Abstract
Objective
Fibromyalgia (FM) represents a complex disorder characterized by widespread pain and tenderness and frequently accompanied by additional somatic and cognitive/affective symptoms. Genetic risk factors are known to contribute to the etiology of the syndrome, but few specific genetic variants have been identified to date and still require replication. In this study, a large scale candidate gene approach was used to examine over 350 genes for association with FM.
Methods
Four hundred ninety-six FM patients were included in the study as cases with a total of 348 chronic pain-free controls. Genotyping was performed using a dedicated gene array chip, the Pain Research Panel, which assays variants characterizing over 350 genes known to be involved in biological pathways relevant to nociception, inflammation, and mood. Association testing was performed using logistic regression.
Results
Significant differences in allele frequencies between cases and controls were observed for three genes: GABRB3 (rs4906902, p = 3.65×10−6), TAAR1 (rs8192619, p = 1.11×10−5) and GBP1 (rs7911, p = 1.06×10−4). These three genes, and seven other genes with suggestive evidence for association, were examined in a second, independent cohort of FM patients and controls genotyped using the Perlegen 600K platform. Evidence of association in the replication cohort was observed for TAAR1, RGS4, CNR1, and GRIA4 genes.
Conclusion
Variation in these genes may serve as a basis for development of new diagnostic approaches, and these genes’ products may contribute to the pathophysiology of FM and represent potential targets for therapeutic action.
Fibromyalgia (FM) is a syndrome characterized by chronic widespread musculoskeletal pain and joint tenderness. Other complaints and symptoms, such as affective distress, hyperalgesia and allodynia, cognitive disturbance, and gastrointestinal disruption frequently accompany the pain, and the diagnosis of FM is often comorbid with other functional syndromes such as irritable bowel syndrome (IBS), temporomandibular joint disease (TMJD), migraine, and vulvar vestibulitis (1, 2). Despite numerous hypotheses, as of yet no single causative pathway has been discovered to explain the etiology of the syndrome. FM is currently considered a complex disorder, with interplay between sensory abnormalities, central nervous system dysfunction, psychological factors, and environmental triggers (1).
Evidence for familial aggregation (3) indicates genetic risk factors contribute to the etiology of FM. Two recent twin studies of chronic widespread pain have estimated the heritability of chronic pain symptoms at 48–54% (4) and 51% (5). Identifying the genes responsible for this substantial genetic contribution to risk should provide a better understanding of the complex mechanisms underlying FM and other chronic pain diseases.
A number of researchers have employed a candidate gene approach to search for FM-associated genes. The majority of such candidates have been genes involved in catecholaminergic or serotonergic neurotransmission, including receptors and transporters for dopamine, serotonin, norepinephrine, and epinephrine, as well as the catabolic enzymes COMT and MAO. Inconsistent results with these candidate genes have led others to investigate nociceptive pathways potentially involved in chronic pain or affective symptoms, including NOS, cytokines, opioids, and glucocorticoids. To date, no large scale candidate gene panel or genome-wide association studies have been reported for an FM cohort.
The present study was undertaken as a more comprehensive approach to identifying genetic markers that are associated with FM, using a two-step discovery and replication design. We first employed a dedicated candidate gene platform, the Pain Research Panel, to look for associations between FM and single nucleotide polymorphisms (SNPs) representing over 350 genes. Almost all of the genes previously investigated in FM have been included on the Pain Research Panel, allowing us to confirm association findings from these studies. A large number of additional genes related to nociception, inflammation, and mood are also assayed by the Panel, providing a means to discover novel mechanisms that have not yet been explored. Several gene loci initially associated in the discovery cohort were then examined in a second sample of FM patients, genotyped using a commercial genome-wide association chip with greater marker density (Perlegen 600K platform), for confirmation of the primary association.
PATIENTS AND METHODS
Cohorts
Cases were 496 FM patients (study protocols Pfizer A0081056 [n=402] and A2571003 [n=94]) taken from two randomized, controlled clinical trials of analgesic compounds. All subjects were Caucasian females at least 18 years of age that met ACR criteria for FM (widespread pain present for >3 months, and tenderness in at least 11 of 18 tender point sites). Subjects had no other inflammatory, rheumatic, or otherwise painful disease beside FM, and were free of severe depression.
A pool of FM-free control subjects was derived from two sources. One hundred sixty-five healthy controls were taken from a cohort designed as a reference population of psychiatric and neurological normal volunteers (Pfizer GEN-0583 protocol), and included only Caucasian females over 18 years of age with no history of chronic pain or depression. An additional control cohort included pain-free subjects (n=224) from a case-control study of TMJD at the University of North Carolina at Chapel Hill (UNC, recruited under NIH grant R01-DE16558 as described previously (6)). The 183 Caucasian female subjects from UNC (not including 41 non-Caucasians that were included for purposes of racial clustering), ranging in age from 18–45, were merged with the GEN subjects as a single control group. All genotyped subjects voluntarily gave a blood sample (9 mL volume, collected in plastic EDTA tubes) and written informed consent for anonymized genetic analysis. Study protocols for all Pfizer and UNC cohorts were approved by their respective Institutional Review Boards/Independent Ethical Committees to be in accordance with the Declaration of Helsinki.
Genotyping
DNA extracted from whole blood was genotyped by Cogenics Inc. (Morrisville, NC, now Beckman-Coulter Genomics) using the Algynomics Pain Research Panel. This platform is a dedicated array for the targeted assessment of genes involved in acute and chronic pain conditions, utilizing the Affymetrix MegAllele platform. The panel genotypes 3295 single nucleotide polymorphisms (SNPs) corresponding to over 350 genes that represent three overlapping domains: (i) genes that mediate the transmission of pain signals by sensory nerve fibers and by CNS neural pathways that mediate the perception of pain; (ii) genes that mediate peripheral and central inflammatory responses to tissue injury or psychological stress; (iii) genes that influence mood and affective states associated with chronic pain conditions.
Genotyping results were returned for 517 FM cases (A0081056 n=415, A2571003 n=102) and 411 control subjects (GEN-0583 n=187, UNC n=224). Genotyping calls were checked for quality and filtered using utilities implemented in PLINK version 1.04 (7). Samples were removed from analysis due to non-Caucasian ancestry (n=38 in the Pfizer cohorts, Supplemental Fig. 1), and a high degree of genetic similarity indicating duplicated samples or cryptic relatedness (n=15). Samples were also removed after applying a filter for overall genotype call rate <95% (n=31). SNPs were filtered based on call rate >95% (n=176); minor allele frequency >1% in either case or control group (n=292); and agreement across 107 repeated samples >99% (n=27).
