Abstract
Angiotensin-I-converting enzyme (ACE) is known to be associated with human cardiovascular and psychiatric pathophysiology. We have undertaken a global survey of the haplotypes in ACE gene to study diversity and to draw inferences on the nature of selective forces that may be operating on this gene. We have investigated the haplotype profiles reconstructed using polymorphisms in the regulatory (rs4277405, rs4459609, rs1800764, rs4292, rs4291), exonic (rs4309, rs4331, rs4343), and intronic (rs4340; Alu [I/D]) regions covering 17.8 kb of the ACE gene. We genotyped these polymorphisms in a large number of individuals drawn from 15 Indian ethnic groups and estimated haplotype frequencies. We compared the Indian data with available data from other global populations. Globally, five major haplotypes were observed. High-frequency haplotypes comprising mismatching alleles at the loci considered were seen in all populations. The three most frequent haplotypes among Africans were distinct from the major haplotypes of other world populations. We have studied the evolution of the two major haplotypes (TATATTGIA and CCCTCCADG), one of which contains an Alu insertion (I) and the other a deletion (D), seen most frequently among Caucasians (68%), non-African HapMap populations (65–88%), and Indian populations (70–95%) in detail. The two major haplotypes among Caucasians are reported to represent two distinct clades A and B. Earlier studies have postulated that a third clade C (represented by the haplotypes TACATCADG and TACATCADA) arose from an ancestral recombination event between A and B. We find that a more parsimonious explanation is that clades A and B have arisen by recombination between haplotypes belonging to clade C and a high-frequency African haplotype CCCTTCGIA. The haplotypes, which according to our hypothesis are the putative non-recombinants (PuNR), are uncommon in all non-African populations (frequency range 0–12%). Conversely, the frequencies of the putative recombinant haplotypes (PuR) are very low in the Africans populations (2–8%), indicating that the recombination event is likely to be ancient and arose before, perhaps shortly prior to, the global dispersal of modern humans. The global frequency spectrum of the PuR and the PuNR is difficult to explain only by drift. It appears likely that the ACE gene has been undergoing a combination of different selective pressures.
Electronic supplementary material
The online version of this article (doi:10.1007/s11568-011-9153-6) contains supplementary material, which is available to authorized users.
Keywords: Haplotype diversity, Recombinant, Non-recombinant, Selection
Introduction
Angiotensin II (Ang II) is a potent vasoactive and fibrogenic peptide. It is derived from conversion of Ang I by the angiotensin-I-converting enzyme encoded by the ACE gene (MIM 106180). The gene coding for ACE has two promoter sites that code for two major isoforms, somatic ACE (sACE) and testis ACE (tACE) (Testut et al. 1993). Variations in the ACE gene, many of which influence levels of the protein, are associated with diseases such as hypertension, myocardial infarction, Alzheimer’s disease, major depression, type II diabetes, and various quantitative precursors of these disorders (Rigat et al. 1990; Tiret et al. 1992; Villard et al. 1996; Daniel et al. 2006; Kehoe et al. 2003; Hadjadj et al. 2007). One of the most important and widely studied variations in ACE is a 287-base pair (bp) Alu insertion/deletion (I/D) polymorphism in intron 16 (rs4340). The D allele was found to raise ACE plasma levels in Europeans (McKenzie et al. 1995; Rigat et al. 1990; Tiret et al. 1992). A large number of studies have found significant associations of the I/D polymorphism with different pathophysiological conditions (Ueda et al. 1995; Murphey et al. 2000; Giner et al. 2000), not all of which were confirmed in replicate studies (Lachurie et al. 1995; Jeunemaitre et al. 1992; Schmidt et al. 1993). Other polymorphisms 5′ to the I/D locus were identified and found to be in strong linkage disequilibrium with the I/D locus (Soubrier et al. 2002). There are strong indications that the impact of the ACE gene on cardiovascular and psychiatric diseases is through the joint—not individual—effect of alleles at the various polymorphic loci in this gene (Keavney et al. 1998; Villard et al. 1996; Zhu et al. 2000, 2001; McKenzie et al. 2001; Cox et al. 2002). Therefore, haplotype-based association studies have been recommended (Soubrier et al. 2002; Sayed-Tabatabaei et al. 2006). A total of 52 haplotypes were identified by resequencing ACE in Europeans and African-Americans with high divergence in respect of haplotypes and their frequencies (Rieder et al. 1999). There is however a paucity of systematic studies on the nature and extent of haplotype diversity across populations, and on inferring selective pressures and evolution of the ACE gene. In this study, we examine global frequency profiles of haplotypes reconstructed from nine polymorphic loci in the ACE gene, including in 15 ethnic populations of India for which data have been generated by us refine existing models pertaining to the evolution of ACE haplotypes and draw inferences on selective forces that may have acted to maintain these haplotypes and their frequencies.
