Abstract
The immunoglobulin heavy (Igh) chain locus is subject to precisely regulated processes, such as variable region gene formation through recombination of variable (VH), diversity (DH), and joining (JH) segments, class switching and somatic hypermutation. The 3’ regulatory region (3’ RR) is a key regulator of the Igh locus, and, as revealed by deletions in mouse plasma cell lines and mice, is required for IgH expression as well as class switching. One of the mechanisms by which the 3’ RR regulates its targets is through long-range physical interactions. Such interactions between elements of the 3’ RR and a target site in the IgH transcription unit have been detected in plasma cells, and in resting and switching B cells, where they have been associated with IgH expression and class switching, respectively. Here, we report that lentiviral shRNA knockdown of transcription factors, CTCF, Oct-2, or OBF-1/OCA-B, had no discernible defects in loop formation or H chain expression in plasma cells. JH-3’ RR interactions in pre-B cell lines were specifically associated with IgH expression. JH-3’ RR interactions were not detected in either Pax5-deficient or RAG-deficient pro-B cells, but were apparent in an Abelson-derived pro-B cell line. These observations imply that the 3’ RR has different loop interactions with target Igh sequences at different stages of B cell development and Igh regulation.
Keywords: Chromosome conformation capture (3C), immunoglobulin gene rearrangements, immunoglobulin heavy chain gene expression, long-range enhancer interactions, lentivirus-mediated shRNA, B cell development
1. Introduction
During B cell maturation and activation, immunoglobulin heavy chain (IgH) genes are subject to DNA rearrangements required for VDJ joining and class switch recombination (CSR), and to VH gene somatic hypermutation (SHM) (reviewed in (Dudley et al., 2005). These processes are tightly regulated by cis elements of the Igh locus, including V region promoters, I/switch region promoters, the intronic enhancer (Eμ) and the hs sites of the 3’ regulatory region (3’ RR), several of which are associated with the four known enhancers (hs3a, hs1.2, hs3b and hs4) (Cogne and Birshtein, 2004). Additional 3’ RR hs sites contain binding sites for Pax5 and CTCF and display their accompanying insulator activity (hs5-7 and “38”) (Fig. 1A) (Chatterjee et al., 2011; Garrett et al., 2005). Functionally, the 3’ RR is important for immunoglobulin heavy chain gene expression in plasma cells and for class switching in activated B cells, as revealed by mouse knockout models (Pinaud et al., 2001; Vincent-Fabert et al., 2010). A contribution of the 3’ RR to SHM was revealed by a reduction in SHM resulting from a deletion of the entire 3’ RR enhancer region from a BAC transgenic mouse (Dunnick et al., 2009).
Fig. 1. Effect of CTCF on Ig expression and 3C interactions in MPC11 plasma cells.
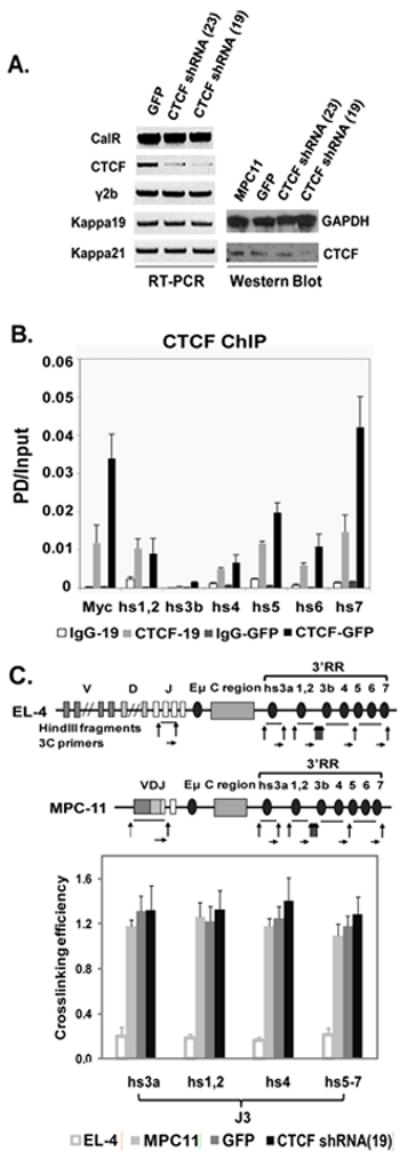
A. CTCF mRNA (RT-PCR) and protein levels (Western blot) in CTCF shRNA treated MPC11 plasma cells, with GFP- infected MPC11 cell lines as a control. MPC11 H (γ2b) and L(kappa 19 and 21) chain expression were unchanged. Calreticulin (CalR) housekeeping gene and GAPDH were used for normalization of mRNA and protein levels, respectively. B. Effect of CTCF KD on ChIP analysis of CTCF binding to 3’ RR enhancers and insulator region. CTCF binding, as measured by PD (pulldown)/input, was reduced by lentiviral shRNA, as shown in the gray bars. Note that the predominant CTCF binding sites are in hs5-7 (Chatterjee et al., 2011). The 5’ end of the c-myc gene was used as a positive control. Levels of binding with non-CTCF specific IgG are also shown. C. Effect of CTCF KD on 3C interactions involving JH3 and 3’ RR. Schematic maps of immunoglobulin heavy chain (IgH) loci in EL-4 T cells and MPC11 plasma cells used for 3C analysis. Vertical arrows indicate HindIII cleavage sites. Horizontal arrows indicate the primers for 3C assay. 3C interactions between J3 and 3’ RR elements were analyzed in EL-4 T cells, MPC11 cells, and MPC11 cells infected with GFP or CTCF shRNA 19. No changes were observed with CTCF KD.