As the GEN and UNC control groups were genotyped in separate batches 6 months apart, missingness rates were compared before combining them, as a test for SNPs most sensitive to technical bias. SNPs were also checked for agreement with Hardy-Weinberg equilibrium, in cases and controls separately.
The final cleaned dataset included 2760 SNPs in 496 FM cases and 348 controls, with an average call rate of 99.8%. The data cleaning steps are summarized in the Supplementary Materials (Table 1).
Replication cohort
A replication study was used to confirm suggestive results from the primary analysis. Cases for this study were self-reported Caucasians drawn from four additional Pfizer FM clinical trial cohorts, recruited from North America (three studies) and Europe (one study). Controls were drawn from healthy subjects from two Genetic Association Information Network (GAIN) studies: one of psoriasis, recruited in the United States (8); and one of Major Depressive Disorder (MDD), from the Netherlands (9). All genotyping was performed on the genome-wide Perlegen 600K platform by Perlegen Sciences. Samples were removed due to duplication, call rate <90%, or evidence of being an outlier with respect to ethnicity or genotype heterozygosity rates. Filters for low quality SNPs were also applied: call rate <95%; minor allele frequency <2% in any cohort; differential missingness p<10−5; Hardy-Weinberg equilibrium p<10−5; discordance with HapMap minor allele frequencies >33%. Only SNPs (n=543) from the selected replication genes were used for association analysis in this step. The final cleaned dataset included 1004 FM cases and 3725 controls, with an overall genotyping rate of 99.5%.
Association Analyses
The case/control analysis was performed using logistic regression as implemented in PLINK. Case status was considered as a binary dependent variable, with the number of minor alleles as the independent variable in a test of an additive inheritance model. Subject age was introduced as a covariate term in the regression model. Odds ratios (OR) with 95% confidence intervals (95% CI) were calculated in order to describe the effect of the minor allele on disease risk. As a simple Bonferroni correction is overly conservative, we used spectral decomposition (10) to estimate the number of effectively independent SNPs tested by the Pain Research Panel after accounting for the LD between neighboring SNPs, in order to generate a p-value threshold to retain an experiment-wide α=0.05.
The number of SNPs to carry forward into the replication step is an important issue. One common approach is to consider some arbitrary p-value cut-off. However, p-value distribution for associated SNPs depends on the sample size, thus different cut-off values have to be used for studies with different sample sizes. In addition, p-values do not directly reflect the magnitude of an association effect. A different approach for choosing the number of follow-up SNPs is to specify a minimum effect size of interest for true associations. Then, given the sample size of the original study, a number of SNPs with the smallest p-values can be determined to carry forward into the replication study. That number is calculated based on a desired probability for these SNPs to contain one or at least one of several true associations. To this end, the method of Kuo and Zaykin (11) was used to quantify the number of rank-ordered SNPs revealed in our initial case/control study needed to have 80% probability to detect a single true association from among these top results, given an average risk allele frequency of 0.2 and a minimum detectable relative risk (RR) of 1.5.
Eleven genes from the initial case/control association test were tested in the second replication cohort (Table 1). Logistic regression associated fibromyalgia case status with all Perlegen-genotyped SNPs in each gene locus surrounding the observed association from the discovery analysis.
Table 1.
Top association results carried forward to replication phase
| SNP | Gene | Chr | Base-pair Position |
MAF cases |
MAF controls |
P | Odds Ratio |
95% Confidence Interval |
|---|---|---|---|---|---|---|---|---|
| rs4906902 | GABRB3 | 15 | 24570861 | 0.14 | 0.22 | 3.6E-06 | 0.46 | 0.33 – 0.64 |
| rs8192619 | TAAR1 | 6 | 133008041 | 0.08 | 0.03 | 1.1E-05 | 3.76 | 2.08 – 6.78 |
| rs7911 | GBP1 | 1 | 89290708 | 0.41 | 0.32 | 1.1E-04 | 1.66 | 1.28 – 2.14 |
| rs4234955 | NPY1R | 4 | 164479726 | 0.23 | 0.29 | 0.001 | 0.64 | 0.49 – 0.84 |
| rs7537937 | GBP2 | 1 | 89355278 | 0.36 | 0.30 | 0.001 | 1.53 | 1.18 – 1.98 |
| rs10754261 | GBP2 | 1 | 89363670 | 0.36 | 0.30 | 0.001 | 1.53 | 1.18 – 1.98 |
| rs4453447 | GABRB3 | 15 | 24554565 | 0.11 | 0.15 | 0.002 | 0.55 | 0.38 – 0.80 |
| rs694066 | GAL | 11 | 68209561 | 0.06 | 0.09 | 0.002 | 0.47 | 0.30 – 0.76 |
| rs10799897 | RGS4 | 1 | 161309712 | 0.46 | 0.41 | 0.002 | 1.47 | 1.15 – 1.89 |
| rs757228 | GPX4 | 19 | 1052992 | 0.44 | 0.48 | 0.002 | 0.69 | 0.54 – 0.88 |
| rs1329119 | GBP2 | 1 | 89346614 | 0.34 | 0.28 | 0.003 | 1.48 | 1.14 – 1.92 |
| rs4656093 | GBP2 | 1 | 89344563 | 0.34 | 0.28 | 0.004 | 1.47 | 1.14 – 1.91 |
| rs4656095 | GBP2 | 1 | 89351396 | 0.34 | 0.28 | 0.004 | 1.47 | 1.14– 1.91 |
| rs7154455 | ESR2 | 14 | 63806413 | 0.33 | 0.30 | 0.005 | 1.43 | 1.12 – 1.84 |
| rs2302109 | GPX4 | 19 | 1061829 | 0.47 | 0.50 | 0.005 | 0.71 | 0.56 – 0.90 |
| rs1256030 | ESR2 | 14 | 63816923 | 0.45 | 0.49 | 0.007 | 0.73 | 0.59 – 0.92 |
| rs6454674 | CNR1 | 6 | 88929649 | 0.29 | 0.33 | 0.007 | 0.69 | 0.53 – 0.91 |
| rs3783736 | ESR2 | 14 | 63821125 | 0.42 | 0.39 | 0.007 | 1.38 | 1.09 – 1.74 |
| rs1271572 | ESR2 | 14 | 63831670 | 0.45 | 0.49 | 0.007 | 0.73 | 0.58 – 0.92 |
| rs642544 | GRIA4 | 11 | 105242230 | 0.38 | 0.43 | 0.009 | 0.72 | 0.57 – 0.92 |
Association results for top 20 ranked SNPs from the primary case/control study used to select genes for replication (SNPs that failed quality checks and ancestry-informative markers have been removed).
