Abstract
Epigenetic mechanisms integrate signals from diverse intracellular transduction cascades and in turn regulate genetic readout. Accumulating evidence has revealed that these mechanisms are critical components of ongoing physiology and function in the adult nervous system, and are essential for many cognitive processes, including learning and memory. Moreover, a number of psychiatric disorders and syndromes that involve cognitive impairments are associated with altered epigenetic function. In this review, we will examine how epigenetic mechanisms contribute to cognition, consider how changes in these mechanisms may lead to cognitive impairments in a range of disorders and discuss the potential utility of therapeutic treatments that target epigenetic machinery. Finally, we will comment on a number of caveats associated with interpreting epigenetic changes and using epigenetic treatments, and suggest future directions for research in this area that will expand our understanding of the epigenetic changes underlying cognitive disorders.
Keywords: learning and memory, neurodegeneration, neuroprotection, drug discovery, development, epigenetics
INTRODUCTION
Changes to the epigenome are a key mechanism by which cells initiate, maintain, and terminate gene expression programs, making them potent modulators of cell function and activity. Recent evidence suggests that epigenetic modifications within the CNS are critical for short and long-term behavioral adaptation to a wide range of environmental stimuli. As such, epigenetic modifications represent candidate mechanisms for the creation and maintenance of behavioral memories at multiple levels. These and other studies have generated interest in the therapeutic potential of drugs capable of enhancing or impairing these reactions in cognitive disorders. Recent advances in our understanding of the role of epigenetic molecular mechanisms in CNS development and function have allowed the conceptualization of epigenetically based CNS disorders. It is now clear that mutations to epigenetic machinery are the root cause of a number of clinical syndromes, including Rett syndrome, Rubenstein–Taybi syndrome, Fragile X mental retardation, and Angelman syndrome (Driscoll et al, 1992; Petrij et al, 1995; Amir et al, 1999; Alarcon et al, 2004; Penagarikano et al, 2007; Urdinguio et al, 2009). Moreover, a number of psychiatric disorders, such as drug addiction, schizophrenia, and Alzheimer's disease, are associated with aberrant epigenetic changes, suggesting that epigenetic modifications may hold promise as potential therapeutic targets in these disorders (Tsankova et al, 2007; Renthal and Nestler, 2008; Graff and Mansuy, 2009; Bredy et al, 2010; Penner et al, 2010a, 2010b).
Epigenetic mechanisms are broadly defined as a set of regulatory modifications that influence gene expression above the level of the genome itself, that is, without changes in gene sequence. Within a cell nucleus, DNA is condensed within a three-dimensional structure called chromatin that regulates the ability of transcriptional machinery to access a specific gene site and initiate transcription (Russo et al, 1996). Thus, this complex forms a general template for the activation and repression of genetic material. The basic structural unit of chromatin is the nucleosome, which is made up ∼147 bp of DNA wrapped around four pairs of basic histone proteins (H2A, H2B, H3, and H4). These histone proteins contain amino terminal ‘tails' that protrude from the nucleosomal complex and contain multiple sites for potential modification (Kouzarides, 2007). Canonically, epigenetic mechanisms would include post-translational modification at the tails of these histone proteins, as well as direct chemical modification of DNA itself in which a methyl group is added to cytosine nucleotides (Russo et al, 1996). In mammals, DNA methylation can occur at CpG sites in the genome, which are vastly underrepresented in general but which are found in clusters (termed CpG islands) near ∼60% of gene promoters (Weber et al, 2005, 2007).
Although both histone modifications and DNA methylation were once viewed as inherently stable mechanisms incapable of rapid change, a wealth of recent data suggests that this is not the case, even for putatively irreversible modifications such as DNA methylation (Swank and Sweatt, 2001; Weaver et al, 2004; Miller and Sweatt, 2007; Kangaspeska et al, 2008; Lubin et al, 2008; Metivier et al, 2008; Koshibu et al, 2009; Gupta et al, 2010; Miller et al, 2010). These findings build upon the long appreciated role for these mechanisms and their upstream regulators in transcriptional output and suggest that epigenetic mechanisms in non-mitotic and terminally differentiated neurons may represent a final common pathway for the alterations in neuronal function that produce long-term behavioral change.
