Abstract
In this study, the reactions of electrons with N-acetyl-proline are investigated by electron spin resonance (ESR) spectroscopy and DFT theory. Electrons are produced by gamma irradiation or by photo-ionization of K4Fe(CN)6 in a neutral 7.5 M LiCl-D2O aqueous glasses at low temperatures with identical results. Electrons are found to add to both the peptide bond and the carboxyl group of the acetyl-proline moiety at 77K. On annealing both the electron adducts undergo fragmentation of the peptide bond between the nitrogen and the alpha carbon of the peptide structure. However the peptide bond electron adduct radical reacts much more rapidly than the carboxyl group electron adduct radical. The DFT calculations predict that the carboxyl adduct is substantially more stable than the peptide bond adduct, with the activation barrier to N-Cα cleavage 3.7 kcal/mol for the amide electron adducts and 23 kcal/mol for the carboxyl adducts in agreement with the relative reactivity found by experiment.
Keywords: electron spin resonance, density functional theory, peptide electron adducts, peptide bond cleavage, cyclic amino acid
1. Introduction
Low energy and solvated electrons are a principle product of water radiolysis induced by high energy radiation (X or γ).1 On addition to DNA and protein structures such electrons cause damage chiefly by bond cleavage reactions.2–4 For example, in peptides electron attachment usually causes fragmentation of the N-terminal amine group or the peptide bond. However, proline which has a cyclic structure is unusual in its behavior: in mass spectrometry experiments carried out for two dipeptides, Gly-Pro and Pro-Gly, no fragmentation was observed for the first case, whereas the characteristic dissociation products of the N-Cα bond were observed for the latter one. This “proline effect” was ascribed to proline ring opening on N-Cα cleavage which still leaves the peptide connected in Gly-Pro but not in Pro-Gly.5
A number of previous studies has investigated radicals formed by the reaction of electrons with amino acids, N-acetyl amino acids and peptides or model systems in a neutral aqueous glasses6–8 and alkaline glasses9–11 and showed that on attachment electrons induce a number of fragmentation reactions in these aqueous systems. Studies of direct gamma irradiation of N-acetyl amino acids in frozen aqueous solutions12 resulted in radicals from both electron and holes in the structure. The electron induced reactions in the frozen ices followed the similar chemistry to those in aqueous glasses.
In current studies we investigate the reactions of electrons with N-acetylproline (N-AcPro) as it is the smallest proline-like structure containing a peptide bond and acts as the simplest model for the proline in a protein backbone (Scheme 1). In an allied work electron attachment to N-AcPro in the gas phase was investigated by photoelectron spectroscopy and while the valence anion and its valence electron affinity was found no evidence for cleavage was observed.13 In this work electron reactions are investigated in aqueous glasses at low temperatures and theoretical DFT calculations of the electron adducts and reaction products were performed for comparison to experiment. Results clearly show that cleavage of the N-Cα bond is the dominant pathway in aqueous media. However, proline’s cyclic nature prevents full cleavage of the protein chain which is an important characteristic. Since proline is an important amino acid in structural components such as cartilage this characteristic of requiring two cleavages to break a protein chain is likely instrumental in maintaining the integrity of these biological structures.
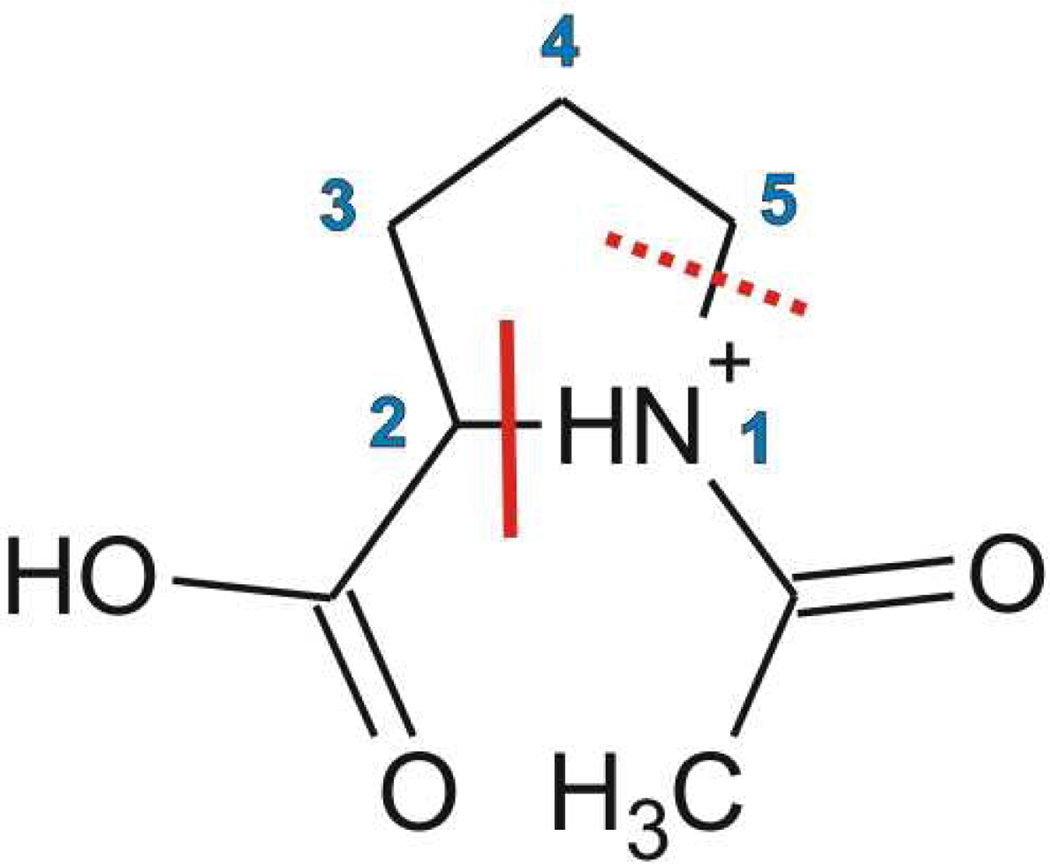
Scheme 1.