Abbreviations: Chr (chromosome), MAF cases (minor allele frequency in cases), MAF controls (minor allele frequency in controls).
Due to the limited overlap between SNPs genotyped by the Pain Research Panel and the GAIN Perlegen platform, we used genotypes from the HapMap reference population to impute additional SNPs at the replication gene loci. A set of HapMap SNP genotypes (Phase 3, release 1, build 36) filtered to include only SNPs with MAF >0.01 and genotyping rate >0.95 was retrieved from the HapMap website in PLINK.ped/.map format. Chi-square statistical tests using the “–proxy-assoc” command in PLINK assessed association between imputed SNPs in the clinical trial cohort and fibromyalgia status. Results were regarded as reliable only if the imputation algorithm had sufficient information to make a valid call; i.e., if the information content metric INFO> 0.80. The Pain Panel datasets were not imputed.
RESULTS
Evaluation of potential bias
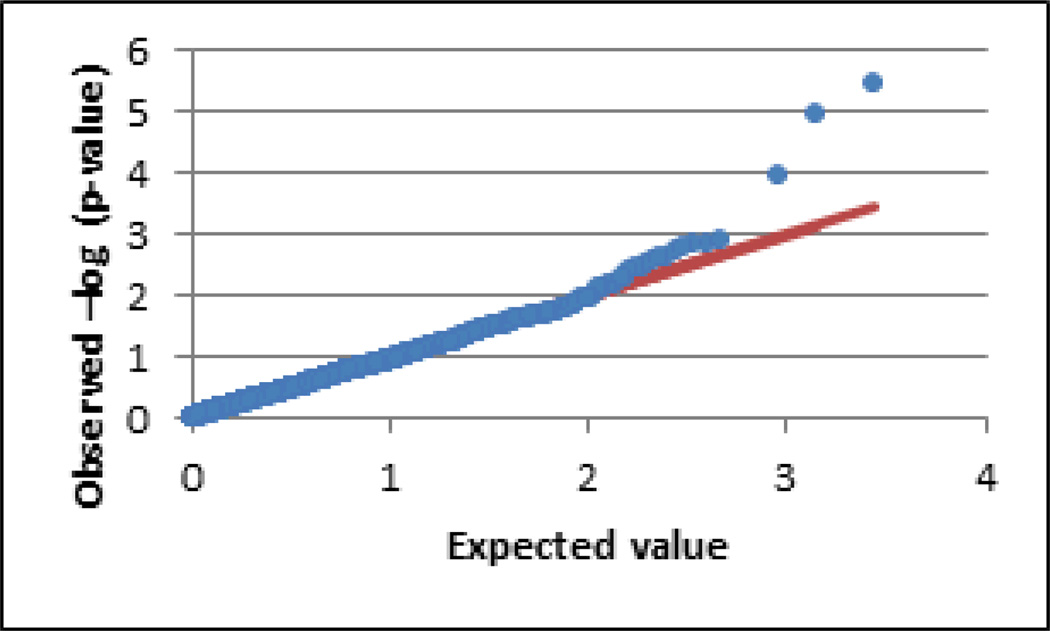
The overall genotyping rate in the final dataset for the primary case/control study using the Pain Research Panel was 99.8%, indicating a very low probability that differential missingness rates in cases and controls might influence group allele frequencies. All of the subjects in this study were Caucasian by self-report and by genetic clustering (see Supplemental Figure 1), but the potential remains for bias due to subtle differences in ethnic makeup between case and control populations. Examination of the distribution of test statistics, by Q-Q plot (Fig. 1) and by the genomic inflation factor statistic lambda, suggested no systematic deviation from their expected distribution under the null that would indicate population stratification. The quality control metrics of all SNPs with p-values <0.01, including HWE p-values and missingness rates, showed no indication that these associations are due to technical errors in sample quality or genotype calling accuracy (Table 1).
Figure 1.
Q-Q plot of FM case/control test. The observed (−log10) p-value distribution (blue dots) is plotted against the expected distribution under the null hypothesis (red line). Systematic elevation from chance over the range of expected values indicates underlying population structure that can bias associations. The genomic inflation factor statistic lambda suggested no such inflation (λ = 0.91).
Primary case/control associations
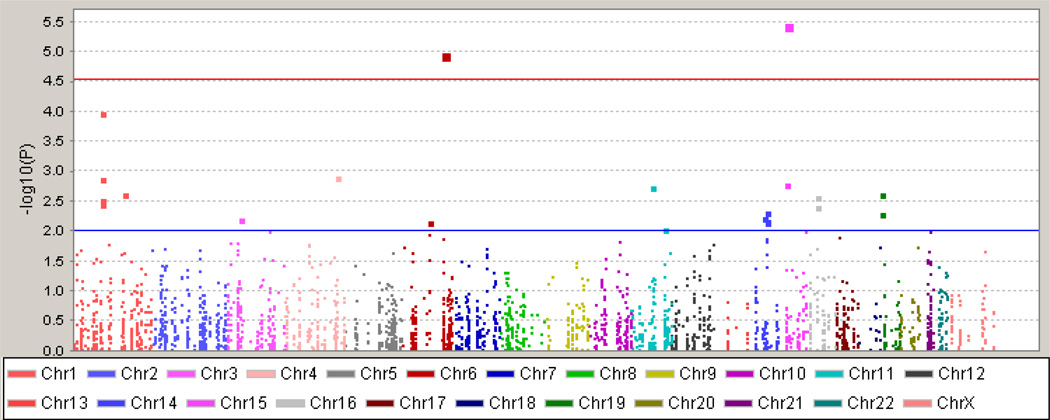
Results for the regression test contrasting FM cases against the control sample are plotted in Fig. 2. The threshold for significance, adjusted for the number of SNPs tested and linkage disequilibrium between them, was set to p<2.8×10−5. We observed two significant SNPs. SNP rs4906902 is in the promoter region of the GABA-A β receptor gene, GABRB3, with the minor allele of this SNP (−897 T>C) displaying a protective effect against FM in our cohort (p=3.65×10−6, OR=0.46, 95% CI 0.33–0.64). This SNP is situated in a highly evolutionarily conserved region (see USCS Human Genome Browser at http://genome.ucsc.edu/), and the variant alters the binding sequence of the transcription factor N-Oct-3, putatively leading to reduced expression of GABRB3. Mice with a deletion of Gabrb3 display thermal hyperalgesia, tactile allodynia, and attenuated analgesic responses to GABA receptor agonists (12).