Chromosome conformation capture (3C) studies have identified loops that juxtapose 3’ RR enhancers with target Igh sequences, such as JH sequences associated with the IgH transcription unit. In switching cells, additional interactions of the 3’ RR with I/switch region promoters have been identified (Wuerffel et al., 2007; Yan et al., 2011). That JH-3’ RR interactions occur in splenic B cells but not in splenic T cells indicates that these interactions are B cell-specific. Loss of H chain expression in a plasma cell line (Ju et al., 2007) or defects in CSR (Wuerffel et al., 2007), each resulting from deletions of 3’ RR enhancers, showed concomitant defects in loop formation. Hence, loop interactions involving the 3’ RR are implicated in these roles for expression and recombination. Additional 3’ RR loop interactions have been identified, one through DICE elements at the VH promoter, which is also associated with Igh expression in a plasma cell line (Ren et al., 2011), and a second with CTCF sites upstream of DFL16.1 in pro-B cells that is associated with Igh locus contraction (Degner et al., 2011) and predicted to impact on VDJ joining.
The transcription factor CTCF has been shown to contribute to loop formation in various loci. Initially identified as a regulator of the chicken c-myc promoter (Lobanenkov et al., 1990), CTCF was subsequently found to be involved in insulation of enhancers from promoters, potentially regulating boundaries between active and silent chromatin (rev. in (Phillips and Corces, 2009)). CTCF sites identified in the imprinted control region (ICR) regulate H19/Igf2 imprinting through loop-formation as measured by chromosome conformation capture (3C) experiments (Kurukuti et al., 2006), and similarly, contribute to MHC locus regulation (Majumder and Boss, 2010). CTCF also plays a role in looping in the β-globin locus, but apparently without a critical role in gene expression (Splinter et al., 2006). Recently, CTCF has been shown to affect Igh locus contraction apparently through an impact on loop interactions (Degner et al., 2011).
Octamer binding motifs are found in many Igh regulatory elements, including V gene promoters, Eμ and the 3’ RR. Together with the co-factor OBF-1, Oct-1 or Oct-2 binds to octamer motifs in B cells and exerts its function. Oct-1 is ubiquitously expressed while Oct-2 is expressed only in B cells and neuronal cells. Although B cell development in fetal liver is not affected in Oct-2 knockout mice, these mice die soon after birth for unclear reasons (Corcoran et al., 1993). OBF-1 (also known as OCA-B and Bob-1) expression is B cell-specific, and OBF-1 knockout mice show reduced levels of IgG isotypes and no germinal center formation (Schubart et al., 1996). Oct-2 and OBF-1 have been shown to regulate 3’ RR activity in B cells (Tang and Sharp, 1999). In human t(14;18) lymphoma cells, the bcl2 gene is involved in a translocation with the Igh locus, and there is an interaction between the translocated bcl2 gene and the 3’ RR. The contribution of Oct-2 to the bcl2-3’ RR interaction was revealed by siRNA knockdown of Oct-2 (Duan et al., 2007). Recent studies have shown a role for OBF-1/OCA-B in interactions between hs4 and DICE sequences downstream of the VH promoter (Ren et al., 2011)
In this report, we find that loop interactions involving the 3’ RR and JH target sequences in a plasma cell line, although critical for Igh expression, do not rely on CTCF, Oct-2 or OBF-1. JH-3’ RR loop interactions detected on the single VDJ rearranged and expressed allele in a pre-B cell line imply their association with Igh gene expression. The analysis of JH-3’RR interactions in pro-B cell sources gives insight into the ontogeny of 3’ RR interactions with the IgH transcription unit during early stages of B cell development. Collectively, these studies monitor the engagement of the 3’ RR with the IgH transcriptional unit during B cell development and suggest that the 3’ RR participates in a variety of functional loops.
2. Materials and Methods
2.1. B cell lines
Murine EL-4 T cells (ATCC #TIB-39) were used as a control for cell type specificity. The mouse pro-B cell line 63-12 (Shinkai et al., 1992), pre-B cell lines 3-1 and 70Z/3 (Nelson et al., 1983; Paige et al., 1978) were maintained in RPMI-1640 medium with 10% fetal calf serum; plasma cell line MPC11 (Laskov and Scharff, 1970) was cultured in DMEM medium with 10-15% fetal calf serum. All cells were grown at 37°C in an atmosphere of 5% CO2. 63-12 cells reconstituted with RAG-2 protein were a generous gift from Dr. Kathryn Calame (Columbia University) (Angelin-Duclos and Calame, 1998). Pro-B cells derived from a Pax5-/- mouse and transduced with a Pax5 expression construct coupled with estrogen receptor (ER) (Pax5-ER (B1.1)) (Nutt et al., 1998) were a generous gift from Dr. Meinrad Busslinger (Research Institute of Molecular Pathology, Vienna, Austria), and were received from Dr. Paolo Norio (Montefiore Medical Center, Bronx, NY). Pax5-ER cells were maintained on a layer of ST2 stromal cells (irradiated at 1100 rad) in the presence of IL-7 (1 ng/ml). Upon induction with 4-hydroxytamoxifen (H7904, Sigma), Pax5 protein was expressed. After 48 hr of culture, ~40% CD19+ cells (CD19 is a Pax5 target gene) were detected by FACS (FACScan, BD Biosciences).