Materials and methods
Haplotype data for three African [Nigerians, Afro-Americans, Jamaicans (Bouzekri et al. 2004), and two Caucasian [British (Keavney et al. 1998) and French (Villard et al. 1996)] populations were collated. These haplotypes were collapsed to include the 9 polymorphic loci rs4277405, rs4459609, rs1800764, rs4291, rs4292(all regulatory), rs4309, rs4331, rs4343 (all exonic), and rs4340(intronic) to make the data comparable (Supplementary Materials & Methods). (rs4340 is the Alu I/D locus). Data on four of these SNPs rs4459609, rs1800764, rs4309, and rs4343 for eleven HapMap populations [ASW: African ancestry in Southwest USA, CEU: Utah residents with Northern and Western European ancestry from the CEPH collection, CHB: Han Chinese in Beijing, China, CHD: Chinese in Metropolitan Denver, Colorado, GIH: Gujarati Indians in Houston, Texas, JPT: Japanese in Tokyo, Japan, LWK: Luhya in Webuye, Kenya, MEX: Mexican ancestry in Los Angeles, California, MKK: Maasai in Kinyawa, Kenya, TSI: Tuscans in Italy, YRI: Yoruba in Ibadan, Nigeria] were downloaded from the HapMap3 database (http://www.hapmap.org). Data on four SNPs (rs4277405, rs4291, rs4292and rs4340) were not available for HapMap samples. Information on the ancestral allele was downloaded from the NCBI SNP database.
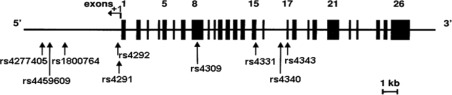
We genotyped these nine loci (Fig. 1) in a total of 450 healthy normal individuals, self-reported to be unrelated to the first cousin level, randomly drawn from 15 ethnic groups representing the geographical, social and linguistic diversity of India. Names of the populations, their linguistic affiliations, sample sizes, and the geographical locations of sampling are presented in Table 1. About 10 ml of blood was collected by venipuncture, with informed consent, from each study participant. DNA was isolated using the salting out protocol described by Miller et al. (1988). Genotypes at all loci were determined by the PCR–RFLP method, using standard protocols (Supplementary Table 1).
Fig. 1.
Schematic representation of the ACE gene depicting the polymorphic SNPs typed in this study
Table 1.
Names of study populations, geographical locations of habitat, and socio-linguistic information
| Population name (code) | Linguistic affiliation | Geographical region (state within India) |
|---|---|---|
| Agharia (AGH) | Indo-European | East (Orissa) |
| Bagdi (BAG) | Indo-European | East (W Bengal) |
| Ho (HO) | Austro-Asiatic | East (Orissa) |
| Irula(ILA) | Dravidian | South (Tamil Nadu) |
| Iyengar (IYN) | Dravidian | South (Tamil Nadu) |
| Iyer (IYR) | Dravidian | South (Tamil Nadu) |
| Jamatia (JAM) | Tibeto-Burman | Northeast (Tripura) |
| Kadar (KAD) | Dravidian | South (Tamil Nadu) |
| Kamma Naidu (KMN) | Dravidian | South (Andhra Pradesh) |
| Mizo (MZO) | Tibeto-Burman | Northeast (Mizoram) |
| Muria (MUR) | Dravidian | Central (Chhattisgarh) |
| Pallan (PLN) | Dravidian | South (Andhra Pradesh) |
| Saryupari Brahmin (SBR) | Indo-European | Central (Chhattisgarh) |
| Satnami (SNM) | Indo-European | West (Chhattisgarh) |
| Toto (TTO) | Tibeto-Burman | East (W Bengal) |
The number of individuals sampled from each population was 30
Haplotype frequencies were estimated using HAPLOVIEW (http://www.broad.mit.edu/mpg/haploview) ab initio from new genotypes generated for the Indian populations and from genotypes of the HapMap populations which were downloaded from the HapMap3 database. The haplotypes taken from literature and our own data source are all from the same (forward) strand. Cluster analysis was done to compute distances between pairs of populations based on haplotype frequencies by the neighbor-joining algorithm by DISPAN (Ota 1993). Pairwise Fst for the four SNPs, rs4459609, rs1800764, rs4309, and rs4343, common with the 11 populations included in Hapmap3, was calculated under the hypothesis of no difference between the populations. The null distribution of pairwise Fst values is obtained by permuting haplotypes between populations using ARLEQUIN (Schneider et al. 2000; http://lgb.unige.ch/arlequin/).
Multiple alignment of human and primate ACE gene sequences (hg19.chr17:61548890–61573900) was performed using Multiz and other tools in UCSC Genome Browser or in Penn State Bioinformatics comparative genomics alignment pipeline. Phylogenetic tree relating primate and human haplotypes was constructed by the “drawtree” program using maximum parsimony method (“dnapars”) in the PHYLIP 3.69 package of programs (Felsenstein 1989) (http://evolution.gs.washington.edu/phylip.html).
Results
Haplotype distribution among Africans and non-Africans
The frequency distributions of haplotypes vary markedly between populations. Populations of African ancestry exhibit a larger number and greater diversity of haplotypes than the non-African populations. Five major (high frequency) haplotypes were present in global populations (Table 2). Their frequencies showed statistically significant differences (P < 0.0001) across populations (Supplementary Table 2). In the African populations, three major haplotypes CCCTTCGIA (12–19%), TACATCADG (16–36%), and TACATCADA (25–29%) were present (Bouzekri et al. 2004). The two major haplotypes in the non-African populations were TATATTGIA (38–72%) and CCCTCCADG (19–40%). It may be noted that these the two haplotypes (a) have mismatching alleles at each locus and (b) are strikingly different in allelic composition compared to the African high-frequency haplotypes We denote the major haplotypes among non-Africans, i.e., TATATTGIA and CCCTCCADG as PuR1 and PuR2, respectively, and PuRs collectively. Similarly, we denote the major haplotypes among the Africans, i.e., TACATCADG, TACATCADA, and CCCTTCGIA, as PuNR1, PuNR2 and PuNR3, respectively, and PuNRs collectively. We observe that populations with African ancestry (Jamaicans and Afro-Americans) have very low frequency (2% among Nigerians to 8% among Jamaicans and Afro-Americans) of the PuRs. Jamaicans and African-Americans are populations admixed between West Africans slaves brought to the new world and people of European descent. The genomes of these admixed populations are primarily African with variable, but usually small proportion of European ancestry (Zakharia et al. 2009). The overall low frequency of the PuRs in Jamaicans and African-Americans is consistent with the low contribution of the European gene pool to these populations.