The structure of N-acetylproline (N-AcPro) showing the preferred cleavage site at N-Cα(N1-C2) after electron addition which occurs at both the carboxyl and acetyl groups. The second dashed line shows an alternative cleavage site not found in this work.
2. Materials and methods
Experiment
N-AcPro used in these experiments was purchased from Sigma-Aldrich as were the lithium chloride, ≥99% (LiCl) and deuterium oxide, 99.9% (D2O), used to prepare 7.5M LiCl/D2O. The employed gamma (Co-60) irradiator was GR-9 producing 1.0 kGy/hr. The UV source was a low pressure Xe-Hg lamp producing most of its intensity in the 254 nm range. All samples were irradiated at 77K. The ESR (electron spin resonance) spectrometer was E-9 Century Series with dual cavities. Suprasil quartz tubes (4mm in diameter) were used to place the samples in and were purchased from Wilmad Labglass. Potassium ferrocyanide (K4Fe(CN)6•3H2O) was purchased from Mallinckrodt.
Sample Preparation
Approximately 5 milligrams (50mM) of the N-Acetyl-proline•HCl was dissolved in 0.5ml of 7.5M LiCl/D2O. Sample pH was near 2; however, on adjusting to pH 7 and 9 similar results were found. Approximately 2 milligrams (10mM) of potassium ferrocyanide (K4Fe(CN)6) was added for samples intended to be UV radiated.8 After the solution was fully dissolved, it was bubbled with nitrogen gas to remove oxygen from the sample. The sample was then drawn into a 4 mm quartz tube and cooled to 77K in liquid nitrogen to form a clear glass. To eliminate the possibility that the Cl2− formed on gamma irradiation of the aqueous glass was contributing to radical formation in N-AcPro on annealing, in several samples K4Fe(CN)6 was added to the 7M LiCl solution before irradiation. In these samples Cl2− is scavenged on annealing of samples to 150K by one electron oxidation of [Fe(CN)6]4− as shown below.
We found the same results with and without added ferrocyanide which suggests attack by Cl2− did not contribute to the radical production on annealing.
Radical formation
The samples were gamma irradiated with Co-60 source for ca. 60 krads or UV irradiated with 254nm light from a low pressure helical mercury vapor lamp for 1–2 mins. After irradiation, an ESR spectrum was taken to view radicals produced from gamma irradiation or UV radiation. Annealing of the sample at temperatures of 152K–170K was performed to view thermally induced radical reactions. Continued UV irradiation for long exposure times results in photolysis of the radicals and therefore photolysis times were restricted to 2 mins.