2.2. Primary cells
CD19+ pro-B cells were isolated from the bone marrow of RAG1-/- mice (The Jackson Laboratory) by positive selection with anti-CD19 magnetic beads (Miltenyi Biotec). The purity of the pro-B cells was ~90%, as detected by FACS. C57BL/6 splenic B cells and DBA splenic B cells were purified by CD43 depletion (Miltenyi Biotec). The purity of both splenic B cell populations (B220+) was ~95%, as detected by FACS.
2.3. Gene knockdown by lentiviral shRNA
106 293T (human kidney epithelial) cells were transfected with packaging plasmids (pMDLg/pRRE, pRSV/Rev and pMD2.VSV.G) and vector plasmid pLKO.1 (Sigma Aldrich), which contains a specific shRNA (Mission). For each desired targeting, multiple shRNA’s were tested, and the most efficacious were used. These were: CTCF: TRCN0000039019, termed 19 in text, NM_007994.1- 10301c1, CCGGGCAGAGAAAGTAGTTGGTAATCTCGAGATTACCAACTACTTTCTCTGCTTTTTG; Oct-2: RCN0000081518, termed 1518 in text, NM_011138.1-1503s1c1, CCGGCTTAGTAACCTCGCCTCTCTTCTCGAGAAGAGAGGCGAGGTTACTAAGTTTTTG, and OBF-1/OCA-B: TRCN0000086513, termed 6513 in text, NM_011136.1-1089s1c1, CCGGGCGGCAAATGATAGTGATATTCTCGAGAATATCACTATCATTTGCCGCTTTTTG. Calcium phosphate-precipitated plasmid DNA was applied to 293T cells that have been treated by chloroquine. Supernatants containing lentiviral particles were collected 24 hr and 48 hr after transfection. A GFP lentiviral shRNA was generated as a control following the same procedure. Virus was concentrated by ultra-centrifugation (SW28) at 30,000 rpm for 2.5 hrs. Concentrated virus was stored at -80°C, and a small aliquot of GFP virus was used to calculate viral titer. Low speed spinoculation was used for lentiviral infection at an MOI (multiplicity of infection) of 20:1. After 48 hr of infection, puromycin was added and cells were maintained for as long as two weeks to obtain a stable cell line. The minimum concentrations of puromycin to kill all the uninfected cells were determined for each cell source in advance of lentivirus infection.
2.4. RT-PCR
2 μg total RNA was applied in each reverse transcription reaction using the Superscript III First-Strand Synthesis System (Invitrogen). After reverse transcription, 50 ng cDNA was used for PCR amplification for 25 to 27 cycles. Primer sequences are listed in Table I.
Table I.
Primer sequences
RT-PCR primers:
|
3C primers:
|
2.5. Western blot
15 μg nuclear extracts was loaded onto a 10% pre-cast mini acrylamide gel (Bio-Rad), and subjected to electrophoresis for 2 hrs at 100 v prior to transfer to nitrocellulose membrane (Amersham Bioscience). After blocking with 10% fat-free milk in tris-buffered saline tween-20 (TBS-T), the membrane was incubated with primary antibodies: anti-CTCF, Upstate 07-729; anti-GAPDH, sc25778, Santa Cruz Biotechnology; anti-OBF-1, rabbit specific antiserum, a generous gift from Dr. Robert Roeder, Rockefeller University, through Dr. Hilda Ye, Albert Einstein College of Medicine; or anti-TBP (TFIID), sc-273, Santa Cruz Biotechnology. The membrane was washed twice with TBS-T, incubated with goat anti-rabbit IgG-HRP-conjugated secondary antibody, sc2004, Santa Cruz Biotechnology, and washed four times with TBS-T. The membrane was developed using the Amersham ECL Western Blot Analyzing System (GE healthcare).
2.6. Chromatin immunoprecipitation (ChIP)
ChIP assays were performed and quantitated as described previously (Garrett et al., 2005). Briefly, cells were fixed with formaldehyde at RT after which cells were lysed and nuclei were sonicated to yield ~200-100bp fragments. Pre-cleared chromatin from ~4×106 cells was taken for each pull down. ChIPs were done with 4 mg of antibodies to CTCF (Upstate 07-729) with 4 mg of rabbit IgG (sc-2027) used as a negative control. 30 ml of salmon sperm protein A+G agarose beads (Upstate) were used to immunoprecipitate the antibody-chromatin complex. The complex was washed several times prior to elution of bound chromatin by elution with SDS and reversal of protein-DNA crosslinks. Chromatin was then digested with proteinase-K and DNA was extracted by Qiagen PCR Purification kit (Cat # 28104). DNA was diluted to a concentration of 4.95 ng (3000 copies)/ml, and a standard curve of CT vs the log of copy number from successive 5-fold dilutions was obtained for each primer using Real Time PCR analysis. Primers for the 3’ RR and controls were previously described (Garrett et al., 2005). For assay of ChIP, a 2% dilution of chromatin was used as input and each pull down (PD) and corresponding input were analyzed in triplicate and calculated for copy number. The results were plotted as the ratio of PD/input (Y axis). ChIP experiments were normalized by comparison to a positive control whenever possible.