Table 2.
Percentage frequencies of haplotypes (major haplotypes in bold) in Indian, Africans, and Caucasian population
| No | Populations→ | AGH | BAG | HO | TTO | JAM | MZO | SBR | SNM | MUR | IYR | IYN | KAD | KMN | PLN | Caucasian | Nigerian | Jamaican | Afro-Am | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype↓ | Language→ Location→ |
IE | IE | AA | TB | TB | TB | IE | IE | DR | DR | DR | DR | DR | DR | Europe (Britain) | W Africa (Nigeria) | Caribbean Islands | USA | |
| East India | Northeast India | Central India | South India | |||||||||||||||||
| 1 | TATATTGIA | 38 | 60 | 72 | 55 | 65 | 55 | 52 | 50 | 55 | 45 | 53 | 68 | 42 | 52 | 40 | 2 | 3 | 8 | |
| 2 | CCCTCCADG | 36 | 19 | 18 | 40 | 32 | 35 | 38 | 33 | 33 | 25 | 33 | 18 | 40 | 27 | 28 | 2 | 5 | 6 | |
| 3 | TATATTADG | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 9 | 0 | 5 | 2 | 5 | 0 | 0 | 0 | 0 | |
| 4 | TATATCADG | 5 | 7 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 3 | 0 | 0 | 2 | 0 | 9 | 0 | 0 | 0 | |
| 5 | CCCTTCGIA | 3 | 6 | 5 | 0 | 0 | 0 | 2 | 0 | 2 | 10 | 2 | 0 | 3 | 2 | 0 | 19 | 14 | 12 | |
| 6 | TATATTGDA | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7 | TATTCCADG | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 8 | CCCACCGDA | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 9 | CCCTTCADG | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 10 | CCCTCTADG | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 11 | TACATTGIA | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 12 | TACATCADG | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 5 | 0 | 5 | 7 | 3 | 2 | 2 | 7 | 16 | 32 | 36 | |
| 13 | CCCTTTGIA | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| 14 | CCCTCCGDG | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 15 | TATATCGIA | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | |
| 16 | CCCTCCGIA | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | |
| 17 | TATATCADA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| 18 | CACTCCADG | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | |
| 19 | CCTATTGIA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 19 | TACATCADA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 2 | 0 | 0 | 29 | 23 | 25 | |
| 20 | CCCTCCAIG | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| 21 | TATATTGDG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 22 | CCCTCTGIA | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | |
| 23 | CCTTCCADG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| 24 | CCCACCGIA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| 25 | TATTTCADG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| 26 | TATATTAIA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| 27 | TACATCGIA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 3 | |
| 28 | CATATTGIA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | |
| 29 | TCCATTGIA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | |
IE Indo-European, DR Dravidian, AA Austro-Asiatic and TB Tibeto-Burman refer to the linguistic affiliations of the Indian populations
The percentages do total to 100 as haplotypes with frequency below 1% were excluded
The two major haplotypes PuR1 and PuR2 (TATATTGIA and CCCTCCADG) in non-African populations were also the major haplotypes in the fifteen Indian groups. The frequency of PuR1 varied from 38 to 71% across groups, while that of PuR2 varied from 8 to 39% (Supplementary Figure 1). The phylogenetic relationships (Fig. 2) between broad population clusters of India (based on language, which is confounded by geographical separation of habitats) and global populations based on ACE haplotype frequencies agree with earlier inferences (Basu et al. 2003; Indian Genome Variation Consortium 2008). However, at the level of individual ethnic populations, the phylogenetic relationships (Fig. 3) did not correlate with socio-cultural or geographical contiguity, which is discordant with some previous studies (Indian Genome Variation Consortium 2008; Majumder 2001; Majumder et al. 1999), possibly due to differences in selective pressures and single locus behavior of the multi-locus haplotypes due to linkage disequilibrium among the multiple loci or simply because of smaller number of individuals genotyped in each ethnic population. Populations of Africa and the African ancestry cluster together. The Indo-Europeans (IE) and Dravidians (DV) are close to Caucasians as shown in many previous studies, while the Austro-Asiatics are closer to the Tibeto-Burman (Basu et al. 2003; Reich et al. 2009; Indian Genome Variation Consortium 2008).
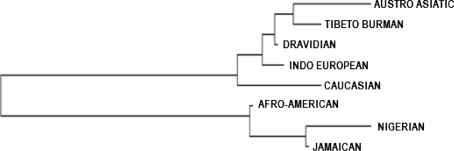
Fig. 2.
Neighbour-joining tree depicting genetic affinities among 15 Indian populations belonging to various linguistic groups (Indo-European, Dravidian, Austro-Asiatic and Tibeto-Burman), one Caucasian, and three African populations (Afro-American, Nigerian and Jamaican) based on nine-locus haplotype frequencies
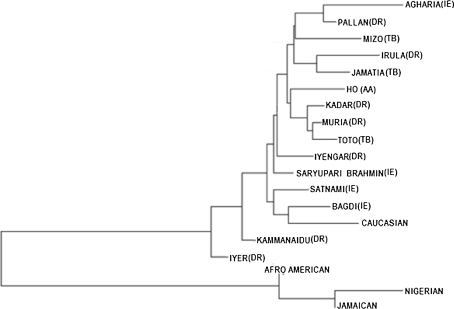
Fig. 3.