Computations
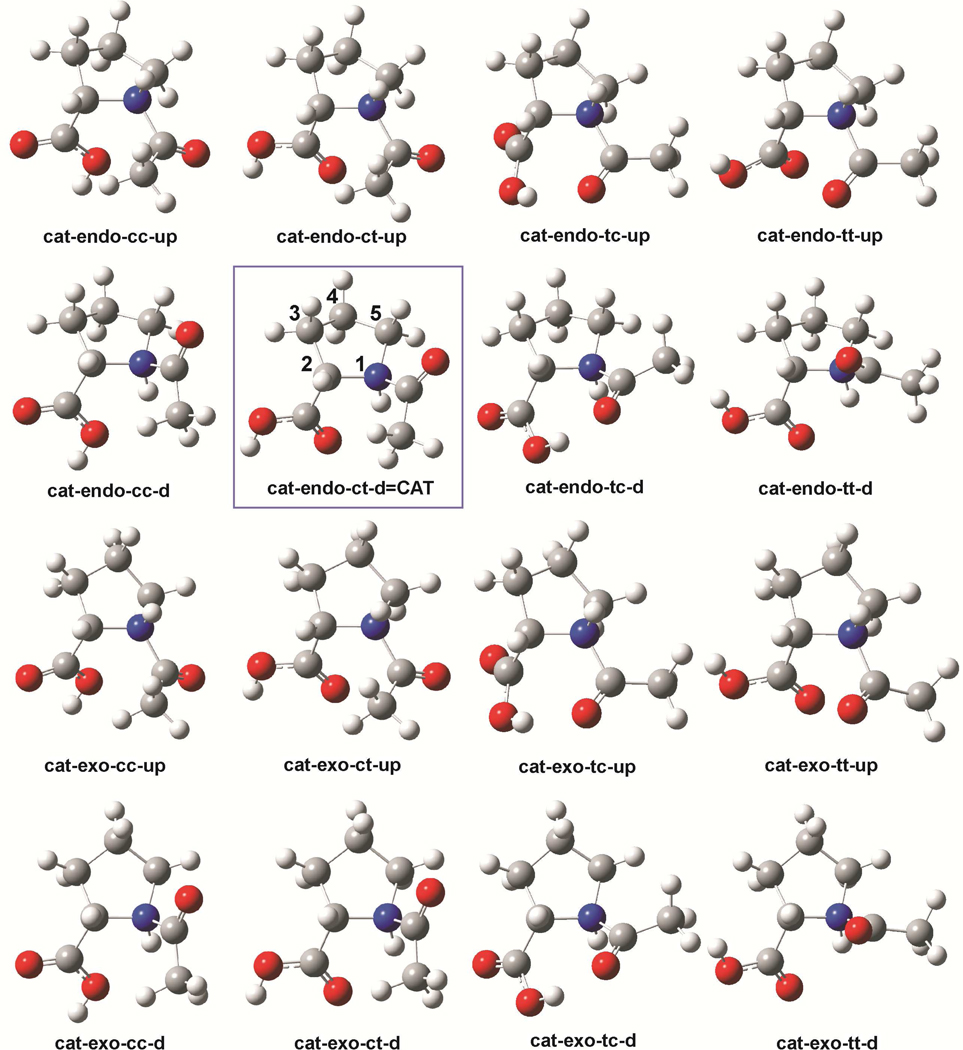
The names of N-AcPro cations are preceded with a prefix “cat”. Since N-AcPro may exist in several conformations, as demonstrated by Aliev et al. in their conformational NMR studies,14–15 “-endo-“ and -exo-” are used to differentiate between the Cγ-endo and Cγ-exo ring conformers, respectively (see Figure 1). Furthermore, the cis and trans rotamers of N-AcPro (related to the acetyl group rotation with respect to the N-AcPro ring) are indicated by a suffix “-c*-“ and ”-t*-“, respectively. Cis and trans position of the carboxyl group is indicated by a suffix "-*c-" or "-*t-", respectively (see Figure 1). Finally, a suffix at the end of the name is related to proton attached in acid conditions, directed upwards (“-up”) or downwards (“-d”) with respect to pyrrolidine’s ring. For instance, "cat-endo-ct-up" stands for a cation conformer in which the pyrolidine ring, the acetyl, the carboxyl group and the acid proton assume Cγ-endo, cis, trans and upwards conformations, respectively.
Figure 1.
Geometries of the N-AcPro (N-acetyl Proline) cationic conformers. The most thermodynamically stable one is framed, and has the pyrrolidine ring numbering scheme.
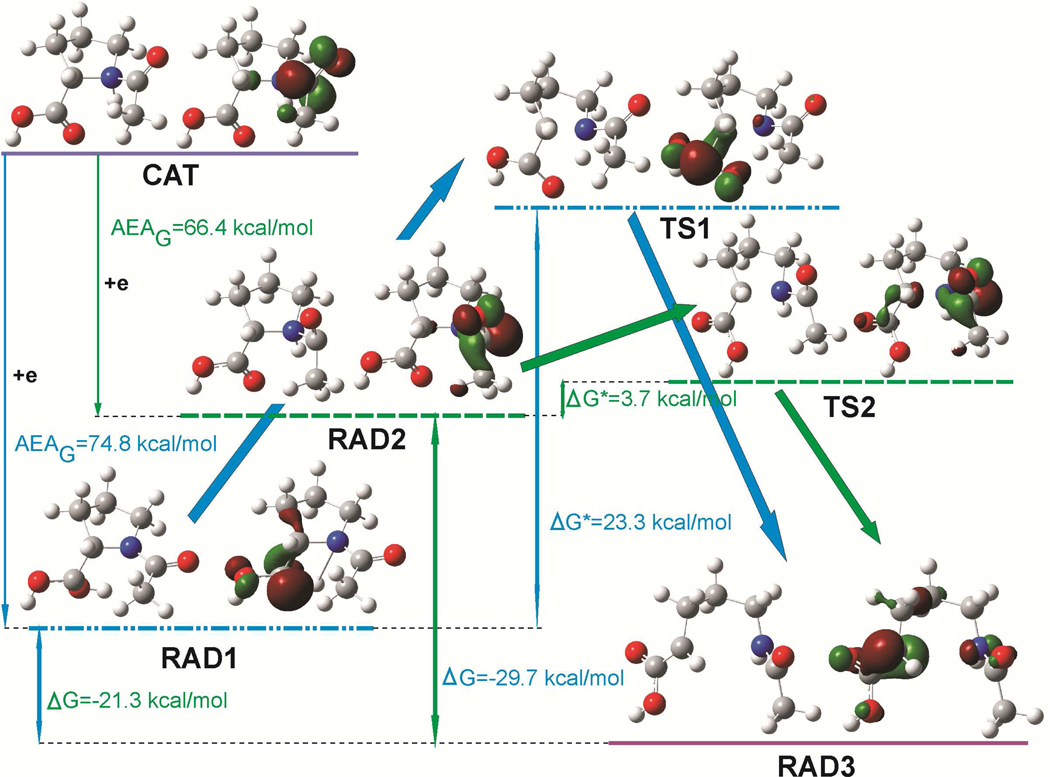
There are two possible radicals, created by electron attachment to the cation: carboxyl type radical, named “RAD1” and acetyl type radical, named “RAD2”. Transition states, leading from these radicals to the broken ring products (“RAD3”), are marked with “TS1” and “TS2”, respectively (see Figure 2).