2.7. 3C assay
The procedure was performed as previously described (Ju et al., 2007). Briefly, 107 cells were cross-linked with formaldehyde for 10 min at room temperature. Nuclei were prepared, and non-cross-linked protein was removed by SDS. Triton X-100 was added to sequester the SDS, after which samples were digested with 1,500 units of HindIII overnight at 37°C with gentle shaking. The digestion was terminated in the presence of SDS at 65°C. Digested nuclei (106 nuclei) were subjected to intramolecular ligation with T4 DNA ligase. Samples were then treated with Proteinase K and RNase A, and DNA was purified by phenol:chloroform extraction and isopropanol precipitation. One hundred nanograms of ligated DNA samples were analyzed by 32 to 34 cycles of PCR (94°C/30 s, 55°C/30 s, and 72°C/30 s) in a 25 μl reaction system. These conditions give amplifications in linear range (Ju et al., 2007). 3C primer sequences are listed in Table I. A control template was generated by mixing DNA from BACs that contain sequences of interest with fragments of the calreticulin (CalR) gene in equal molar ratios. This control template was subjected to HindIII digestion and to DNA ligation and was used to normalize PCR efficiency among different primer pairs. PCR products were subjected to electrophoresis on a 2% gel, and gel signals (S) were analyzed by SYNGENE GENE genius BioImaging System. Crosslinking efficiency between two HindIII fragments was calculated as previously, namely (SIgH/SCalR in selected cell type)/(SIgH/SCalR in control template).
3. Results and Discussion
3. 1 Neither CTCF nor Oct-2 or OBF-1 is essential for 3’ RR-JH long-range interactions of IgH expression in MPC11 cells
3.1.1 CTCF
To address whether CTCF is essential for 3’ RR-JH interactions and for IgH expression in the MPC11 plasmacytoma cell line, we utilized lentivirally-cloned shRNA to silence CTCF expression. Of the two shRNAs tested, the 19 shRNA (Materials and Methods) was the more efficient at silencing, achieving an ~70% knockdown as shown by RT-PCR and Western blot (Fig. 1A). ChIP analysis of wild-type and CTCF-KD cells showed considerable although incomplete reduction of CTCF binding to the 3’ RR (Fig. 1B). However, no significant change in either heavy chain or light chain expression was detected in CTCF-KD cells (Fig. 1A), and 3C data showed that there was no alteration in loop-formation (Fig. 1C). It should be noted that knockdowns of CTCF at these levels with comparable residual binding to the 3’ RR as measured by ChIP resulted in measurable differences in locus contraction and 3C interactions between the 3’ RR/CTCF sites and DFL16.1/CTCF sites (Degner et al., 2011). We concluded that 3’ RR-JH loops in MPC11 cells are not critically dependent on these levels of CTCF, implying that these interactions require less CTCF than those between this region and DFL16.1/CTCF, or that proteins other than or in addition to CTCF are essential. Of interest are studies that show that it is not CTCF (Splinter et al., 2006) but rather cohesin (Chien et al., 2011), which binds at both CTCF and non-CTCF sites in the β-globin LCR and globin target genes, that is essential for β-globin expression.
3.1.2 Oct-2 and OBF-1/OCA-B
We addressed the contribution of Oct-2 to loops involving the 3’ RR and the IgH transcription unit in the mouse Igh locus in MPC11 by using a lentiviral shRNA technique to KD Oct-2 expression. RT-PCR showed that Oct-2 mRNA expression was considerably reduced by 1518 shRNA (see Materials and Methods) (Fig. 2A). However, neither IgH nor Igκ expression was affected. In addition, there were no detectable changes in Oct-1 expression. Considering that Oct-1 might functionally compensate for the loss of Oct-2, we treated MPC11 cells with shRNA to target OBF-1/OCA-B (see Materials and Methods), a coactivator of both Oct-1 and Oct-2 and essential for VH promoter-driven expression (Luo and Roeder, 1995). RT-PCR and Western blot analysis showed that OBF-1 mRNA expression was substantially reduced by 6513 shRNA (see Materials and Methods) (Fig. 2B); and as was shown in Oct-2 KD cells, neither IgH nor Igκ expression was affected. 3C experiments also showed no significant differences in loop formation in either Oct-2- or OBF-1- silenced MPC11 cells (Fig. 2C). The level of KD achieved in our studies was estimated to be ~85% (Fig. 2B), enabling us to infer that normal IgH expression and loop formation are achieved by as low as 15% of normal levels of OBF-1/OCA-B and Oct-2.
Fig. 2. Effect of Oct-2 and OBF-1 on Ig expression and 3C interactions in MPC11 plasma cell line.
