages of pegylating polyplexes and incorporating cell binding ligands for in vivo applications have been well established (Beyerle et al., 2010; Ogris M, 1999; Owens and Peppas, 2006; Sagara and Kim, 2002; Webster et al., 2007; Yamaoka et al., 1994, 1995). Furthermore, not only did pegylation have no adverse effect on knockdown efficiency, but it also reduced toxicity. This reduction in toxicity was slightly offset by the mannosylation but not significantly so. In addition to observing a reduction in cellular toxicity, we anticipate that pegylation of the polyplexes will also reduce toxicity at the systemic level by reducing aggregation of the polyplexes and therefore the capillary embolism that has been associated with the use of unmodified PEI (Intra and Salem, 2008; Ogris M, 1999).
One aspect of mannosylated pegylated PEI delivery systems that has not been explored before is the importance of the location in which mannose is bound to the construct. In our studies, PEI-PEG-mannose/siRNA polyplexes resulted in higher toxicity and higher knockdown efficiency than mannose-PEI-PEG. Furthermore, qualitatively we observe that PEI-PEG-mannose/siRNA polyplexes have more stable uptake by RAW264.7 cells than mannose-PEI-PEG/siRNA polyplexes. This observation is not clearly understood but could be explained by the structural difference between two constructs. The PEI-PEG-mannose construct has mannose moieties exposed at the tip of PEG chain whereas the mannose in mannose-PEI-PEG could be hindered by the PEG chains. Thus the mannose ligand-receptor interaction could be obstructed by the shielding effect of PEG chains. The use of a PEG chain can impair cell binding by shielding not only PEI but also the ligand (Kunath et al., 2003a). We carried out cellular uptake time-lapse studies at various time points including 0.5, 1, 2, 4, 8, and 24 hours (data not shown). Throughout the course of experiment, we observed that PEGylated PEI had delayed endocytosis relative to PEI alone. The reason that PEG-PEI without mannosylaton at an N:P ratio of 3 still has better luciferase knockdown than either the mannose-PEI-PEG or PEI-PEG-mannose constructs could be attributed to its better cytotoxicity profile or the use of an optimized degree of pegylation for the PEI-PEG construct.
The endogenous knockdown generated by our modified PEIs was significantly less effective than RNAiMax at targeted mRNA reduction. In addition, in our hands, TransIT-TKO can routinely provide 80–90% knockdown, which is significantly more effective than the modified PEIs (data not shown).
The modified PEIs were more efficient at luciferase knockdown than that of another commercial transfection reagent, siLentFect, which was used following the manufacturers guidelines. Moreover, both mannose and PEG have well established attributes for in vivo delivery such as selected cell binding, reduced systemic toxicity and enhanced circulation. In particular, mannose ligands have shown significant potential for binding antigen presenting cells (APCs) such as mouse macrophages and dendritic cells. Furthermore, PEGylated PEI/siRNA complexes have been reported to have decreased random uptake into non-specific organs including liver and spleen compared to unmodified PEI.
Finally, the determination of the optimal location of cell binding ligands in delivery construct in this study is expected to have important implications in the design of several plasmid DNA, oligonucleotide and siRNA delivery systems currently in development that utilize alternative cell binding ligands and alternative cationic backbones such as chitosan.
Figure 1.
PsiCHECK™-2 vector map. It is a specifically designed 6.3kb pDNA for RNAi experiments. It has two different reporter genes, firefly (hluc+) luciferase with HSV-TK promoter and Renilla (hRluc) luciferase with SV40 promoter, and a synthetic poly(A) sequence in between to reduce the potential for recombination events. This firefly reporter sequence has been specifically designed as an intraplasmid transfection normalization reporter, thus the Renilla luciferase signal can be normalized to the firefly luciferase signal.