Figure 2.
Manhattan plot of FM case/control test. The observed (−log10) p-values of all tested SNPs are plotted against their chromosomal position. The red line shows the significance threshold to retain an experiment-wide α = 0.05, adjusting for the estimated number of independent SNPs. The blue line represents a nominal significance value of p = 0.01, the threshold corresponding to the top 25 rank-ordered SNPs revealed in our initial case/control study needed to have 80% probability to detect a single true association from among these top results considering an average risk allele frequency of 0.2 and an estimated relative risk RR = 1.5. This threshold was then used to select genes to carry forward to the replication step.
The second significant association was observed at the SNP rs8192619, in the TAAR1 (trace amine-associated receptor 1) gene that results in a synonymous amino acid substitution (Cys265Cys) in the single exon of the TAAR1 locus (p=1.11×10−5, OR=3.8, 95% CI 2.1–6.8). Although this SNP was not assessed in the replication study, a number of nearby genotyped and imputed SNPs showed strong association effects in the replication cohort, suggesting that rs8192619 tags a haplotype associated with increased risk in both cohorts, confirming our initial finding. The TAAR family of receptors binds a range of biogenic amines comprising <1% of endogenous amines in most brain areas with generally uncharacterized effects, although it has been proposed that they modulate dopaminergic activity (13).
An alternative method for determining significance identifies SNPs that are elevated above the chance “expected” line of the p-values in a Q-Q plot. Fig. 1 shows that while the distribution of tested SNPs followed the expected line, three SNPs clearly deviated from it, also including rs7911, in the 3’-untranslated region of the GBP1 guanylate binding protein 1 (p=1.06×10−4, OR=1.7, 95% CI 1.3–2.1), with the minor C allele (T918C) increasing risk for FM. This gene is induced by interferon and other cytokines, and may contribute to inflammatory diseases including psoriasis (14) and inflammatory bowel disease (IBD) (15). No pain phenotypes arising from genetic variation in GBP1 have been previously observed.
Next, to identify SNPs to carry forward into the replication step we used the method of Kuo and Zaykin (11). This method estimates the number of top SNPs to contain a true association with a desired probability. We calculated that a true association with RR≥1.5 would be found among the top 25 SNPs, ranked by p-value, with 80% probability. These 25 SNPs corresponded to a threshold of p≤0.01 in our results, implicating 8 additional genes to test in the replication experiment (after removing 5 SNPs that failed quality checks or were ancestry-informative markers): NPY1R (neuropeptide Y receptor 1); GBP2 (although this may simply reflect the signal from the nearby GBP1, due to linkage disequilibrium); CNR1 (cannabinoid receptor 1); GRIA4 (glutamate receptor, ionotrophic, AMPA 4); GAL (galanin); RGS4 (regulator of G-protein signaling 4); GPX4 (glutathione peroxidase 4 [phospholipid hydroperoxidase]); and ESR2 (estrogen receptor 2 [beta]).
Replication Study
The 11 genes corresponding to the top 25 ranked SNPs were then examined in the second cohort, using densely situated genotypes from the Perlegen platform, with the exception of GPX4 for which data was not available. Where Perlegen data were missing, we attempted to impute SNPs based on HapMap haplotype frequencies. P-values from the two cohorts were compared, taking into consideration direction of effect and strand orientation, to confirm previously observed associations. The distribution of the p-values from the genes deviated substantially from that expected under the null hypothesis, which is likely explained by non-random selection of the gene loci and enrichment for loci contributing to FM (Supp. Fig. 3).
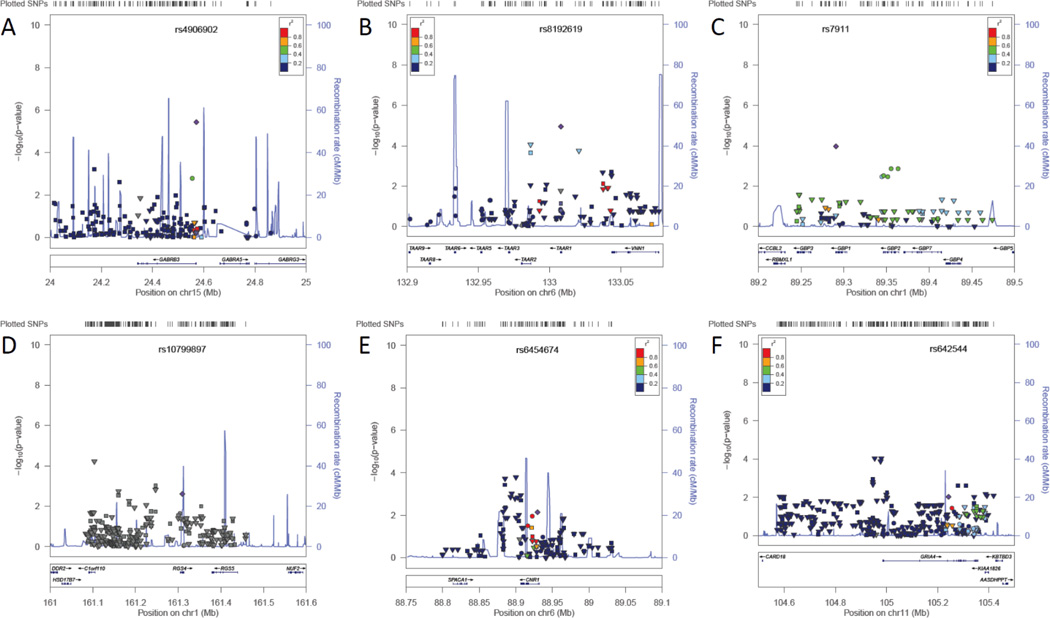
The strongest association in the original case/control study was SNP rs4906902, in the GABRB3 gene (Fig. 3A). This SNP was not significantly associated with FM status in the replication cohort (p=0.49). No other nearby SNPs demonstrated an association signal in this cohort.