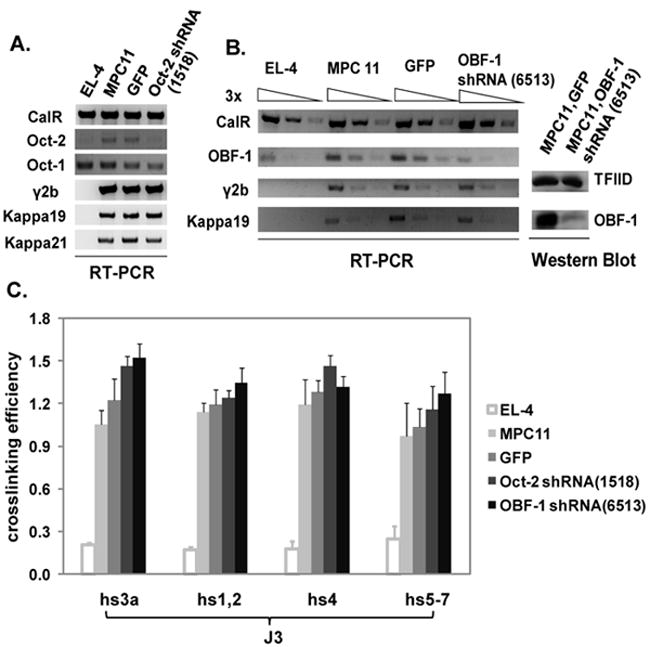
A. RT-PCR detection of Oct-2 mRNA levels in GFP or Oct-2 shRNA-infected MPC11 cells. The levels of Oct-1 expression in the GFP- or Oct-2 shRNA treated cells are equivalent and both reduced as compared to the untreated MPC 11 cells. The levels of MPC11 H (γ2b) and L(kappa 19 and 21) chain expression were unchanged. Calreticulin (CalR) housekeeping gene was used for normalization of mRNA levels. EL-4 T cells were used for comparison. B. OBF-1 mRNA and protein levels were assessed in OBF-1 shRNA-treated MPC11 cells by RT-PCR (by three-fold dilutions) and Western blot, respectively. TFIID was used for protein normalization. C. 3C interactions were measured in EL-4 T cells, MPC11 cells, and GFP or Oct-2/OBF-1 shRNA infected MPC11 cells. No differences in interactions were observed with the KDs.
3.1.3. Other studies involving OBF-1 in IgH expression in a plasma cell line
Recent studies have reported that KD of OBF-1/OCA-B in MPC11 cells resulted in a reduction of Igh expression and 3C interactions (Ren et al., 2011), in clear contrast to our observations. The most likely explanation is differences in the efficiency of the KDs. Our studies used single shRNA’s, respectively, to KD OBF-1/OCA-B and Oct-2, and involved considerable cell death during the process of obtaining puromycin-resistant cell lines. Studies by Ren et al used sequential shRNAs (X. Ren and R. Roeder, personal communication). Hygromycin-resistant cell lines with the lowest OBF-1/OCA-B expression were initially selected and then reinfected by lentivirus carrying a second shRNA with puromycin as the selectable marker. Doubly resistant cell lines with the lowest OBF-1/OCA-B expression level were chosen for analyses. Although levels of KD reported by Ren appear to have been similar to ours (Ren et al., 2011), a double KD might be predicted to result in additional reduction in OBF-1/OCA-B expression, thereby crossing a threshold for an effect. It is interesting to consider the possibility that the double KD affected not only the well-characterized OBF-1/OCA-B, but also a predicted and partially redundant OCA-B2 (R. Roeder, personal communication). Notably, even with the double KD approach, there was substantial remaining IgH expression and loop formation, requiring real-time RT-PCR to detect changes in IgH gene expression (X. Ren and R. Roeder, personal communication).
Reporter assays in human B cells have shown that hs4 is regulated by different contributions of NFκB and octamer-binding proteins in different cell lines (Michaelson et al., 1996). In accord with a model proposed by Ren et al (Ren et al., 2011), residual IgH expression in double KD MPC11 cells could result from compensation through the NFκB-mediated pathway: partially in the double KD and essentially fully in the single KD. In this respect, the MPC11 cell line has frequently been shown to generate drug-induced or spontaneous variants that can affect H chain expression (Birshtein et al., 1974; Koskimies and Birshtein, 1976) or loop formation (Ju et al., 2007). In both instances, i.e. as reported here, and by Ren et al (Ren et al., 2011), there was considerable selection pressure, potentially resulting in variant formation.
Collectively, these observations suggest that loop formation and high levels of IgH transcription in MPC11 cells are dependent on synergy or cooperation among multiple factors, such as CTCF, Oct-2 or OBF-1/OCA-B (or Oct-1), or require other, as yet, unidentified, factors.
3.2 Long-range JH-3’ RR interactions occur on the expressed but not unexpressed allele in a pre-B cell line
As noted in the introduction, 3’ RR loop interactions important for IgH expression in plasma cells have been reported (Ju et al., 2007) as have those for class switch recombination in B cells (Wuerffel et al., 2007). To determine whether long range JH-3’ RR interactions associated with IgH expression occurred before the B cell stage of maturation, we analyzed the 3-1 pre-B cell line. Our experiments showed that JH-3’ RR interactions in 3-1 cells were equivalent in magnitude to those detected in mature B cells (Fig. 3A). Analysis of the 70Z/3 pre-B cell line permitted us to determine whethr loop interactions were associated with IgH expression. The 70Z/3 pre-B cell line contains an expressed, VDJ rearranged allele, and a non-expressed, DJ recombined allele. 70Z/3 was generated from C57BL/6 X DBA mice (Fig. 3). The expressed IgH allele (C57BL/6) has a VDJH1 rearrangement, together with a spontaneous deletion of the region containing 3’ RR enhancers hs3a and hs1,2. The IgH non-producing allele, derived from DBA mice, has a DHJH2 rearrangement, and possesses an intact 3’ RR (Saleque et al., 1999). Based on these findings, we were able to identify primer pairs to uniquely track the individual alleles. Because JH1 is retained only on the expressed IgH allele, JH1-hs3b or -hs4 interactions are specific for the expressed VDJ allele; and because hs3a and hs1.2 are present only on the unexpressed allele, JH3-hs3a or JH3-hs1.2 interactions are specific for the non-expressed DJ rearranged allele. 3C data showed that JH3-hs3a or JH3-hs1,2 interactions in 70Z/3 cells (both deriving from the non-expressed allele) were at a low level similar to those detected in splenic T cells and substantially less than those detected in splenic B cells from DBA mice (Fig. 3B). In contrast, there are significant JH1-hs4 interactions (deriving from the expressed allele), which are at even higher levels than those detected in normal B cells. The relative reduction in signal in normal B cells likely reflects JH usage and the location of HindIII sites in the JH genomic regions, HindIII being the enzyme that is used to separate genomic fragments as part of the 3C method. JH1 and JH2 are separated by one HindIII site, and JH3 and JH4 by a second HindIII site. Thus, a JH3 primer amplifies VDJ rearrangement to both JH2 and JH3 during PCR, while the JH1 primer amplifies only a VDJH1 rearrangement.