Figure 2.
Geometries and singly occupied molecular orbitals (SOMO) distribution in stationary points (radical substratum – RAD1 and RAD2, transition states – TS1 and TS2 and product – RAD3) on the reaction paths, concerning the N1-C2 bond splitting in the cationic N-AcPro, after electron attachment to cationic N-AcPro (CAT). ΔG, ΔG* and AEAG denote the free energy of reaction, the activation free energy and adiabatic electron affinity in free energy scale, respectively. SOMO orbitals plotted with a contour value of 0.06 bohr−3/2.
We have applied the density functional theory method with the Becke’s three-parameter hybrid functional (B3LYP)16–18 and the 6-31++G(d,p) basis set.19–20 Additionally, to simulate an aqueous environment, the polarized continuum model (PCM)21 with the UAHF solvation radii22 was employed.
All geometries presented here were fully optimized without any geometrical constraints, and the analysis of harmonic frequencies proved that all of them are either structures at energetic minima (all force constants positive) or first-order saddle points (all but one force constants positive). The relative energies (ΔE) and free energies (ΔG) of cations were defined with respect to the most stable one (cat-endo-ct-d=CAT). Moreover, the difference in Gibbs free energy between the most stable cation entity and its radicals both in their correspondingly fully relaxed structures is denoted by AEAG. The relative energies (ΔE) and free energies (ΔG) as well as activation energies (ΔE*) and free energies (ΔG*) are calculated with respect to RAD1 and RAD2.
All quantum chemical calculations were carried out with the GAUSSIAN0323 code on dual Intel Itanium 2 nodes at the Academic Computer Center in Gdansk (TASK) and the pictures of molecules and orbitals were plotted with the GaussView 5 package.24
3. Results and Discussion
Experiment
Acetyl-DL-Proline under gamma irradiation
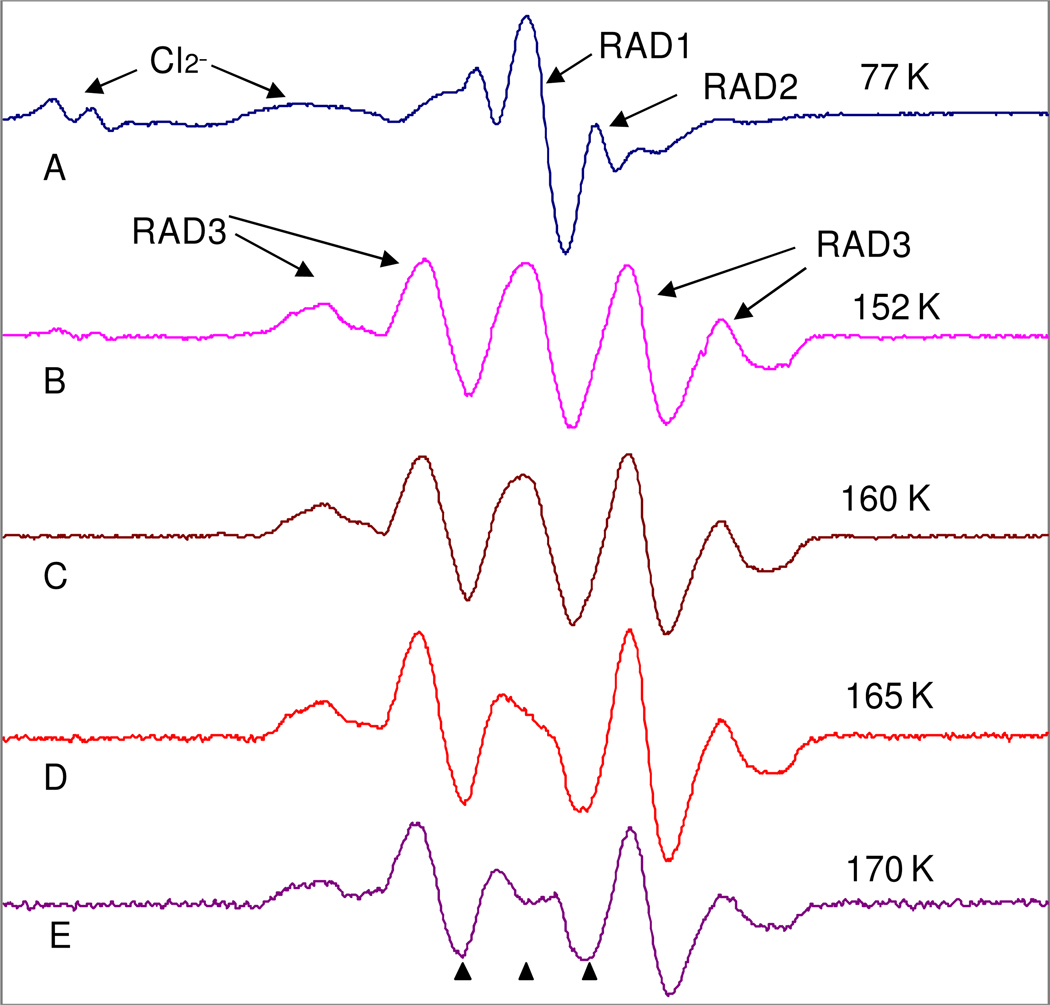
Results of gamma irradiation of N-Acetyl-proline•HCl in 7.5 M LiCl (D2O) at pH 2 is shown in Figure 3. The large central peak is assigned to the carboxyl electron adduct radical (RAD1) and the 24 G doublet is assigned to the peptide bond electron adduct radical (RAD2). In addition we see some initial reaction of the electron adducts to form cleavage product (RAD3, peaks in wings). The low