Figure 3.
Annotated results in genomic context for top associated genes in the discovery step (A, B, C) and additional replication supported genes (D, E, F). In the upper section of each figure, transformed −log10(p-values) for each SNP are graphed on the y-axis against their genomic location (scale in Mb) on the x-axis. Association results from the discovery study are indicated by circles; results from genotyped SNPs in the replication study are indicated by squares, and well-imputed SNPs in this study are indicated by triangles. The lower section of each figure shows gene loci aligned to the same coordinates. The top associated SNP from the discovery phase is labeled and indicated by a purple diamond; the color of the other SNP symbols indicates LD with this SNP (information from the HapMap CEU population), in r2 values as indicated by the legend. A. GABRB3, B. TAAR1, C. GBP1, D. RGS4, E. CNR1, F. GRIA4.
The second-ranked SNP, rs8192619 in the TAAR1 gene (Fig. 3B), was not genotyped in the replication set, and could not be imputed since it was not included in the HapMap dataset. However, a number of flanking SNPs were strongly associated with fibromyalgia in the replication cohort, including rs4129256 (p=2.20×10−4, OR=1.3 or with missing values imputed p=8.63×10−5, OR=1.3), located in an intron of the neighboring TAAR2 gene, and the imputed SNP rs2745428 (p=1.74×10−4, OR=1.3, INFO=0.95).
The strongest associated SNP in the GBP1 gene, rs7911, was not genotyped in the replication cohort and was only poorly imputed, with a p-value that barely exceeded significance (uncorrected one-tailed test, p=0.03, OR=1.1, INFO=0.78, Fig. 3C). A second region of suggestive association in the GBP1 gene was located about 55kb downstream, with p-values between 1.3–3.5×10−3. This region showed no similar evidence for association in the replication cohort.
Several other genes showed evidence for association only in the replication cohort at levels greater than expected by chance. Genes containing SNPs that exceeded a corrected p-value threshold of p≤1.63×10−4 (for 306 independent SNPs tested in the replication study) included RGS4, CNR1, and GRIA4. The other genes examined in the replication step, including GBP2, NPY1R, GAL, and ESR2, did not confirm the original SNP associations, and showed weak to no association at other SNPs.
Located in an intron of the RGS4 (regulator of G-protein signaling 4, Fig. 3D) gene, the A158G SNP was suggestively associated with FM (rs10799897, p=2.4×10−3, OR=1.47, 95% CI=1.1–1.9), but was neither genotyped nor imputed in the replication cohort. Two other SNPs showing association in the replication cohort (rs2842003, p=9.6×10−4, OR=1.2; SNP rs2805050, imputed p=6.0×10−5, OR=3.4, INFO=1.18) likely represent distinct risk markers rather than replication, since they are not in LD with rs10799897.
The CB-1 cannabinoid receptor gene CNR1 (Fig. 3E) had one SNP that qualified for the replication study, an intronic A>C substitution (rs6454674, p=7.2×10−3, OR=0.7, 95% CI 0.5–0.9). This SNP was not genotyped in the replication cohort and was not found in the HapMap dataset used for imputation. However, several tightly linked promoter region SNPs (about 25 kb upstream) showed strong associations with FM, suggesting that a haplotype near the original SNP harbors the true risk polymorphism. These SNPs included a G>A polymorphism (rs1078602, p=3.7×10−4, OR=0.8) and an imputed A>G substitution (rs10485171, p=1.6×10−4, OR=0.8, INFO=0.90), although the functional consequences of these variants are not known.
The lowest p-value SNP in the GRIA4 gene (Fig. 3F), which codes for the AMPA ionotropic glutamate receptor 4 subunit, was a protective intronic T4086G change (rs642544, p=9.3×10−3, OR=0.72, 95% CI=0.57–0.92). This SNP was not genotyped in the replication dataset, and was only poorly imputed (p=0.14). However, a group of genotyped SNPs in LD with rs643544 (D’=1.0, r2=0.67) but with lower MAF showed even stronger evidence for association, including rs17104711 (p=2.4×10−3, OR=1.3). It should be noted that the direction of effect for the minor allele for these SNPs in the replication cohort is reversed compared to rs642544. Another region upstream from the GRIA4 locus not in LD with the original associated SNP was also observed to show strong evidence for association with FM, including the genotyped SNP rs2510177 (p=1.1×10−3, OR=1.3), and two imputed SNPs in complete LD with each other, rs10895837 and rs10502058 (p=9.2×10−5, OR=1.3, INFO=0.98). Given the complex LD structure of this gene and the lack of overlapping SNPs in both cohorts, it is difficult to decode an association pattern and contribution of each SNP without further dense genotyping of this locus; however, it seems that genetic variants of GRIA4 contribute to FM.
Multiple SNPs in each gene were tested in the replication sample. Therefore, the largest odds ratio estimates obtained for each gene are not unbiased. A method by Ghosh et al. (16) was used to obtain bias reduced estimates of odds ratios. The corrected odds ratio values were 2.62 (rs2805050, RGS4); 0.95 (rs10485171, CNR1); 1.12 (rs4129256, TAAR1) and 1.11 (rs10895837, GRIA4). Corrected odds ratios for other genes did not significantly differ from one.
Previously associated FM genes
Several genes have been investigated for polymorphisms that increase risk for FM and chronic widespread pain, with a number of implicated genetic variants. The Pain Research Panel assesses many of these genes, often with more extensive coverage than the original candidate gene studies. For the majority of these candidates, no evidence of association with FM was detected in our cohort.
Serotonergic genes have been extensively investigated for risk variants for FM. The long allele of a 44-base pair insertion/deletion in the promoter region of the 5-HTT transporter gene SLC6A4 has been associated with FM in German (17) as well as Israeli cohorts, but not in Turkish subjects (18), nor was it associated with FM pain severity in a recent report of Canadian patients (19). Although the Pain Research Panel does not include this variant, no SNPs from this gene exceeded a nominal significance threshold of p<0.01. FM and pain severity have also been associated with the silent T102C (rs6313) SNP of the serotonin 2A receptor gene HTR2A (20). The Pain Research Panel SNP rs4941573 is a useful tag for rs6313 (tight LD r2=1.0), and no association was observed for this marker in our cohort (p=0.86).