Acknowledgments
We gratefully acknowledge support from the American Cancer Society (RSG-09-015-01-CDD), the National Cancer Institute at the National Institutes of Health (1R21CA13345-01/1R21CA128414-01A2/UI Mayo Clinic Lymphoma SPORE), and the Pharmaceutical Research and Manufacturers of America (PhRMA) Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas AO, Donovan MD, Salem AK. Formulating poly(lactide-co-glycolide) particles for plasmid DNA delivery. Journal of pharmaceutical sciences. 2008;97:2448–2461. doi: 10.1002/jps.21215. [DOI] [PubMed] [Google Scholar]
- Abe T, Goda K, Futami K, Furuichi Y. Detection of siRNA administered to cells and animals by using a fluorescence intensity distribution analysis polarization system. Nucleic Acids Research. 2009;37 doi: 10.1093/nar/gkp131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner A. Applications of RNA interference: current state and prospects for siRNA-based strategies in vivo. Applied microbiology and biotechnology. 2007;76:9–21. doi: 10.1007/s00253-007-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner A, Fischer D, Merdan T, Brus C, Kissel T, Czubayko F. Delivery of unmodified bioactive ribozymes by an RNA-stabilizing polyethylenimine (LMW-PEI) efficiently down-regulates gene expression. Gene Ther. 2002;9:1700–1707. doi: 10.1038/sj.gt.3301839. [DOI] [PubMed] [Google Scholar]
- Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. The Journal of clinical investigation. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyerle A, Merkel O, Stoeger T, Kissel T. PEGylation affects cytotoxicity and cell-compatibility of poly(ethylene imine) for lung application: Structure-function relationships. Toxicology and Applied Pharmacology. 2010;242:146–154. doi: 10.1016/j.taap.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Biswal BK, Debata NB, Verma RS. Development of a targeted siRNA delivery system using FOL-PEG-PEI conjugate. Molecular Biology Reports. 2010;37:2919–2926. doi: 10.1007/s11033-009-9853-3. [DOI] [PubMed] [Google Scholar]
- Bonsted A, Wagner E, Prasmickaite L, Hogset A, Berg K. Photochemical enhancement of DNA delivery by EGF receptor targeted polyplexes. Methods Mol Biol. 2008;434:171–181. doi: 10.1007/978-1-60327-248-3_11. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130:57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Brus C, Petersen H, Aigner A, Czubayko F, Kissel T. Physicochemical and biological characterization of polyethylenimine-graft-poly(ethylene glycol) block copolymers as a delivery system for oligonucleotides and ribozymes. Bioconjugate chemistry. 2004;15:677–684. doi: 10.1021/bc034160m. [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kursa M, Wagner E, Cotten M, Zenke M. Mannose polyethylenimine conjugates for targeted DNA delivery into dendritic cells. The Journal of biological chemistry. 1999a;274:19087–19094. doi: 10.1074/jbc.274.27.19087. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Lehrmann H, Kursa M, Wagner E, Cotten M, Zenke M. Efficient Gene Delivery into Human Dendritic Cells by Adenovirus Polyethylenimine and Mannose Polyethylenimine Transfection. Human Gene Therapy. 1999b;10:775–786. doi: 10.1089/10430349950018535. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Plank C, Cotten M, Wagner E, Zenke M. Mannose receptor-mediated gene delivery into antigen presenting dendritic cells. Somatic cell and molecular genetics. 2002;27:65–74. doi: 10.1023/a:1022975705406. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- Fishburn CS. The pharmacology of PEGylation: balancing PD with PK to generate novel therapeutics. Journal of pharmaceutical sciences. 2008;97:4167–4183. doi: 10.1002/jps.21278. [DOI] [PubMed] [Google Scholar]
- Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. Journal of Controlled Release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Biodegradable long-circulating polymeric nanospheres. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- Handwerger RG, Diamond SL. Biotinylated Photocleavable Polyethylenimine: Capture and Triggered Release of Nucleic Acids from Solid Supports. Bioconjugate chemistry. 2007;18:717–723. doi: 10.1021/bc060280t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Morimoto M, Saimoto H, Shigemasa Y, Yanagie H, Eriguchi M, Sato T. Gene Transfer by DNA/mannosylated Chitosan Complexes into Mouse Peritoneal Macrophages. Biotechnology Letters. 2006;28:815–821. doi: 10.1007/s10529-006-9006-x. [DOI] [PubMed] [Google Scholar]
- Hobel S, Aigner A. Polyethylenimine (PEI)/siRNA-mediated gene knockdown in vitro and in vivo. Methods Mol Biol. 2010;623:283–297. doi: 10.1007/978-1-60761-588-0_18. [DOI] [PubMed] [Google Scholar]
- Intra J, Salem AK. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo. J Control Release. 2008;130:129–138. doi: 10.1016/j.jconrel.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen KY, Gewirtz AM. Suppression of gene expression by targeted disruption of messenger RNA: available options and current strategies. Stem Cells. 2000;18:307–319. doi: 10.1634/stemcells.18-5-307. [DOI] [PubMed] [Google Scholar]
- Jiang HL, Kim YK, Arote R, Jere D, Quan JS, Yu JH, Choi YJ, Nah JW, Cho MH, Cho CS. Mannosylated chitosan-graft-polyethylenimine as a gene carrier for Raw 264.7 cell targeting. International journal of pharmaceutics. 2009a;375:133–139. doi: 10.1016/j.ijpharm.2009.03.033. [DOI] [PubMed] [Google Scholar]
- Kang JH, Tachibana Y, Kamata W, Mahara A, Harada-Shiba M, Yamaoka T. Liver-targeted siRNA delivery by polyethylenimine (PEI)-pullulan carrier. Bioorganic & Medicinal Chemistry. 2010;18:3946–3950. doi: 10.1016/j.bmc.2010.04.031. [DOI] [PubMed] [Google Scholar]
- Katayose S, Kataoka K. Water-soluble polyion complex associates of DNA and poly(ethylene glycol)-poly(L-lysine) block copolymer. Bioconjugate chemistry. 1997;8:702–707. doi: 10.1021/bc9701306. [DOI] [PubMed] [Google Scholar]
- Kawakami S, Hashida M. Targeted delivery systems of small interfering RNA by systemic administration. Drug metabolism and pharmacokinetics. 2007;22:142–151. doi: 10.2133/dmpk.22.142. [DOI] [PubMed] [Google Scholar]
- Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. LHRH receptor-mediated delivery of siRNA using polyelectrolyte complex micelles self-assembled from siRNA-PEG-LHRH conjugate and PEI. Bioconjugate chemistry. 2008;19:2156–2162. doi: 10.1021/bc800249n. [DOI] [PubMed] [Google Scholar]
- Kim TH, Jin H, Kim HW, Cho MH, Cho CS. Mannosylated chitosan nanoparticle-based cytokine gene therapy suppressed cancer growth in BALB/c mice bearing CT-26 carcinoma cells. Molecular Cancer Therapeutics. 2006;5:1723–1732. doi: 10.1158/1535-7163.MCT-05-0540. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharmaceutical research. 2009;26:657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- Kunath K, Merdan T, Hegener O, Häberlein H, Kissel T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. The Journal of Gene Medicine. 2003a;5:588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- Kunath K, von Harpe A, Fischer D, Kissel T. Galactose-PEI-DNA complexes for targeted gene delivery: degree of substitution affects complex size and transfection efficiency. J Control Release. 2003b;88:159–172. doi: 10.1016/s0168-3659(02)00458-3. [DOI] [PubMed] [Google Scholar]