This manuscript will review how epigenetic modifications occur and how drugs that target these modifications may be useful in the treatment of cognitive disorders. We will start by examining histone modifications, then turn to DNA methylation, and finally mention non-canonical mechanisms like proteins with prion-like activity and microRNAs. Finally, we will suggest some future directions and potential challenges for the study of epigenetics in cognitive disorders.
HISTONE MODIFICATIONS
Histone Acetylation
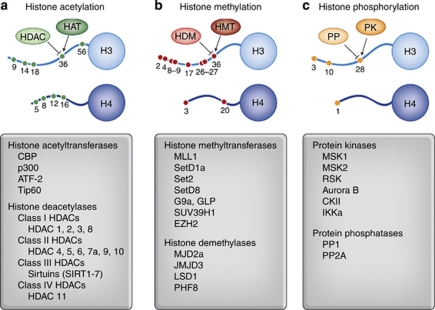
Perhaps the most well-understood epigenetic modification is the acetylation of lysine residues on histone N-terminal tails. This modification is typically associated with transcriptional activation, as it can physically relax the positive charge between the histone tail and the DNA backbone, enabling chromatin to unravel and transcriptional machinery to gain access to DNA (Russo et al, 1996; Clayton et al, 2006; Kouzarides, 2007). Additionally, the acetylation of lysine residues can lead to binding of bromodomain-containing proteins, which can recruit transcriptional activators (Dyson et al, 2001). Histone acetylation occurs via the activity of histone acetyltransferases (HATs), which include CREB-binding protein (CBP) and p300 (see Figure 1) (Ogryzko et al, 1996; McManus and Hendzel, 2003). Acetyl groups are removed from lysine residues by histone deacetylases (HDACs), a large family of proteins, which are organized into four different classes that differ in expression profiles across brain regions (Kouzarides, 2007; Renthal et al, 2007).
Figure 1.
Summary of well-understood histone modifications and histone-modifying enzymes. (a) Histone acetylation at numerous lysine residues on histone tails is catalyzed by histone acetyltransferases (HATs) and removed by histone deacetlyases (HDACs). Histone acetylation is generally a transcriptionally permissive mark. Different HAT and HDAC enzymes are listed below. Importantly, specific HDACs isoforms are differentially expressed across brain structures and appear to uniquely regulate different aspects of cognition. (b) Histone methylation at lysine and arginine residues on histone tails is catalyzed by histone methyltransferases (HMTs) and removed by histone demethylases (HDMs). Histone methylation at different amino acid residues has been linked to both transcriptional activation and transcriptional repression. Methylation can occur in mono-, di-, or even tri-methylated states. Many HDMs and HMTs are specific for modifications at individual amino acids on histone tails or even a specific number of methyl groups. (c) Histone phosphorylation at serine residues is catalyzed by protein kinases (PKs) such as mitogen- and stress-activated protein kinase 1 (MSK1), whereas phosphorylation marks are removed by protein phosphatases such as protein phosphatase 1 (PP1). Histone phosphorylation is generally linked to transcriptional activation.
A wealth of research suggests that histone acetylation has a critical role in cognitive abilities such as learning and memory. Thus, behavioral paradigms that induce learning increase histone acetylation in the brain areas that regulate those specific types of learning. For example, exposing rodents to a contextual fear conditioning paradigm induces a significant increase in histone acetylation of multiple lysine residues on H3 and H4 within the hippocampus (Levenson et al, 2004; Chwang et al, 2006; Peleg et al, 2010). Impairing histone acetylation by knocking out the HAT CBP produces an impairment in memory formation as well as its cellular correlate, long-term potentiation (LTP) (Alarcon et al, 2004). Likewise, increasing histone acetylation by blocking the activity of HDACs improves memory formation and enhances the development of LTP in hippocampal slices (Levenson et al, 2004; Vecsey et al, 2007; Stefanko et al, 2009; McQuown et al, 2011). Importantly, these changes occur in concert with signal transduction mechanisms and transcription factors that are known to regulate learning and memory. Thus, blocking extracellular signal-regulated kinase (ERK) prevents increases in histone acetylation following contextual fear learning, whereas activation of NMDA receptors increases histone acetylation in an ERK-dependent manner (Levenson et al, 2004). Likewise, the CREB-binding protein CBP possesses HAT activity, and proper CREB function is required for the HDAC inhibitor trichostatin-A (TSA) to enhance fear memory and hippocampal LTP (Vecsey et al, 2007).