Genes relevant to catecholaminergic neurotransmission have also been examined in FM, including the dopamine transporter gene SLC6A3 (21) and the dopamine D4 receptor DRD4 (22); or with painful symptoms in the D3 dopamine receptor DRD3 (23), and the β2- and α1A-adrenergic receptor genes ADRB2 and ADRA1A (24). We observed no evidence for association with these genes in our cohort, although no DRD4 SNPs passed genotype quality inspection. Other studies have investigated the role of MAO and COMT, which metabolize catecholamines and influence the bioavailability of these neurotransmitters. Although variants of the two MAO genes MAOA and MAOB have not been implicated in FM risk (25, 26), the low-activity variant of the COMT Val158Met (rs4680) polymorphism occurs more often in FM patients in some studies (27–29), but not in others (30, 31). In our FM cohort, we observed no association with a tag SNP proxy (rs4633) for the Val158Met polymorphism (p=0.54), nor the functional haplotypes of COMT (32).
DISCUSSION
A large scale candidate gene approach was used to evaluate over 350 genes known to be involved in nociception, inflammation, and affect. While not an exhaustive survey of all pathways potentially involved in the pathophysiology of FM, the study provided a comprehensive and in-depth evaluation of the majority of genes currently known to influence persistent pain conditions. Three unsuspected genes, GABRB3, TAAR1, and GBP1, harbored SNPs that differed in frequency between FM patients and healthy controls; TAAR1 displayed evidence for association in a second cohort. The available meager evidence suggests that receptors for trace amines might influence the development of chronic pain by modulating activity of dopaminergic receptors (33). TAAR-induced alterations in dopamine bioavailability and function may increase pain sensitivity, a hallmark of FM (34). This receptor presents an interesting target for the development of novel TAAR-selective compounds with analgesic properties (35).
Three additional genes—RGS4, CNR1, and GRIA4—were suggestively implicated in two different cohorts, indicating convergent evidence for their involvement in FM susceptibility. Like TAAR1, two of these genes play important roles in the modulation of analgesic pathways.
RGS4, expressed in CNS regions such as the locus coeruleus (LC), the bed nuclei of the stria terminalis, and the dorsal horn of the spinal cord (36), negatively regulates G protein signaling and may therefore play a modulatory role in descending inhibition of pain perception. Within the noradrenergic nucleus of the LC, overexpression of RGS4 down-regulates μ-opioid receptor function (37), and in the spinal cord RGS4 is upregulated up to 230% in a rat model of neuropathic pain, resulting in a substantial attenuation of morphine analgesia (38).
The analgesic properties of the CB-1 cannabinoid receptor encoded by CNR1 have been well-documented, and the cannabinoid agonist nabilone has demonstrated improvements in pain and life quality in patients with FM (39). However, the question remains whether endocannabinoid activity contributes to the etiology of FM. Variants within the CNR1 locus have been associated with other painful chronic conditions, such as irritable bowel syndrome (IBS) (40), migraine (41) and post-traumatic stress disorder (PTSD) (42). FM patients have an increase in circulating anandamide, an endocannabinoid (43), which may be a marker of a “clinical endocannabinoid deficiency,” hypothesized to underlie idiopathic pain conditions such as FM (44). It also raises the possibility of the existence of a specific clinical cluster of FM patients with deficiency in the cannabinoid system that could be identified by a genetic test and would benefit from drugs like nabilone.
In contrast to the effects of TAAR1, RGS4, and CNR1 genetic variants on analgesic mechanisms, the association of GRIA4 with FM might be due to production of central sensitization, a hallmark of FM and related conditions (45). GRIA4 encodes the AMPA-sensitive, ionotropic glutamate receptor subunit GluR4, which mediates fast excitatory transmission of nociceptive signals in the CNS. Little is known about the specific contribution of this receptor subunit to the development of hypersensitivity, but AMPA receptors in the brain and spinal cord are integral to synaptic plasticity and the development of persistent pain (46). AMPA receptors in analgesia-related brain regions, such as the rostral ventromedial medulla (RVM), are also involved in descending modulation of pain perception (47). Spinal AMPA receptors have also been implicated in the production of visceral hyperalgesia (48). Collectively, these observations suggest that alterations in AMPA receptors are likely to contribute to the complex signs, symptoms, and comorbidities associated with FM.
FM patients have been classified by patterns of symptom presentation (49, 50); it may be useful to incorporate genotypic information into such clustering exercises to account for inherited susceptibility to certain intermediate phenotypes, and to associate specific risk factors with clinical outcomes. By implicating a set of genes known to influence nociceptive and analgesic pathways, this study provides evidence for multiple biological mechanisms underlying the pathophysiology of FM, and suggests novel avenues of investigation into the roles that these systems play in the development of persistent pain conditions. Dysfunction within descending inhibitory circuitry, originating from deficiencies in GABAergic, aminergic, or cannabinoid signaling, may lead to an insufficient analgesic response that predisposes certain individuals to chronic pain following trauma or stress. Others might be predisposed to central sensitization due to hyperexcitability of glutamatergic receptors or an increased response to inflammatory cytokines. Future research should consider the heterogeneity of FM and the diversity of instigators of persistent pain, which will increase the power of genetic studies to identify risk determinants, and will improve the prospects for an individualized approach to treating FM and other chronic pain diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Pfizer and Algynomics, and by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (DZ). An NIH/NIDRC supported grant (RO1-DE16558 [LD, WM]) was used to collect a portion of the data used in the study.
Footnotes
Conflict of Interest Statement: This study was conceived jointly by Pfizer, Inc., and Algynomics, Inc., and funded by Pfizer. Genotyping and analysis was directed by Algynomics, which holds patent claims on diagnostic use of genes investigated in the study. Authors are employees and/or stock holders of their respective institutions.