Neighbour-joining tree depicting genetic affinities among 15 Indian, one Caucasian, and three African populations (Afro-American, Nigerian, and Jamaican) based on nine-locus haplotype frequencies. The linguistic affiliation of each population is give in parenthesis (IE Indo-European, DR Dravidian, AA Austro-Asiatic and TB Tibeto-Burman)
Cladistic model of ACE haplotypes: an alternative proposal
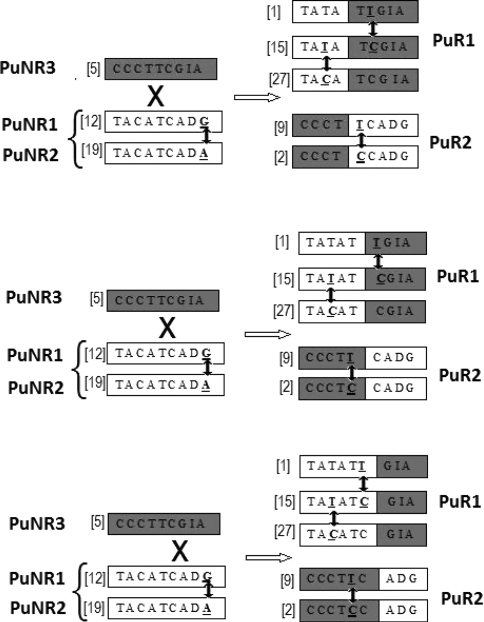
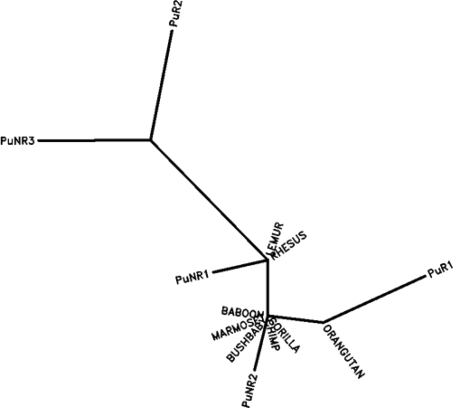
Keavney et al. (1998) and Farrall et al. (1999) named the two high-frequency haplotypes present in Europe as clade A (TATATTGIA) and clade B (CCCTCCADG). They also proposed that the clade C (TACATCADG and TACATCADA) which has a lower frequency in Europe, but a high frequency in Africa, arose from a recombination event between clades A and B. Their inference was based on the relative abundance of clades A and B in Europe compared to the frequency of the suggested recombinant haplotype (clade C). When we assessed this proposal against the frequency and spread of the ACE haplotypes across world populations, it appeared unlikely to be a parsimonious explanation. We define the clade A and B as the putative recombinant haplotypes (PuRs) and clade C and the haplotype CCCTTCGIA as the putative non-recombinant haplotypes (PuNRs). We suggest an alternative model where both clades A and clade B arose by pairwise recombination among the frequent African haplotypes CCCTTCGIA, TACATCADG, and TACATCADA. This appears to be more reasonable as it also accounts for the other haplotype PuNR3 (CCCTTCGIA) along with clade C haplotypes which has arisen as a consequence of the recombination event. The original hypothesis of Keavney et al. (1998) also explains the CCCTTCGIA (PuNR3) being the other haplotype formed by recombination between TATATTGIA and CCCTCCADG. We have illustrated the possible chronology of events in Fig. 4. The pattern of mutations and recombinations is equally parsimonious and so cannot be used to decide between the two options (mutation and recombination) being responsible for the change in nucleotide at any position. Comparison with the five major human haplotypes with the Chimp Ancestral haplotype (TANATCGDA) also supports the hypothesis that the African non-recombinant haplotypes are more ancient. The corresponding regions of ACE gene from 8 primate species Chimp, Gorilla, Orangutan, Rhesus, Baboon, Marmoset, Mouse lemur, Bushbaby were downloaded (Table 3) and along with the five major human haplotypes [PuR (1 and 2) and PuNR (1,2,3)] an unrooted phylogenetic tree was estimated, by maximum parsimony approach, using PHYLIP (Fig. 5). Among the primates, the Lemur and Rhesus are close but separated from the remaining primate species (Chimp, Gorilla, Orangutan, Baboon, Marmoset, and Bushbaby) that cluster together. The PuNR1 and PuNR2 haplotypes are, respectively, closer to the {Mouse lemur, Rhesus} and the {Chimp, Gorilla, Orangutan, Baboon, Marmoset, and Bushbaby} cluster. The results indicate that the PuNR2 haplotype is likely to be the most ancestral haplotype. The multi-species cladogram provides good support, except for the fact that PuNR3 does not seem particularly ancient as it stands out distantly as a single point cluster (Fig. 4). The PuNR3, which is absent in the primate species, probably arose after the divergence of human from primates.
Fig. 4.
Proposed recombination events have resulted in different common haplotypes. The putative recombinants (PuR1 and PuR2) represent clades A and B proposed by Keavney et al. (1998), while the putative non–recombinants PuNR1 and PuNR2 represent the clade C. The putative non-recombinants PuNR1, PuNR2, and PuNR3 is the major haplotype in the African populations. The arrow (↕) represents possible mutation events. Numbers within crotchets indicate the haplotype number from Table 2
Table 3.