field lines are from Cl2− which do not affect the g=2 region significantly. RAD1 has a beta proton but does not show a significant coupling because the geometry places it in the nodal plane of the carboxyl carbon p-orbital which is the dominant site of the unpaired spin (see Figure 2). RAD2 shows only one coupling from the methyl group because it is in a locked configuration at 77K (see Figure 2 and vide infra). We note that the electron adducts of the peptide bond (pKa =ca. 13)25 and the carboxyl group (pKa=9.5)25 will protonate to form neutral radicals even in neutral solutions. Experiments at pH 7 and 9 gave similar results but increased the formation of RAD2 over RAD1. Annealing of the sample to 152K for fifteen minutes increases the amount of RAD3 with the complete loss of RAD2 and partial loss of RAD1. Subsequent gradual annealing results in the continued loss of RAD1 until at 170K only RAD3 is found (Figure 3E). The last spectrum shows a 5 line pattern (large 44 G doublet split by a triplet of 20 G separation) resulting from 1 beta proton with 44 G coupling and two protons with 20 G coupling, one of which is assigned to the alpha proton at the radical site and the second – to the beta proton coupling on the side chain. In Figure 3E a small sharp peak between the second and third markers at g =2.000 is from a quartz background signal from gamma irradiation of the quartz tube and not a part of the N-AcPro spectrum (Figure 4E in which UV light was employed does not show this peak). The overall fragmentation of the electron adduct is shown in Figure 2. The possible cleavage of the N1-C5(side chain) (for atom numbering see Figure 1) bond to form the •CH2CH2CH2-radical was considered but no evidence for this species was found.
Figure 3.
ESR spectra of N-AcPro in 7.5 M LiCl after 45 minutes of gamma irradiation at 77K. A) Sample at 77K showing electron adducts RAD1 and RAD2. B) Sample after annealing to 152K for 15 minutes which converts RAD1 and RAD2 to RAD3. Gradual annealing from 152 K (B) to 170 K (E) results in further loss of RAD1 and formation of RAD3. After annealing to 170K for 10 minutes the spectrum is entirely due to RAD3. The three markers are separated by 13.09 G each. The middle marker is at g=2.0056. All spectra were recorded at 77K after annealing to temperatures shown in the figure.