Fig. 3. Analysis of 3C interactions in pre-B cell lines.
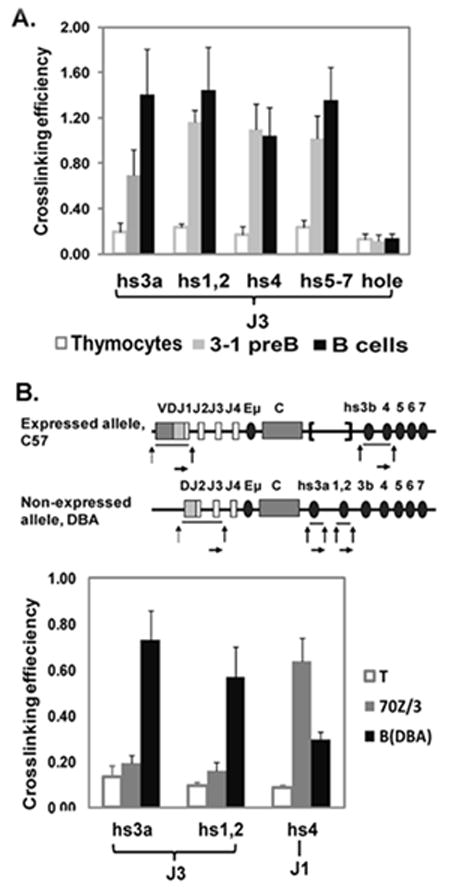
A. JH3-3’RR interactions were analyzed in thymocytes, 3-1 pre-B cells, and splenic B cells. Hole, the nearest non-Ig gene downstream of the 3’ RR, was used as a negative control. B. A schematic description of DJ/VDJ rearrangements on expressed (C57BL/6) and non-expressed (DBA) IgH alleles in 70Z/3 pre-B cell line. The J1 and hs4 primers were paired to detect 3C interaction only on the expressed allele. J3 and hs3a/hs1,2 were paired to detect 3C interactions only on the non-expressed allele. 3C interactions were measured in T cells from DBA mice (negative control), 70Z/3 pre-B cells, and DBA splenic B cells (positive control). Specific interactions were observed on the expressed allele of 70Z/3 pre-B cells.
These data indicating that long-range interactions occur on the expressed allele, but not on the non-expressed allele in 70Z/3 pre-B cells, suggest that JH-3’ RR interactions are associated with transcription rather than recombination.
3.3 Long range JH-3’ RR interactions are detected in a pro-B cell line, but not in primary pro-B cells
We then analyzed long-range JH-3’ RR interactions in an Abelson virus-induced pro-B cell line, 63-12, derived from a RAG-2 deficient mouse (Shinkai et al., 1992). JH-3’ RR interactions were clearly evident in this cell line (Fig. 4A), which because it lacks RAG proteins, cannot undergo DJ or VDJ joining. To determine whether RAG protein expression might increase loop formation, we compared 63-12 with 63-12 cells that were reconstituted with RAG-2 (Angelin-Duclos and Calame, 1998) (Fig. 4A). The levels of interactions were unchanged.
Fig. 4. Analysis of JH-3’ RR interactions involving the 3’ RR in pro-B cells.