Since these discoveries, a key challenge has been to understand how HDAC inhibitors like trichostatin-A, sodium butyrate, and suberoylanilide hydroxamic acid (SAHA) enhance memory and whether they globally increase expression of all genes or are limited in action to specific genes. Given that both long-lasting hippocampal LTP and long-term memory require gene transcription and new protein synthesis (Frey et al, 1988, 1996; Alberini, 2008), it is not surprising that HDAC inhibitors require gene transcription to be an effective enhancer of LTP in the hippocampus (Levenson et al, 2004; Vecsey et al, 2007). Although it may be expected that treatment with HDAC inhibitors before behavioral conditioning would indiscriminately enhance all gene products (or at least boost a number of memory-associated genes), this does not appear to occur in vivo. Rather, treatment with TSA increases expression of a limited set of memory-induced genes, including the inducible nerve growth factor-B (Vecsey et al, 2007).
Importantly, the efficacy of HDAC inhibitors may also be caused by action at specific HDAC isoforms. Thus, compared with HDAC1, HDAC2 binding is enhanced at multiple memory and plasticity-associated genes such as BDNF, Egr1, and CREB, and CaMKII. Accordingly, overexpression of HDAC2 (but not HDAC1) in mice impairs hippocampal LTP and memory formation (Guan et al, 2009). Likewise, genetic deletion of Hdac2 increases dendritic spine density, enhances hippocampal LTP, and improves fear memory formation (Guan et al, 2009). Nevertheless, additional studies have shown that focal deletion or pharmacological inhibition of HDAC3 (a class 1 HDAC) in the dorsal hippocampus results in increased expression of Nr4a2 and c-Fos genes, and improves long-term memory retention on an objection location memory task (McQuown et al, 2011). In contrast, other reports have indicated that the sirtuins, a separate family of NAD+-dependent HDACs, may positively regulate learning and memory. Thus, mice lacking SIRT1 activity in the brain exhibit impaired performance on a number of learning and memory tasks, as well as impaired hippocampal LTP (Gao et al, 2010). This deficit occurs because SIRT1 normally represses expression of a microRNA, miR-134, that when expressed leads to downregulation of the memory-related genes CREB and BDNF (Gao et al, 2010). Together, these data reveal that multiple HDAC isoforms contribute to memory formation and do so in unique ways.
A number of learning and memory disorders and syndromes are associated with impaired histone acetylation. For example, Rubenstein–Taybi syndrome, a disorder characterized by numerous physical defects and behavioral deficits, is caused by mutations in the gene for CBP (Petrij et al, 1995). Moreover, the results discussed above indicate that HDAC inhibitors enhance memory formation and could therefore potentially form the basis of pharmacotherapies for disorders of learning and memory, including age-associated cognitive decline and Alzheimer's disease (Abel and Zukin, 2008; Chuang et al, 2009; Penner et al, 2010a). Although this latter possibility is only beginning to be explored in human patients, the validity of this idea has already been confirmed in animal models. Thus, in a rodent model of spatially restricted neurodegeneration, the HDAC inhibitor sodium butyrate (as well as an enriched environment) boosted associative and spatial learning and memory, and even improves access to memories formed before neurodegeneration (Fischer et al, 2007). Similarly, the aging process induces a significant learning impairment in mice, which is associated with a decrease in acetylation at lysine 12 on H4 in the hippocampus and an inability to generate learning-related increases in expression of memory-related genes. However, intrahippocampal infusions of the HDAC inhibitor SAHA restored H4K12 acetylation and significantly improved memory function (Peleg et al, 2010). Moreover, a variety of HDAC inhibitors have also been shown to reverse learning deficits in a mouse model of Alzheimer's disease (Kilgore et al, 2010). In contrast, activation of the HDAC SIRT1 by resveratrol prevents learning deficits and signs of neurodegeneration and tauopathy in a separate Alzheimer's mouse model (Kim et al, 2007). This result is consistent with findings that SIRT1 boosts (rather than impairs) memory formation (Gao et al, 2010), and reveals a distinct pathway for therapeutic manipulation in the context of learning and memory disorders (Bonda et al, 2011).