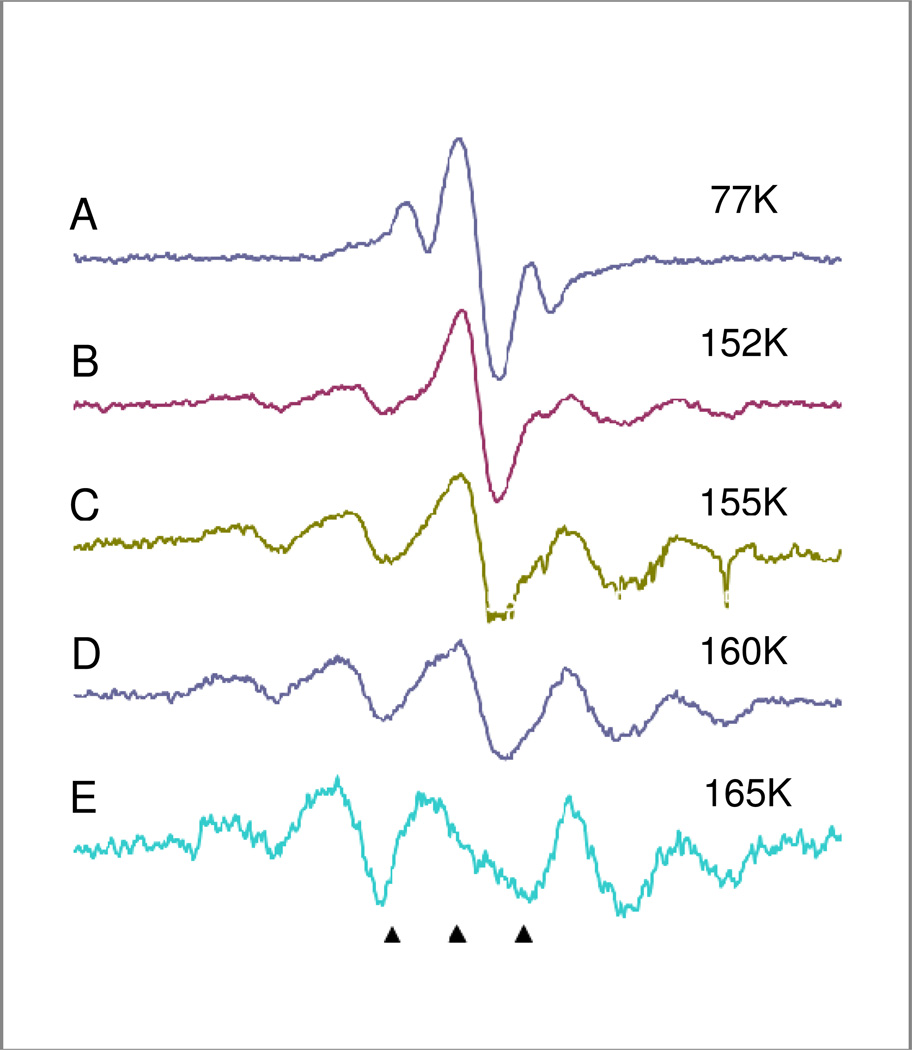
Figure 4.
ESR spectra of N-AcPro in 7.5M LiCl with K4Fe(CN)6 after 1 minute of 254 nm UV photoionization of K4Fe(CN)6 at 77K to produce electrons. A) Sample immediately after UV radiation at 77K; the photoejected electrons added to N-AcPro to form the electron adducts, RAD1( the singlet in the center) and RAD2 (the doublet ). B) Sample annealed to 152K resulting in loss of RAD2 and formation of RAD3. C) Sample annealed to 155K which results in the loss of RAD1 and increased formation of RAD3. D) Sample after annealing to 160K. E) Sample after annealing to 165K at which point all RAD1 has reacted. Some loss of intensity occurs due to radical – radical recombination. All spectra were recorded at 77K after annealing to temperatures shown in the figure.
UV irradiation of N-Acetyl Proline
The comparison of UV radiation and gamma radiation was of interest to eliminate the possible role of Cl2− in radical production. Figure 4 shows spectra of radicals produced by 1 minute of 254 nm UV photolysis of a sample of N-AcPro in 7.5M LiCl with K4Fe(CN)6. The electrons photoejected from K4Fe(CN)6 add to N-AcPro to form the electron adducts. The radicals produced this way were identical to those induced by gamma radiation. The initial radicals (at 77K) were again the carboxyl radical (RAD1) and the peptide radical (RAD2) but little cleavage reaction was found at 77K. As found for gamma irradiation, on progressive annealing both RAD1 (by 152K) and RAD2 (by 165K) are lost with the corresponding appearance of RAD3. The UV irradiated sample contains no Cl2− as it is formed only in gamma irradiated samples.
Molecular mechanism of N-AcPro degradation
Cations
Neutral forms of N-acetylproline (N-AcPro) are easily protonated in acidic conditions, giving the respective cations. These cations can exist in various conformer forms, differing in pyrrolidine ring shape as well as in acetyl and carboxyl groups and proton orientations (see Figure 1). Among the various conformers, the most stable in terms of free energy, ΔG, is one of the endo-type, i.e. cat-endo-ct-d = CAT (see Table 1 and Figure 1). This conformer was, therefore, taken under consideration in the detailed investigations.
Table 1.
Energetic characteristics of the cation conformers of N-AcPro calculated at the B3LYP/6-31++G** level. ΔE and ΔG stand for the relative electronic energy and free energy, respectively, calculated with respect to the cat-endo-ct-d structure (the lowest energy conformer); all values given in kcal/mol.