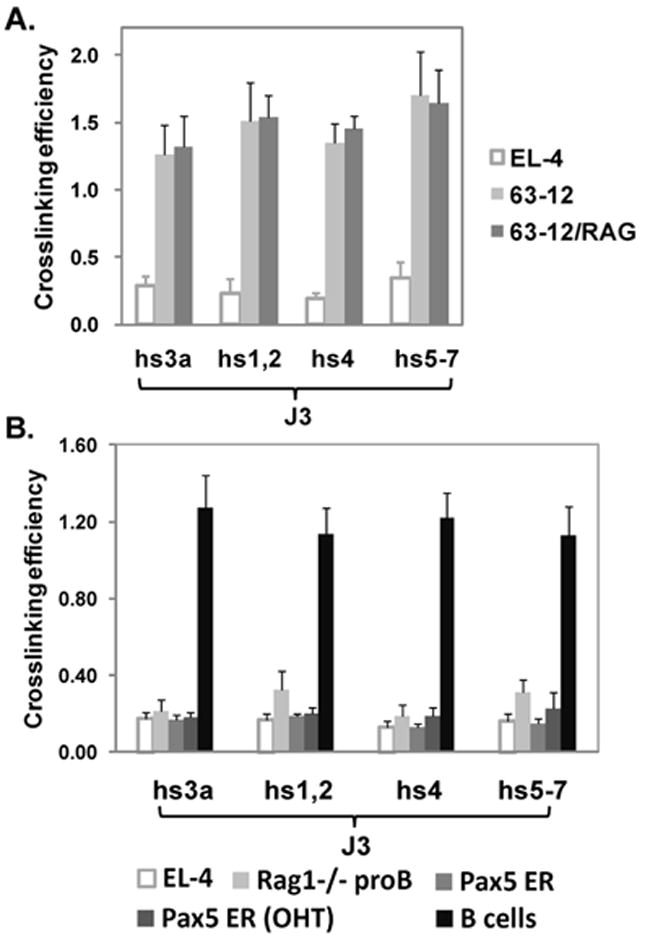
A. 3C interactions in 63-12 RAG2 -/- pro-B cell line, and in 63-12 cells reconstituted with RAG2 protein. B. JH3-3’RR interactions measured in EL-4 T cells, RAG1-/- pro-B cells, Pax5 ER (B1.1 cells), Pax5 ER (OHT), and B cells. Interactions were observed in B cells.
However, JH-3’ RR interactions were not detected in other sources of pro-B cells. CD19+ pro-B cells isolated from bone marrow of RAG1-/- mice showed only baseline levels of interaction similar to those detected in EL-4 thymocytes (Fig. 4B). Pax5 deficient mice provide another source of pro-B cells. In Pax5 knockout mice (Urbanek et al., 1994), B cells do not develop at all in fetal liver, and B cell development in the bone marrow is arrested at the pro-B cell stage at the proximal VH to DJ joining step prior to locus contraction and accessibility of distal VH genes (Fuxa et al., 2004). Similar to RAG1-/- pro-B cells, an estrogen-sensitive Pax5 inducible pro-B cell line, Pax5-ER (see Materials and Methods), showed only baseline levels of JH3-3’ RR interactions in the absence of Pax5 expression (Fig. 4B); and these levels were unchanged even after induction of nuclear localization and expression of Pax5-ER with tamoxifen (Nutt et al., 1998) when ~40% of the assayed cells were CD19 positive.
4. Overview
Other long-range interactions involving 3’ RR sequences have been observed in pro-B cells. In addition to the especially high density of CTCF sites in hs5-7 of the 3’ RR (Garrett et al., 2005), CTCF sites are located throughout the V region and are associated with the DFL16.1 segment (Degner et al., 2011; Degner et al., 2009; Featherstone et al., 2010). CTCF-dependent interactions between the 3’ RR/CTCF region and CTCF sites in DFL16.1 (Degner et al., 2011) have been detected both in E2a-/- pre-pro B cells and RAG1-/- pro-B cells. DFL16.1 also interacts with Eμ in the IgH transcription unit at a lower level and in a CTCF-independent manner (Degner et al., 2011). These data raise the possibility that different sets of interactions are present in different subsets of pro-B cells. 3’ RR-DFL16.1 interactions have been associated with locus contraction and are predicted to impact on VDJ joining (Degner et al., 2011). We propose that the interaction between JH and 3’ RR is independently regulated and related to the architecture needed for H chain expression.
In the 70Z/3 pre-B cell line where we can distinguish the expressed (VDJ) allele from the unexpressed (DJ) allele, JH-3’RR interactions are detected only on the expressed allele, and are, therefore, predicted to be associated with H chain expression. Similar interactions are associated with IgH expression in plasma cells (Ju et al., 2007). The finding of 3’ RR-JH interactions in 63-12 pro-B cells raises the possibility that interactions can occur prior to VDJ joining and provide a structure for supporting subsequent IgH transcriptional expression once VDJ joining has taken place. It should be noted that analyses carried out in cell lines and normal cells may show differences that relate to cellular transformation of cultured cells or to particular stages of differentiation that are captured by the culture adaptation process.
Collectively, these studies identify several different loop interactions involving the 3’ RR as an anchor. 3’ RR-DFL16.1 interactions associated with locus contraction and potentially with VDJ joining are detected in pro-B cells, while 3’ RR-JH interactions associated with heavy chain transcription are detected in the 63-12 Abelson-virus transformed pro-B cell line, and in pre-B and plasma cells. Interactions with I/switch regions are important for class switch recombination (Chatterjee et al., 2011; Wuerffel et al., 2007; Yan et al., 2011). The identification of proteins that facilitate these interactions is in progress.
Highlights.
>JH-3’RR interactions are essential for IgH expression in plasma cells. >These interactions can be independent of CTCF or Oct-2/OBF-1. > JH-3’RR interactions on the expressed allele in a pre-B cell line. >JH3-3’RR interactions detected in Abelson virus-transformed pro-B cell line. >JH3-3’RR interactions not detected in RAG-/- or Pax5-/- pro-B cells.