The ACE haplotypes for 8 primate species and the reference human downloaded using the UCSC Genome Browser by multiple alignment of human and primate ACE gene sequence (hg19.chr17:61548890-61573900)
| Species | rs4277405 | rs4459609 | rs1800764 | rs4291 | rs4292 | rs4309 | rs4331 | rs4340 | rs4343 |
|---|---|---|---|---|---|---|---|---|---|
| Human | C | C | C | T | C | C | A | D | G |
| Chimp | T | A | ? | A | T | C | G | D | A |
| Gorilla | T | A | ? | A | T | C | G | D | G |
| Orangutan | T | A | C | A | T | T | G | D | A |
| Rhesus | ? | ? | ? | A | T | C | G | D | G |
| Baboon | ? | ? | C | A | T | C | G | D | G |
| Marmoset | ? | ? | ? | A | T | C | A | D | A |
| Mouse lemur | ? | ? | ? | A | T | C | G | D | G |
| Bushbaby | ? | ? | ? | A | T | C | G | D | A |
“?” refers to the nucleotide at that position which is missing on multiple alignment of the human and primate ACE sequence by the UCSC Genome Browzer
Fig. 5.
Phylogenetic tree relating primate haplotypes with the five major human haplotypes
Among Indian populations, the frequencies of the PuRs were strikingly high, even higher than among Europeans. With the exception of AGH (74%), BAG (79%), IYR (70%), and PLN (79%), the two major haplotypes (PuR1 and PuR2) accounted for more than 80% haplotypes in each population; in some populations (TTO and JAM), these two haplotypes accounted for more than 95% (Table 2).
ACE haplotype distribution in HapMap populations
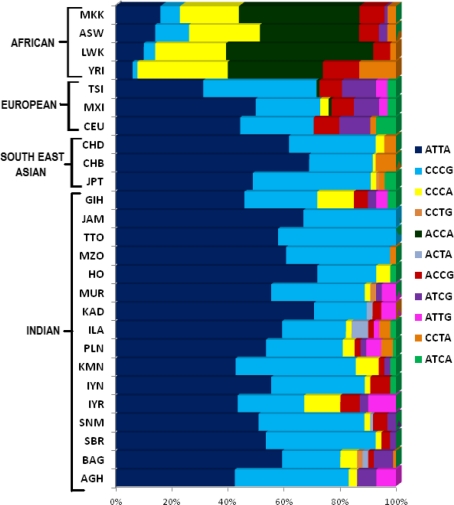
In order to re-examine our model in a larger set of world populations, we constructed haplotypes using four SNPs (rs4459609, rs1800764, rs4309, and rs4343), which were available in the HapMap database and were chosen from either side of the proposed recombination breakpoint. It was easy to derive one-to-one correspondence between the set of longer (9 loci) haplotypes and the set of shorter (4 loci) haplotypes. The corresponding putative recombinant (PuRs) (ATTA and CCCG) and putative non-recombinant (PuNRs) (CCCA, ACCA, ACCG) haplotype frequencies were estimated. As expected, the corresponding PuNR haplotypes turned out to be the major haplotypes in all African populations (YRI, LWK, MKK, ASW), while the PuR haplotypes were major in the other world populations including the 15 Indian populations. As we observed previously, the major (PuR) haplotypes of non-African populations occur at very low frequencies in the African populations. Similarly the major (PuNR) African haplotypes occurred but at a low frequencies in the non-African populations (Supplementary Table 3). The frequency of the PuNR haplotype (CCCA) was present at a frequency of 13% in the GIH, which may be due to their small effective size.
Figure 6 displays the estimated frequencies of each inferred haplotype present in the eleven HapMap and fifteen Indian populations. In this figure, the order of the haplotypes was set by the relative frequency of the PuR, PuNR, and the rarer haplotypes, and was retained for all the same order in the 26 populations. The observed difference in frequency distributions of haplotypes between the African and non-African populations was consistent with two high-frequency haplotypes PuRs in non-Africans populations from the West (CEU, MEX, TSI) to India (GIH), China (CHB and CHD), and the Japan (JPT) in the East and three major PuNR haplotypes in Africans (YRI, LWK, MKK, ASW) with very little haplotype sharing with non-Africans. The smaller ACE haplotypes studied by Rebai et al. (2006) in the North African Tunisian population showed that both the PuR haplotypes (0.522) and the PuNR haplotypes (0.385) were found in high frequencies. It is likely due to its geographical location favoring admixture with the non-Africans (Rebai et al. 2006).
Fig. 6.
Haplotype frequencies in samples from eleven HapMap and fifteen Indian populations arranged by their frequencies. Patterns represent 4 locus reconstructed haplotypes of the ACE gene. (See supplementary Table 4 for estimated haplotype frequencies in each population)
Discussion
The dispersal and variation of the ACE haplotypes among various populations of the world seem enigmatic. Zhang et al. (2003) observed that “Analysis of common haplotypes in 62 random genomic loci and 85 gene coding regions in humans shows that the proportion of the genome spanned by perfectly mismatching yin-yang haplotypes is 75–85%.” In the ACE gene too, we observe a similar pattern in all the populations that we have studied the total frequency of only two or three haplotypes exceed 65% in all the population. In some populations, the frequency of the mismatching haplotypes together account for more than 90%. Zhang et al. (2003) initially interpreted this pattern as suggestive of deep population splitting or maintenance of ancient lineages by selection. Further analysis, including coalescent simulation, revealed that the yin-yang phenomenon can be more parsimoniously explained by strictly neutral evolution (Zhang et al. 2003). However, some have argued that balancing selection might have also helped in shaping and maintenance of these patterns along the genome, particularly in the genic regions (Hollox and Armour 2008; Cagliani et al. 2010a; Mukherjee et al. 2009; Fumagalli et al. 2009).