| acetyl and carboxyl groups conformations |
ring conformation |
|||
|---|---|---|---|---|
| endo | exo | |||
| ΔE | ΔG | ΔE | ΔG | |
| cc-up | 5.36 | 5.45 | 5.50 | 5.75 |
| cc-d | 2.22 | 2.80 | 1.94 | 2.34 |
| ct-up | 3.62 | 3.56 | 3.71 | 3.73 |
| ct-d | 0.00 | 0.00 | −0.13 | 0.15 |
| tc-up | 10.63 | 10.00 | 11.26 | 10.66 |
| tc-d | 7.42 | 6.68 | 7.87 | 7.29 |
| tt-up | 7.54 | 7.63 | 6.86 | 6.65 |
| tt-d | 2.33 | 2.61 | 3.16 | 3.49 |
Reaction path
Two types of radicals (carboxyl – RAD1 and acetyl – RAD2; see Figure 2) can be created due to electron attachment to the cationic N-AcPro. The cationic N-AcPro is a strong electron acceptor: for both the above-mentioned radicals adiabatic electron affinity (AEAG) is extremely high (ca. 70 kcal/mol, see Figure 2). While electron attaches to the carboxyl group it loses its planarity. Moreover, a barrier-free intramolecular proton transfer (BFIPT) between the protonated pyrrolidine nitrogen and carbonyl oxygen of the carboxyl group takes place. Such BFIPT process is not possible for the radical formed due to electron attachment to the acetyl group. Thus, BFIPT accounts for significantly larger AEAG observed for RAD1 (74.7 (RAD1) vs. 66.4 (RAD2) kcal/mol; see Figure 2).
Both radicals are thermodynamically stable, however, they can undergo further stabilization related to the dissociation of the ring C2-N1 bond. These reactions are exergonic for both with ΔG = −21.3 kcal/mol for RAD1 and ΔG = −29.7 kcal/mol for RAD2. Both ring opening reactions are thermally activated and the calculated activation barriers found are 23 kcal/mol for RAD1 and only 4 kcal/mol for RAD2 (see Figure 2). The heights of these barriers are quite different since already small C2-N1 bond stretching leads to the transition state configuration for RAD2 while for RAD1 the C2-N1 bond stretching, proton retransferring and the carboxyl group flattening are involved in the reaction coordinate.
ESR Assignments
The suggested mechanism of electron induced N-AcPro degradation is in good agreement with the results of the ESR experiments described previously (see Figures 3,4). As mentioned, the singlet results from the carboxyl radical (RAD1). The electron is localized on the deformed carboxyl group (see Figure 2) and does not couple with any proton magnetic moments because of the molecule geometry.
In the initial doublet from acetyl peptide radical (RAD2) the electron is localized on peptide bond (see Figure 2). One would expect coupling of the unpaired electron to three protons from the methyl group as well as other couplings from nitrogen and the proton at the nitrogen. However, only one large coupling from one proton from the methyl group is found, because at low temperature rotation of the methyl group is hindered and a single orientation is produced.11,26,27 Moreover, when it comes to proton bonded with N atom, it should be remembered, that D2O was used in experiment and deuterium couplings would be too small to be observed. The doublet from RAD2 has been found previously in experiments with electron adducts of acetyl peptides, acetamide, and even acetate at 77K. All three methyl couplings are found in acetamide and acetate only on warming to 180K where the methyl group rotates.11,26,27 No such warming cycle is possible for N-AcPro as RAD2 is unstable to temperature increase. We find the electron adduct to the peptide group of N-AcPro (RAD2) is formed in lower amounts than the RAD1 carboxyl adduct, likely because RAD1 is significantly more thermodynamically stable than RAD2. Theory shows that RAD2 disappears earlier than RAD1 singlet because its lower thermodynamic stability as well as a significantly lower kinetic barrier to formation of RAD3.
The open-ring product RAD3 shows couplings to 1 alpha proton (ca. 20 G) at C2 and two beta protons (44 G and 20 G) at C3 with different orientations to the radical site. These couplings compare well to previous results with similar molecular structures,8 but not for the optimized DFT calculated hyperfine couplings shown in Table 2. Only a change in dihedral angle, θ=Ccarboxyl-C2-C3-C4 (see Scheme 1) places the two β-protons so that the couplings agree (see Table 2). The energy required for this change is small (ca. 1.9 kcal/mol) and is likely a matrix induced minimum structure.
Table 2.
Theoretical and Experimental Proton Hyperfine Couplings for Proline Radicals.a
| Radicals | DFT Calculated/Experimental Proton Couplings |
|||
|---|---|---|---|---|
| C2-H | C3-H1 | C3-H2 | Acetyl-CH3 | |
| RAD1 | 3.4G/<5G | - | - | |
| RAD2 | - | - | - | 26.1G/24Gb (1H) |
| RAD3 | -19.6G/20G -19.6G/20G |
16.7G/44G 47.4G/44G |
6.6/20Gc 20.5G/20Gd |
- |
Theoretical/Experimental values
Only one coupling from the methyl group is experimentally observed. At 77K the methyl group is locked so to produce one large and two small experimentally not observed couplings.
Equilibrium conformation
Couplings for a non-equilibrium conformation with dihedral Ccarboxyl-C2-C3-C4=23° that gives experimental values.