REFERENCES
- 1.Diatchenko L, Nackley A, Slade G, Fillingim R, Maixner W. Idiopathic pain disorders--pathways of vulnerability. Pain. 2006;123(3):226–230. doi: 10.1016/j.pain.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 2.Williams DA, Clauw DJ. Understanding fibromyalgia: Lessons from the broader pain research community. J Pain. 2009;10(8):777–791. doi: 10.1016/j.jpain.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold LM, Hudson JI, Hess EV, Ware AE, Fritz DA, Auchenbach MB, et al. Family study of fibromyalgia. Arthritis Rheumatism. 2004;50(3):944–952. doi: 10.1002/art.20042. [DOI] [PubMed] [Google Scholar]
- 4.Kato K, Sullivan PF, Evengård B, Pedersen NL. Importance of genetic influences on chronic widespread pain. Arthritis Rheum. 2006;54(5):1682–1686. doi: 10.1002/art.21798. [DOI] [PubMed] [Google Scholar]
- 5.Markkula R, Järvinen P, Leino-Arjas P, Koskenvuo M, Kalso E, Kaprio J. Clustering of symptoms associated with fibromyalgia in a Finnish Twin Cohort. Eur J Pain. 2009;13(7):744–750. doi: 10.1016/j.ejpain.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Tchivileva IE, Lim PF, Smith SB, Slade GD, Diatchenko L, McLean SA, et al. Effect of catechol-O-methyltransferase polymorphism on response to propranolol therapy in chronic musculoskeletal pain: a randomized, double-blind, placebo-controlled, crossover pilot study. Pharmacogenet Genomics. 2010;20(4):239–248. doi: 10.1097/FPC.0b013e328337f9ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-[kappa]B pathways. Nat Genet. 2009;41(2):199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry. 2010;16:516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- 10.Nyholt D. A simple correction for multiple testing for SNPs in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo C-L, Zaykin DV. Novel rank-based approaches for discovery and replication in genome wide association studies. Genetics. 2011 doi: 10.1534/genetics.111.130542. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ugarte SD, Homanics GE, Firestone LL, Hammond DL. Sensory thresholds and the antinociceptive effects of GABA receptor agonists in mice lacking the [beta]3 subunit of the GABAA receptor. Neuroscience. 1999;95(3):795–806. doi: 10.1016/s0306-4522(99)00481-9. [DOI] [PubMed] [Google Scholar]
- 13.Lindemann L, Meyer CA, Jeanneau K, Bradaia A, Ozmen L, Bluethmann H, et al. Trace amine-associated receptor 1 modulates dopaminergic activity. J Pharmacol Exp Ther. 2008;324(3):948–956. doi: 10.1124/jpet.107.132647. [DOI] [PubMed] [Google Scholar]
- 14.Lubeseder-Martellato C, Guenzi E, Jorg A, Topolt K, Naschberger E, Kremmer E, et al. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am J Pathol. 2002;161(5):1749–1759. doi: 10.1016/S0002-9440(10)64452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Buhr MF, Mahler M, Geffers R, Hansen W, Westendorf AM, Lauber J, et al. Cd14, Gbp1, and Pla2g2a: three major candidate genes for experimental IBD identified by combining QTL and microarray analyses. Physiol Genomics. 2006;25(3):426–434. doi: 10.1152/physiolgenomics.00022.2005. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh A, Zou F, Wright FA. Estimating odds ratios in genome scans: an approximate conditional likelihood approach. Am J Hum Genet. 2008;82(5):1064–1074. doi: 10.1016/j.ajhg.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Offenbaecher M, Bondy B, Jonge SD, Glatzeder K, Krüger M, Schoeps P, et al. Possible association of fibromyalgia with a polymorphism in the serotonin transporter gene regulatory region. Arthritis & Rheumatism. 1999;42(11):2482–2488. doi: 10.1002/1529-0131(199911)42:11<2482::AID-ANR27>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 18.Gursoy S. Absence of association of the serotonin transporter gene polymorphism with the mentally healthy subset of fibromyalgia patients. Clin Rheumatol. 2002;21(3):194–197. doi: 10.1007/s10067-002-8284-5. [DOI] [PubMed] [Google Scholar]
- 19.Potvin S, Larouche A, Normand E, Souza JBd, Gaumond I, Marchand S, et al. No relationship between the ins del polymorphism of the serotonin transporter promoter and pain perception in fibromyalgia patients and healthy controls. Eur J Pain. 2010;14(7):742–746. doi: 10.1016/j.ejpain.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Bondy B, Spaeth M, Offenbaecher M, Glatzeder K, Stratz T, Schwarz M, et al. The T102C polymorphism of the 5-HT2A receptor gene in fibromyalgia. Neurobiol Dis. 1999;6(5):433–439. doi: 10.1006/nbdi.1999.0262. [DOI] [PubMed] [Google Scholar]
- 21.Ablin J, Bar-Shira A, Yaron M, Orr-Urtreger A. Candidate-gene approach in fibromyalgia syndrome: association analysis of the genes encoding substance P receptor, dopamine transporter and alpha1-antitrypsin. Clin Exp Rheumatol. 2009;27(5) Suppl 56:S33–S38. [PubMed] [Google Scholar]
- 22.Buskila D, Cohen H, Neumann L, Ebstein RP. An association between fibromyalgia and the dopamine D4 receptor exon III repeat polymorphism and relationship to novelty seeking personality traits. Mol Psychiatry. 2004;9(8):730–731. doi: 10.1038/sj.mp.4001506. [DOI] [PubMed] [Google Scholar]
- 23.Potvin S, Larouche A, Normand E, Souza JBd, Gaumond I, Grignon S, et al. DRD3 Ser9Gly polymorphism Is related to thermal pain perception and modulation in chronic widespread pain patients and healthy controls. J Pain. 2009;10(9):969–975. doi: 10.1016/j.jpain.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Vargas-Alarcón G, Fragoso J-M, Cruz-Robles D, Vargas A, Martinez A, Lao-Villadóniga J-I, et al. Association of adrenergic receptor gene polymorphisms with different fibromyalgia syndrome domains. Arthritis & Rheumatism. 2009;60(7):2169–2173. doi: 10.1002/art.24655. [DOI] [PubMed] [Google Scholar]
- 25.Su S-Y, Chen J, Lai C-C, Chen C-M, Tsai F-J. The association between fibromyalgia and polymorphism of monoamine oxidase A and interleukin-4. Clinical Rheumatology. 2007;26(1):12–16. doi: 10.1007/s10067-006-0213-6. [DOI] [PubMed] [Google Scholar]