Our observations in respect of the ACE haplotypes, though superficially similar, are far more complicated. The ACE haplotypes harbor an ancient recombination that resulted in haplotypes with high frequencies among African populations very distinct from the haplotypes found outside of Africa. Keavney et al. (1998) and Farrall et al. (1999) had correctly identified the site of recombination which gave rise to these five major haplotypes around the globe. The perfectly mismatching haplotypes (clades A and B), which they presumed to be ancestral, are present in higher frequencies in populations outside of Africa and in extremely low frequencies within Africa. The present day low frequencies of these so-called ancestral haplotypes in Africa do not inspire confidence that they are truly ancestral. The fact that clade C haplotypes have the highest frequencies in Africa imply, under the hypothesis of Keavney et al. and Farrall et al., that the recombinant clade C replaced, through strong selective effects, the ancestral clades A and B. Such strong selection should then have operated only within Africa, after the last wave of out-of-Africa migration of modern humans, since clades A and B are the predominant haplotypes in all non-African populations. These strong assumptions are required to explain the observations on the spread and frequencies of the ACE haplotypes under the hypothesis of Keavney et al. and Farrall et al. that are likely to be true. A more parsimonious explanation not requiring such strong and likely assumptions is that recombinations between CCCTTCGIA (PuNR3) (the predominant African haplotype) and haplotypes belonging to clade C(PuNR1 and PuNR2) gave rise to the haplotypes belonging to clades A(PuR1) and B(PuR2). The recombination event occurred early during the migration of modern humans out of Africa. It is more likely that the selection and/or bottleneck effects resulted in high frequencies of PuR haplotype in non-Africans, than the alternative that selection for the PuNR happened in multiple African populations after the out-of-Africa event, or that drift meant that the an ancient PuRs went to low frequencies in multiple African populations. In order to determine whether selection might be operating on the ACE gene, we looked into the pairwise Fst values for the four SNPs we have common with the HapMap3 populations. From the permutation p-values, we find that the African populations were significantly different from the rest of the world indicating possible selection (Supplementary Table 5). It appears that clades A and B may not have enjoyed a selective advantage over the ancestral haplotypes within Africa, but when humankind moved out of Africa, the haplotypes belonging to clades A and B may have gained some selective advantage, as has been suggested, for example, for genes determining skin color (Lao et al. 2007) and also for ACE haplotypes (Cagliani et al. 2010b). While it is true that the majority of African populations in this study are of West African origin, the pattern of haplotype frequency distribution remains similar when the other HapMap African populations, including LWK (Luhya in Webuye, Kenya) and MKK (Maasai in Kinyawa, Kenya), are considered.
The ancestral African environment was characterized by salt scarcity. Therefore, humans and non-human primates had enhanced salt avidity (Gleiberman 2001). In support of this, evidence of selection has been shown in two genes that influence blood pressure. AGT (angiotensinogen) and CYP3A5 (cytochrome P450 3A5) genes harbor SNPs with functional alleles that influence salt avidity and blood pressure. For both the genes, the allele that increases salt avidity is the major allele among Africans. In contrast, the alleles that do not increase salt avidity have risen to high frequencies outside of Africa, probably due to selection (Nakajima et al. 2004; Thompson et al. 2004). PuNR1 and PuNR2 that are associated with elevated circulating ACE activity may influence the activity of the renin-angiotensin system thus fostering increase in sodium reabsorption and increased blood pressure (De Wardener and MacGregor 2002). Genetic studies in families of African ancestry suggest that there is a major heritable ACE contribution to ACE enzyme levels and blood pressure (Hajjar and Kotchen 2003). People of African descent in North America are disproportionately affected by hypertension with 1.5–2-fold increase in hypertension prevalence among African-Americans as compared with European-descended Americans. African-Americans are also at higher risk of hypertension (Karter et al. 2002), end-stage renal disease (Exner et al. 2001), poorer response and outcome to anti-hypertensive treatment (Exner et al. 2001; Sehgal 2004), and more likely to experience adverse effects with ACE inhibitor treatment (McDowell et al. 2006) as compared to Caucasians. Whatever may be the reasons for increase in the frequencies of the putative recombinant clades in non-African populations, the post-agriculture population explosion facilitated these haplotypes to become the dominant haplotypes worldwide outside of Africa.
Electronic supplementary material
Acknowledgments
We are grateful to the two anonymous reviewers for their comments and suggestions on an earlier version, which greatly improved the value of the manuscript. This work was supported by grant to S. F. under the women scientist (WOS-A) scheme of Department of Science and Technology, New Delhi. Support provided to P. P. M. by the Department of Biotechnology and Indian Statistical Institute is also gratefully acknowledged. We are grateful to A. Ramesh, M. V. Usha Rani, Samir Sil, Mitashree Mitra, and C. S. Chakrabarti for help in collection of samples from the Indian populations.