- Lecocq M, Wattiaux-De Coninck S, Laurent N, Wattiaux R, Jadot M. Uptake and Intracellular Fate of Polyethylenimine in Vivo. Biochemical and Biophysical Research Communications. 2000;278:414–418. doi: 10.1006/bbrc.2000.3809. [DOI] [PubMed] [Google Scholar]
- Leroux JC, De Jaeghere F, Anner B, Doelker E, Gurny R. An investigation on the role of plasma and serum opsonins on the internalization of biodegradable poly(D,L-lactic acid) nanoparticles by human monocytes. Life Sci. 1995;57:695–703. doi: 10.1016/0024-3205(95)00321-v. [DOI] [PubMed] [Google Scholar]
- Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjugate chemistry. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- Martin SE, Caplen NJ. Applications of RNA interference in mammalian systems. Annual review of genomics and human genetics. 2007;8:81–108. doi: 10.1146/annurev.genom.8.080706.092424. [DOI] [PubMed] [Google Scholar]
- Merkel OM, Beyerle A, Librizzi D, Pfestroff A, Behr TM, Sproat B, Barth PJ, Kissel T. Nonviral siRNA Delivery to the Lung: Investigation of PEG‚àíPEI Polyplexes and Their In Vivo Performance. Molecular Pharmaceutics. 2009a;6:1246–1260. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, De Smedt SC, BÈhÈ M, Kissel T. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly(ethylene glycol) under in vivo conditions: Effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. Journal of Controlled Release. 2009b;138:148–159. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Moore NM, Barbour TR, Sakiyama-Elbert SE. Synthesis and Characterization of Four-Arm Poly(ethylene glycol)-Based Gene Delivery Vehicles Coupled to Integrin and DNA-Binding Peptides. Mol Pharm. 2008;5:140–150. doi: 10.1021/mp700072n. [DOI] [PubMed] [Google Scholar]
- Nimesh S, Chandra R. Polyethylenimine nanoparticles as an efficient in vitro siRNA delivery system. Eur J Pharm Biopharm. 2009 doi: 10.1016/j.ejpb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Ogris MBS, Schüller S, Kircheis R, Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Therapy. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. International journal of pharmaceutics. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Pang S, Fiume MZ. Final report on the safety assessment of Ammonium, Potassium, and Sodium Persulfate. International journal of toxicology. 2001;20(Suppl 3):7–21. doi: 10.1080/10915810152630710. [DOI] [PubMed] [Google Scholar]
- Park IY, Kim IY, Yoo MK, Choi YJ, Cho MH, Cho CS. Mannosylated polyethylenimine coupled mesoporous silica nanoparticles for receptor-mediated gene delivery. International journal of pharmaceutics. 2008a;359:280–287. doi: 10.1016/j.ijpharm.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Park IY, Kim IY, Yoo MK, Choi YJ, Cho MH, Cho CS. Mannosylated polyethylenimine coupled mesoporous silica nanoparticles for receptor-mediated gene delivery. International journal of pharmaceutics. 2008b;359:280–287. doi: 10.1016/j.ijpharm.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Patnaik S, Arif M, Pathak A, Singh N, Gupta KC. PEI-alginate nanocomposites: Efficient non-viral vectors for nucleic acids. International journal of pharmaceutics. 2010;385:194–202. doi: 10.1016/j.ijpharm.2009.10.041. [DOI] [PubMed] [Google Scholar]
- Pearce ME, Mai HQ, Lee N, Larsen SC, Salem AK. Silicalite nanoparticles that promote transgene expression. Nanotechnology. 2008;19 doi: 10.1088/0957-4484/19/17/175103. [DOI] [PubMed] [Google Scholar]
- Peracchia MT, Harnisch S, Pinto-Alphandary H, Gulik A, Dedieu JC, Desmaele D, d’Angelo J, Muller RH, Couvreur P. Visualization of in vitro protein-rejecting properties of PEGylated stealth polycyanoacrylate nanoparticles. Biomaterials. 1999;20:1269–1275. doi: 10.1016/s0142-9612(99)00021-6. [DOI] [PubMed] [Google Scholar]