- 26.Gursoy S, Erdal E, Sezgin M, Barlas I, Aydeniz A, Alasehirli B, et al. Which genotype of MAO gene that the patients have are likely to be most susceptible to the symptoms of fibromyalgia? Rheumatology International. 2008;28(4):307–311. doi: 10.1007/s00296-007-0454-y. [DOI] [PubMed] [Google Scholar]
- 27.Gursoy S, Erdal E, Herken H, Madenci E, Alasehirli B, Erdal N. Significance of catechol-O-methyltransferase gene polymorphism in fibromyalgia syndrome. Rheumatol Int. 2003;23(3):104–107. doi: 10.1007/s00296-002-0260-5. [DOI] [PubMed] [Google Scholar]
- 28.Vargas-Alarcon G, Fragoso J-M, Cruz-Robles D, Vargas A, Vargas A, Lao-Villadoniga J-I, et al. Catechol-O-methyltransferase gene haplotypes in Mexican and Spanish patients with fibromyalgia. Arthritis Research & Therapy. 2007;9(5):R110. doi: 10.1186/ar2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen H, Nuemann L, Glazer Y, Ebstein RP, Buskila D. The relationship between a common catechol-O-methyltransferase (COMT) polymorphism val(158)met and fibromyalgia. Clin Exp Rheumatol. 2009;27(5) Suppl 56:S51–S56. [PubMed] [Google Scholar]
- 30.Hagen K, Pettersen E, Stovner L, Skorpen F, Zwart J-A. No association between chronic musculoskeletal complaints and Val158Met polymorphism in the Catechol-O-methyltransferase gene. The HUNT study. BMC Musculoskeletal Disorders. 2006;7(1):40. doi: 10.1186/1471-2474-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tander B, Gunes S, Boke O, Alayli G, Kara N, Bagci H, et al. Polymorphisms of the serotonin-2A receptor and catechol-O-methyltransferase genes: a study on fibromyalgia susceptibility. Rheumatol Int. 2008;28(7):685–691. doi: 10.1007/s00296-008-0525-8. [DOI] [PubMed] [Google Scholar]
- 32.Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet. 2005;14(1):135–143. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]
- 33.Xie Z, Miller GM. Trace amine-associated receptor 1 as a monoaminergic modulator in brain. Biochem Pharmacol. 2009;78(9):1095–1104. doi: 10.1016/j.bcp.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood PB, Schweinhardt P, Jaeger E, Dagher A, Hakyemez H, Rabiner EA, et al. Fibromyalgia patients show an abnormal dopamine response to pain. Eur J Neurosci. 2007;25(12):3576–3582. doi: 10.1111/j.1460-9568.2007.05623.x. [DOI] [PubMed] [Google Scholar]
- 35.Sotnikova TD, Caron MG, Gainetdinov RR. Trace amine-associated receptors as emerging therapeutic targets. Mol Pharmacol. 2009;76(2):229–235. doi: 10.1124/mol.109.055970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17(20):8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gold SJ, Han M-H, Herman AE, Ni YG, Pudiak CM, Aghajanian GK, et al. Regulation of RGS proteins by chronic morphine in rat locus coeruleus. Eur J Neurosci. 2003;17(5):971–980. doi: 10.1046/j.1460-9568.2003.02529.x. [DOI] [PubMed] [Google Scholar]
- 38.Garnier M, Zaratin PF, Ficalora G, Valente M, Fontanella L, Rhee M-H, et al. Up-regulation of Regulator of G protein signaling 4 expression in a model of neuropathic pain and insensitivity to morphine. J Pharmacol Exp Ther. 2003;304(3):1299–1306. doi: 10.1124/jpet.102.043471. [DOI] [PubMed] [Google Scholar]
- 39.Skrabek RQ, Galimova L, Ethans K, Perry D. Nabilone for the treatment of pain in fibromyalgia. J Pain. 2008;9(2):164–173. doi: 10.1016/j.jpain.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Park JM, Choi M-G, Cho YK, Lee IS, Kim SW, Choi KY, et al. Cannabinoid receptor 1 gene polymorphism and irritable bowel syndrome in the Korean population: a hypothesis-generating study. J Clin Gastroenterol. 2010 doi: 10.1097/MCG.0b013e3181dd1573. Publish Ahead of Print: 10.1097/MCG.0b013e3181dd1573. [DOI] [PubMed] [Google Scholar]
- 41.Juhasz G, Lazary J, Chase D, Pegg E, Downey D, Toth ZG, et al. Variations in the cannabinoid receptor 1 gene predispose to migraine. Neurosci Lett. 2009;461(2):116–120. doi: 10.1016/j.neulet.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 42.Lu AT, Ogdie MN, Järvelin M-R, Moilanen IK, Loo SK, McCracken JT, et al. Association of the cannabinoid receptor gene (CNR1) with ADHD and post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1488–1494. doi: 10.1002/ajmg.b.30693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaufmann I, Schelling G, Eisner C, Richter HP, Krauseneck T, Vogeser M, et al. Anandamide and neutrophil function in patients with fibromyalgia. Psychoneuroendocrinology. 2008;33(5):676–685. doi: 10.1016/j.psyneuen.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Russo E. Clinical endocannabinoid deficiency (CECD): can this concept explain therapeutic benefits of cannabis in migraine, fibromyalgia, irritable bowel syndrome and other treatment-resistant conditions? Neuro Endocrinol Lett. 2004;25(1–2):31–39. [PubMed] [Google Scholar]
- 45.Yunus MB. Fibromyalgia and overlapping disorders: the unifying concept of central sensitivity syndromes. Semin Arthritis Rheum. 2007;36(6):339–356. doi: 10.1016/j.semarthrit.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Bleakman D, Alt A, Nisenbaum ES. Glutamate receptors and pain. Sem Cell Dev Biol Pain. 2006;17(5):592–604. doi: 10.1016/j.semcdb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Guan Y, Terayama R, Dubner R, Ren K. Plasticity in excitatory amino acid receptor-mediated descending pain modulation after inflammation. J Pharmacol Exp Ther. 2002;300(2):513–520. doi: 10.1124/jpet.300.2.513. [DOI] [PubMed] [Google Scholar]
- 48.Peles S, Miranda A, Shaker R, Sengupta JN. Acute nociceptive somatic stimulus sensitizes neurones in the spinal cord to colonic distension in the rat. J Physiol. 2004;560(1):291–302. doi: 10.1113/jphysiol.2004.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turk DC, Okifuji A, Sinclair JD, Starz TW. Pain, disability, and physical functioning in subgroups of patients with fibromyalgia. J Rheumatol. 1996;23(7):1255–1262. [PubMed] [Google Scholar]
- 50.Giesecke T, Williams DA, Harris RE, Cupps TR, Tian X, Tian TX, et al. Subgrouping of fibromyalgia patients on the basis of pressure-pain thresholds and psychological factors. Arthritis Rheum. 2003;48(10):2916–2922. doi: 10.1002/art.11272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.