References
- Basu A, Mukherjee N, Roy S, et al. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzekri N, Zhu X, Jiang Y, et al. Angiotensin I-converting enzyme polymorphisms, ACE level and blood pressure among Nigerians, Jamaicans and African-Americans. EJHG. 2004;12:460–468. doi: 10.1038/sj.ejhg.5201166. [DOI] [PubMed] [Google Scholar]
- Cagliani R, Fumagalli M, Riva S (2010a) Polymorphisms in the CPB2 gene are maintained by balancing selection and result in haplotype-preferential splicing of exon 7. Mol Biol Evol Epub [DOI] [PubMed]
- Cagliani R, Fumagallia M, Rivaa S, et al. Genetic variability in the ACE gene region surrounding the Alu I/D polymorphism is maintained by balancing selection in human populations. Pharmacogenet Genomics. 2010;20:131–134. doi: 10.1097/FPC.0b013e3283333532. [DOI] [PubMed] [Google Scholar]
- Cox R, Bouzekri N, Martin S, et al. Angiotensin-1-converting enzyme (ACE) plasma concentration is influenced by multiple ACE-linked quantitative trait nucleotides. Hum Mol Genet. 2002;11:2969–2977. doi: 10.1093/hmg/11.23.2969. [DOI] [PubMed] [Google Scholar]
- Daniel PK, Grzegorz P, Serena C, et al. Disease haplotype for advanced nephropathy in type 2 diabetes at the ACE locus. Diabetes. 2006;55:2660–2664. doi: 10.2337/db06-0496. [DOI] [PubMed] [Google Scholar]
- Wardener H, MacGregor GA. Sodium and blood pressure. Curr Opin Cardiol. 2002;17:360–367. doi: 10.1097/00001573-200207000-00007. [DOI] [PubMed] [Google Scholar]
- Exner DV, Dries DL, Domanski MJ, Cohn JN. Lesser response to angiotensin-converting-enzyme inhibitor therapy in black as compared with white patients with left ventricular dysfunction. N Engl J Med. 2001;344:1351–1357. doi: 10.1056/NEJM200105033441802. [DOI] [PubMed] [Google Scholar]
- Farrall M, Keavney B, McKenzie C, et al. Fine-mapping of an ancestral recombination breakpoint in DCP1. Nat Genet. 1999;23:270–271. doi: 10.1038/15449. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fumagalli S, Cagliani R, Pozzoli U, et al. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res. 2009;19:199–212. doi: 10.1101/gr.082768.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giner V, Poch E, Bragulat E, et al. Renin-angiotensin system genetic polymorphisms and salt sensitivity in essential hypertension. Hypertension. 2000;35:512–517. doi: 10.1161/01.hyp.35.1.512. [DOI] [PubMed] [Google Scholar]
- Gleiberman L. Salt, hypertension, evolution. Psychosom Med. 2001;63:325–327. doi: 10.1097/00006842-200103000-00021. [DOI] [PubMed] [Google Scholar]
- Hadjadj S, Tarnow L, Forsblom C, et al. Association between angiotensin-converting enzyme gene polymorphisms and diabetic nephropathy case-control, haplotype, and family-based study in three European populations. J Am Soc Nephrol. 2007;18:1284–1291. doi: 10.1681/ASN.2006101102. [DOI] [PubMed] [Google Scholar]
- Hajjar I, Kotchen TA. Trends in prevalence, awareness, treatment, and control of hypertension in the United States, 1998–2000. J Am Med Assoc. 2003;290:199–206. doi: 10.1001/jama.290.2.199. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JAL. Directional and balancing selection in human beta-defensins. BMC Evol Biol. 2008;8:113–127. doi: 10.1186/1471-2148-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indian Genome Variation Consortium Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- Jeunemaitre X, Lifton RP, Hunt SC, et al. Absence of linkage between the angiotensin converting enzyme locus and human essential hypertension. Nat Genet. 1992;1:72–75. doi: 10.1038/ng0492-72. [DOI] [PubMed] [Google Scholar]
- Karter AJ, Ferrara A, Liu JY, Moffet HH, Ackerson LM, Selby JV. Ethnic disparities in diabetic complications in an insured population. J Am Med Assoc. 2002;287:2519–2527. doi: 10.1001/jama.287.19.2519. [DOI] [PubMed] [Google Scholar]
- Keavney B, McKenzie CA, Connell JM, et al. Measured haplotype analysis of the angiotensin-I converting enzyme gene. Hum Mol Genet. 1998;7:1745–1751. doi: 10.1093/hmg/7.11.1745. [DOI] [PubMed] [Google Scholar]
- Kehoe PG, Katzov H, Feuk L, et al. Haplotypes extending across ACE are associated with Alzheimer’s disease. Hum Mol Genet. 2003;12:859–867. doi: 10.1093/hmg/ddg094. [DOI] [PubMed] [Google Scholar]
- Lachurie ML, Azizi M, Guyene TT, et al. Angiotensin-converting enzyme gene polymorphism has no influence on the circulating renin-angiotensin-aldosterone system or blood pressure in normotensive subjects. Circulation. 1995;91:2933–2942. doi: 10.1161/01.cir.91.12.2933. [DOI] [PubMed] [Google Scholar]
- Lao O, Gruijter JM, Duijn K, Navarro A, Kayser M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 2007;71:354–369. doi: 10.1111/j.1469-1809.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- Majumder PP. Ethnic populations of India as seen from an evolutionary perspective. J Biosci. 2001;26:533–545. doi: 10.1007/BF02704750. [DOI] [PubMed] [Google Scholar]
- Majumder PP, Roy B, Banerjee S, et al. Human specific insertion/deletion polymorphisms in Indian populations and their possible evolutionary implications. Eur J Hum Genet. 1999;7:435–446. doi: 10.1038/sj.ejhg.5200317. [DOI] [PubMed] [Google Scholar]