- Peracchia MT, Vauthier C, Passirani C, Couvreur P, Labarre D. Complement consumption by poly(ethylene glycol) in different conformations chemically coupled to poly(isobutyl 2-cyanoacrylate) nanoparticles. Life Sci. 1997;61:749–761. doi: 10.1016/s0024-3205(97)00539-0. [DOI] [PubMed] [Google Scholar]
- Petersen H, Fechner PM, Martin AL, Kunath K, Stolnik S, Roberts CJ, Fischer D, Davies MC, Kissel T. Polyethylenimine-graft-Poly(ethylene glycol) Copolymers: Influence of Copolymer Block Structure on DNA Complexation and Biological Activities as Gene Delivery System. Bioconjugate chemistry. 2002;13:845–854. doi: 10.1021/bc025529v. [DOI] [PubMed] [Google Scholar]
- Sagara K, Kim SW. A new synthesis of galactose-poly(ethylene glycol)-polyethylenimine for gene delivery to hepatocytes. Journal of Controlled Release. 2002;79:271–281. doi: 10.1016/s0168-3659(01)00555-7. [DOI] [PubMed] [Google Scholar]
- Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm G, Belliot SO, Daemen T, Lasic DD. Surface modification of nanoparticles to oppose uptake by the mononuclear phagocyte system. Advanced drug delivery reviews. 1995;17:31–48. [Google Scholar]
- Sun YX, Zeng X, Meng QF, Zhang XZ, Cheng SX, Zhuo RX. The influence of RGD addition on the gene transfer characteristics of disulfide-containing polyethyleneimine/DNA complexes. Biomaterials. 2008;29:4356–4365. doi: 10.1016/j.biomaterials.2008.07.045. [DOI] [PubMed] [Google Scholar]
- Swami A, Kurupati RK, Pathak A, Singh Y, Kumar P, Gupta KC. A unique and highly efficient non-viral DNA/siRNA delivery system based on PEI-bisepoxide nanoparticles. Biochem Biophys Res Commun. 2007;362:835–841. doi: 10.1016/j.bbrc.2007.08.073. [DOI] [PubMed] [Google Scholar]
- Webster R, Didier E, Harris P, Siegel N, Stadler J, Tilbury L, Smith D. PEGylated proteins: evaluation of their safety in the absence of definitive metabolism studies. Drug metabolism and disposition: the biological fate of chemicals. 2007;35:9–16. doi: 10.1124/dmd.106.012419. [DOI] [PubMed] [Google Scholar]
- Yamaoka T, Tabata Y, Ikada Y. Distribution and tissue uptake of poly(ethylene glycol) with different molecular weights after intravenous administration to mice. Journal of pharmaceutical sciences. 1994;83:601–606. doi: 10.1002/jps.2600830432. [DOI] [PubMed] [Google Scholar]
- Yamaoka T, Tabata Y, Ikada Y. Comparison of body distribution of poly(vinyl alcohol) with other water-soluble polymers after intravenous administration. The Journal of pharmacy and pharmacology. 1995;47:479–486. doi: 10.1111/j.2042-7158.1995.tb05835.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Godbey WT. Viral vectors for gene delivery in tissue engineering. Advanced drug delivery reviews. 2006;58:515–534. doi: 10.1016/j.addr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Intra J, Salem AK. Comparative study of poly (lactic-co-glycolic acid)-poly ethyleneimine-plasmid DNA microparticles prepared using double emulsion methods. Journal of Microencapsulation. 2008;25:1–12. doi: 10.1080/02652040701659347. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liu B, Yu X, Zha X, Zhang X, Chen Y, Wang X, Jin Y, Wu Y, Shan Y, Liu J, Kong W, Shen J. Controlled release of PEI/DNA complexes from mannose-bearing chitosan microspheres as a potent delivery system to enhance immune response to HBV DNA vaccine. J Control Release. 2007a;121:200–207. doi: 10.1016/j.jconrel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjugate chemistry. 2008a;19:1448–1455. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]