Acknowledgments
Supported by NIH RO1AI13509. We thank Drs. Sabrina Volpi, Sergio Roa and Alexander V. Emelyanov of Albert Einstein College of Medicine for helpful discussions of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angelin-Duclos C, Calame K. Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment. Mol Cell Biol. 1998;18:6253–64. doi: 10.1128/mcb.18.11.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birshtein BK, Preud’homme JL, Scharff MD. Variants of mouse myeloma cells that produce short immunoglobulin heavy chains. Proc Natl Acad Sci U S A. 1974;71:3478–82. doi: 10.1073/pnas.71.9.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Ju Z, Hassan R, Volpi SA, Emelyanov AV, Birshtein BK. Dynamic changes in binding of immunoglobulin heavy chain 3’ regulatory region to protein factors during class switching. J Biol Chem. 2011;286:29303–12. doi: 10.1074/jbc.M111.243543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk DA, Kong X, Ball AR, Jr, Calof AL, Lander AD, Groudine MT, Yokomori K. Cohesin mediates chromatin interactions that regulate mammalian {beta}-globin expression. J Biol Chem. 2011;286:17870–8. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogne M, Birshtein BK. In: Regulation of class switch recombination. Honjo T, Alt FW, Neuberger M, editors. Elsevier Academic Press; San Diego, CA: 2004. pp. 289–305. [Google Scholar]
- Corcoran LM, Karvelas M, Nossal GJ, Ye ZS, Jacks T, Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993;7:570–82. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108:9566–71. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–8. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Heckman CA, Boxer LM. The immunoglobulin heavy-chain gene 3’ enhancers deregulate bcl-2 promoter usage in t(14;18) lymphoma cells. Oncogene. 2007;26:2635–41. doi: 10.1038/sj.onc.1210061. [DOI] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P, Papavasiliou FN. Switch recombination and somatic hypermutation are controlled by the heavy chain 3’ enhancer region. J Exp Med. 2009;206:2613–23. doi: 10.1084/jem.20091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone K, Wood AL, Bowen AJ, Corcoran AE. The mouse immunoglobulin heavy chain V-D intergenic sequence contains insulators that may regulate ordered V(D)J recombination. J Biol Chem. 2010;285:9327–38. doi: 10.1074/jbc.M109.098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–22. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3’ end of the igh locus involves modular regulation of histone modifications during B-cell development and in vivo occupancy at CTCF sites. Mol Cell Biol. 2005;25:1511–25. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, Volpi SA, Hassan R, Martinez N, Giannini SL, Gold T, Birshtein BK. Evidence for physical Interaction between the immunoglobulin heavy chain variable region and the 3’ regulatory region. J Biol Chem. 2007;282:35169–78. doi: 10.1074/jbc.M705719200. [DOI] [PubMed] [Google Scholar]
- Koskimies S, Birshtein BK. Primary and secondary variants in immunoglobulin heavy chain production. Nature. 1976;264:480–2. doi: 10.1038/264480a0. [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskov R, Scharff M. Synthesis, assembly, and secretion of gamma globulin by mouse myeloma cells. I. Adaptation of the Merwin plasma cell tumor-11 to culture, cloning, and characterization of gamma globulin subunits. J Exp Med. 1970;131:515–541. doi: 10.1084/jem.131.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5’-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–53. [PubMed] [Google Scholar]
- Luo Y, Roeder RG. Cloning, functional characterization, and mechanism of action of the B- cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–24. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Boss JM. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol Cell Biol. 2010;30:4211–23. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JS, Singh M, Snapper CM, Sha WC, Baltimore D, Birshtein BK. Regulation of 3’ IgH enhancers by a common set of factors, including kappa B-binding proteins. J Immunol. 1996;156:2828–39. [PubMed] [Google Scholar]
- Nelson KJ, Haimovich J, Perry RP. Characterization of productive and sterile transcripts from the immunoglobulin heavy-chain locus: Processing of mm and ms mRNA. Mol Cell Biol. 1983;3:1317–1332. doi: 10.1128/mcb.3.7.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Morrison AM, Dorfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 1998;17:2319–33. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige CJ, Kincade PW, Ralph P. Murine B cell leukemia line with inducible surface immunoglobulin expression. J Immunol. 1978;121:641–647. [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogne M. Localization of the 3’ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–99. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- Ren X, Siegel R, Kim U, Roeder RG. Direct Interactions of OCA-B and TFII-I Regulate Immunoglobulin Heavy-Chain Gene Transcription by Facilitating Enhancer-Promoter Communication. Mol Cell. 2011;42:342–55. doi: 10.1016/j.molcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Singh M, Birshtein BK. Ig heavy chain expression and class switching in vitro from an allele lacking the 3’ enhancers DNase I-hypersensitive hs3A and hs1,2. J Immunol. 1999;162:2791–803. [PubMed] [Google Scholar]
- Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–42. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–67. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Sharp PA. Transcriptional regulation of the murine 3’ IgH enhancer by OCT-2. Immunity. 1999;11:517–26. doi: 10.1016/s1074-7613(00)80127-2. [DOI] [PubMed] [Google Scholar]
- Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–12. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogne N, Cogne M, Denizot Y. Genomic deletion of the whole IgH 3’ regulatory region (hs3a, hs1,2, hs3b, hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116:1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S Synapsis during class switch recombination Is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;22:711–22. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Pieretti J, Ju Z, Wei S, Christin JR, Bah F, Birshtein BK, Eckhardt LA. Homologous elements hs3a and hs3b in the 3’ regulatory region of the murine immunoglobulin heavy chain (Igh) locus are both dispensable for class-switch recombination. J Biol Chem. 2011;286:27123–31. doi: 10.1074/jbc.M111.230995. [DOI] [PMC free article] [PubMed] [Google Scholar]