- McDowell SE, Coleman JJ, Ferner RE. Systematic review and meta analysis of ethnic differences in risks of adverse reactions to drugs used in cardiovascular medicine. Br Med J. 2006;332:1177–1181. doi: 10.1136/bmj.38803.528113.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie CA, Julier C, Forrester T, et al. Segregation and linkage analysis of serum angiotensin I-converting enzyme levels: evidence for two quantitative-trait loci. Am J Hum Genet. 1995;57:1426–1435. [PMC free article] [PubMed] [Google Scholar]
- McKenzie CA, Abecasis GR, Keavney B, et al. Trans-ethnic fine mapping of a quantitative trait locus for circulating angiotensin I-converting enzyme (ACE) Hum Mol Genet. 2001;10:1077–1084. doi: 10.1093/hmg/10.10.1077. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Sarkar-Roy N, Wagener DK, Majumder PP. Signatures of natural selection are not uniform across genes of innate immune system, but purifying selection is the dominant signature. Proc Natl Acad Sci. 2009;106:7073–7078. doi: 10.1073/pnas.0811357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey LJ, Gainer JV, Vaughan DE, et al. Angiotensin converting enzyme insertion/deletion polymorphism modulates the human in vivo metabolism of bradykinin. Circulation. 2000;102:829–832. doi: 10.1161/01.cir.102.8.829. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Wooding S, Sakagami T, Emi M, Tokunaga K, et al. Natural selection and population history in the human angiotensinogen gene (AGT): 736 complete AGT sequences in chromosomes from around the world. Am J Hum Genet. 2004;74:898–916. doi: 10.1086/420793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T. DISPAN: in genetic distance and phylogenetic analysis. University Park, PA: Pennsylvania State University; 1993. [Google Scholar]
- Rebai M, Kharrat N, Ayadi I, Rebai A. Haplotype structure of five SNPs within the ACE gene in the Tunisian population. Ann Hum Biol. 2006;33:319–329. doi: 10.1080/03014460600621977. [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–495. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA. Sequence variation in the human angiotensin converting enzyme. Nat Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed-Tabatabaei FA, Oostra BA, Isaacs A, et al. ACE polymorphisms. Circ Res. 2006;98:1123–1133. doi: 10.1161/01.RES.0000223145.74217.e7. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Hooft IM, Grobbee DE, et al. Polymorphism of the angiotensin I converting enzyme gene is apparently not related to high blood pressure: Dutch hypertension and offspring study. J Hypertens. 1993;11:345–348. doi: 10.1097/00004872-199304000-00003. [DOI] [PubMed] [Google Scholar]
- Schneider S, Kueffer JM, Roessli D, Excoffier L. ARLEQUIN: a software for population genetic data analysis. Geneva, Switzerland: University of Geneva; 2000. [Google Scholar]
- Sehgal AR. Overlap between whites and blacks in response to antihypertensive drugs. Hypertension. 2004;43:566–572. doi: 10.1161/01.HYP.0000118019.28487.9c. [DOI] [PubMed] [Google Scholar]
- Soubrier F, Martin S, Alonso A, et al. High-resolution genetic mapping of the ACE-linked QTL influencing circulating ACE activity. Eur J Hum Genet. 2002;10:553–561. doi: 10.1038/sj.ejhg.5200847. [DOI] [PubMed] [Google Scholar]
- Testut P, Soubrier F, Corvol P, et al. Functional analysis of the human somatic angiotensin I-converting enzyme gene promoter. Biochem J. 1993;293:843–848. doi: 10.1042/bj2930843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, et al. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiret L, Rigat B, Visvikis S, et al. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet. 1992;51:197–205. [PMC free article] [PubMed] [Google Scholar]
- Ueda S, Elliott HL, Morton JJ, Connell JM. Enhanced pressure response to angiotensin I in normotensive men with the deletion genotype (DD) for angiotensin-converting enzyme. Hypertension. 1995;25:1266–1269. doi: 10.1161/01.hyp.25.6.1266. [DOI] [PubMed] [Google Scholar]
- Villard E, Tiret L, Visvikis S, Rakotovao R, Cambien F, Soubrier F. Identification of new polymorphisms of the angiotensin I-converting enzyme (ACE) gene, and study of their relationship to plasma ACE levels by two-QTL segregation-linkage analysis. Am J Hum Gene. 1996;58:1268–1278. [PMC free article] [PubMed] [Google Scholar]
- Zakharia F, Basu A, Absher D, et al. Characterizing the admixed African ancestry of African Americans. Genome Biol. 2009;10:R141–R151. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rowe WL, Clark AG, Buetow KH. Genome wide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am J Hum Genet. 2003;73:1073–1081. doi: 10.1086/379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, McKenzie CA, Forrester T, et al. Localization of a small genomic region associated with elevated ACE. Am J Hum Genet. 2000;67:1144–1153. doi: 10.1016/s0002-9297(07)62945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Bouzekri N, Southam L, et al. Linkage and association analysis of angiotensin I-converting enzyme (ACE)-gene polymorphisms with ACE concentration and blood pressure. Am J Hum Genet. 2001;68:1139–1148. doi: 10.1086/320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.