4. Summary
We find both the carboxyl and peptide electron adduct radicals are formed with the carboxyl adduct favored. Annealing to higher temperatures both the carboxyl and peptide electron adducts react to cleave the peptide link at the N1-C2 bond. However the N1-C5 bond to the side chain link remains intact. The peptide electron adduct reacts far more quickly than the carboxyl adduct and this is supported by theory that shows the carboxyl adduct to be the most stable. The method used to produce electrons was also studied, and the radicals produced with both gamma irradiation and UV photoionization of K4Fe(CN)6, were identical. DFT calculations predict the initial radical identities, their stability, reactivity and coupling constants in good accord with experiment.
Supplementary Material
Acknowledgements
The authors greatly appreciate the aid of the Department of Chemistry for granting the Thompson Undergraduate Research Award (J.K.) and the NIH (RO1 CA 045424) for partial support of this research. This work was also supported by the Polish Ministry of Science and Higher Education (MNiSW) Grant No.: DS/8221-4-0140-1 (J.R.). The calculations were performed at the Academic Computer Center in Gdansk (TASK). Support from the NSF under grant number is CHE-1111693 is also acknowledged (KHB). Professor Amitava Adhikary, Alyson Engle and Deepti Khanduri are acknowledged for their generous aid and helpful discussions.
Footnotes
Supporting Information Available: Cartesian coordinates for the constrained RAD3 geometry This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ershov BG, Gordeev AV. Rad. Phys. Chem. 2008;77:928–935. [Google Scholar]
- 2.Boudaiffa B, Cloutier P, Hunting D, Huels MA, Sanche L. Science. 2000;287:1658–1660. doi: 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- 3.Sanche L. Eur. Phys. J. D. 2005;35:367–390. [Google Scholar]
- 4.Syrstad EA, Tureček F. J. Am. Soc. Mass Spectrom. 2005;16:208–224. doi: 10.1016/j.jasms.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Hayakawa S, Hashimoto M, Matsubara H, Tureček F. J. Am. Chem. Soc. 2007;129:7936–7949. doi: 10.1021/ja0712571. [DOI] [PubMed] [Google Scholar]
- 6.Sevilla MD, Brooks VL. J. Phys. Chem. 1973;77:2954–2959. [Google Scholar]
- 7.Sevilla MD, Failor-Koszykowski R. J. Phys. Chem. 1977;81:1198–1200. [Google Scholar]
- 8.Sevilla MD, D’Arcy JB, Suryanarayana D. J. Phys. Chem. 1978;82:2589–2594. [Google Scholar]
- 9.Sevilla MD. J. Phys. Chem. 1970;74:2096–2102. doi: 10.1021/j100909a008. [DOI] [PubMed] [Google Scholar]
- 10.Sevilla MD. Phys. Chem. 1970;74:3366–3372. doi: 10.1021/j100712a010. [DOI] [PubMed] [Google Scholar]
- 11.Sevilla MD. Phys. Chem. 1970;74:669–672. [Google Scholar]
- 12.a. Sevilla MD, D’Arcy JB, Morehouse KM. J. Phys. Chem. 1979;83:2893–2897. [Google Scholar]; b. Sevilla MD, D’Arcy JB, Morehouse KM. J. Phys. Chem. 1979;83(22):2887–2892. [Google Scholar]
- 13.Chomicz L, Rak J, Paneth P, Sevilla MD, Ko YJ, Wang H, Bowen KH. J. Chem. Phys. 2011;135 doi: 10.1063/1.3625957. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aliev AE, Courtier-Murias D. J. Phys. Chem. B. 2007;111:14034–14042. doi: 10.1021/jp076729c. [DOI] [PubMed] [Google Scholar]
- 15.Aliev AE, Bhandal S, Courtier-Murias D. J. Phys. Chem. A. 2009;113:10858–10865. doi: 10.1021/jp906006w. [DOI] [PubMed] [Google Scholar]
- 16.Becke AD. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 17.Becke AD. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 18.Lee C, Yang W, Parr RG. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 19.Ditchfield R, Hehre WJ, Pople JA. J. Chem. Phys. 1971;54:724–728. [Google Scholar]
- 20.Hehre WJ, Ditchfield R, Pople JA. J. Chem. Phys. 1972;56:2257–2261. [Google Scholar]
- 21.Tomasi J, Mennucci B, Cammi R. Chem. Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 22.Barone V, Cossi M, Tomasi J. J. Chem. Phys. 1997;107:3210–3221. [Google Scholar]
- 23.Frisch MJ, et al. Gaussian 03, revision B.05. Pittsburgh, PA: Gaussian Inc.; 2003. [Google Scholar]
- 24.GaussView 5. Frisch Æ, Hratchian HP, Dennington RD, II, Keith TA, Millam J. Wallingford CT: Gaussian, Inc.; 2009. [Google Scholar]
- 25.von Sonntag C. Free-radical-induced DNA Damage and Its Repair (A Chemical Perspective) Berlin, Heidelberg: Springer-Verlag; 2006. [Google Scholar]
- 26.Suryanarayana D, Sevilla MD. J. Phys. Chem. 1979;83:1323–1327. [Google Scholar]
- 27.Suryanarayana D, Sevilla MD. J. Phys. Chem. 1980;84:3045–3049. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.