Histone acetylation has also been implicated in drug addiction, a disorder characterized by chronic and persistent relapse in drug taking despite adverse consequences. Repeated exposure to addictive drugs like cocaine is associated with long-term changes in brain reward circuits, specifically the nucleus accumbens (NAc). Specifically, cocaine produces an increase in histone acetylation at the promoter regions of plasticity genes within the NAc (Kumar et al, 2005), and treatment with HDAC inhibitors concurrent with drug experience enhances the development of conditioned place preferences for cocaine and morphine (Kumar et al, 2005; Sanchis-Segura et al, 2009). Likewise, overexpression of HDAC isoforms (specifically the striatally enriched isoforms HDAC5) impairs the development of drug place preference (Renthal et al, 2007). In contrast, cocaine place preference learning and cocaine self-administration are reduced by intra-NAc infusions of a sirtuin antagonist, indicating that this distinct class of HDACs regulates cocaine reward in a manner distinct from other HDAC classes (Renthal et al, 2009). Although these studies reveal that histone acetylation is potentially important for the development of drug addictions, the goal of treatment would be to reverse drug-related behavioral preferences. Consistent with this idea, a recent study found that HDAC inhibition during extinction of cocaine place preference significantly facilitated behavioral extinction and reduced subsequent cocaine reinstatement of a place preference (Malvaez et al, 2010). Therefore, this result indicates that treatment with HDAC inhibitors, in combination with behavioral therapy, may be useful in preventing drug relapse.
Beyond learning and memory, HDAC inhibitors have also been investigated for a number of other disorders that include cognitive components of psychiatric disorders, including depression, schizophrenia, Parkinson's disease, and anxiety disorders (Tsankova et al, 2007; Chuang et al, 2009; Bredy et al, 2010). However, there are also several potential issues concerning the use of HDAC inhibitors for cognitive disorders. First and foremost, the systemic use of these drugs can produce considerable side effects (Bruserud et al, 2007). This may be due to the fact that most available HDAC inhibitors are not isoform-selective, but bind with roughly equal affinity to distinct HDAC proteins in the same family (Kilgore et al, 2010). As discussed above, the memory-enhancing effects of HDAC inhibition is likely isoform-specific, as HDAC2 and HDAC3 have larger roles in the regulating memory formation than HDAC1 (Guan et al, 2009; McQuown et al, 2011). Additionally, side effects may occur as a result of off-target effects, as HDAC inhibitors (in addition to inhibiting acetylation of histones) also act to reduce acetylation at a host of other non-histone proteins (Drummond et al, 2005). Going forward, it will be important to develop new isoform-specific HDAC inhibitors, which will allow for specific functions to be targeted while others remain unperturbed. This is presently an area of intense focus in pharmaceutical drug discovery.
Histone Methylation
Unlike histone acetylation, lysine, and arginine methylation on histone tails has been associated with both transcriptional activation and transcriptional repression, depending largely on the specific lysine residue and the histone protein (ie, H3 or H4). Histone methyltransferases (HMTs), such as G9a and SUV39H1, catalyze histone methylation, but not in a global manner as appears to be the case for HATs (Greiner et al, 2005). Thus, G9a methylates lysine 9 on H3 (H3K9), which is generally a mark of repressed transcription (Tachibana et al, 2001, 2008). Likewise, MLL1 methylates H3K4, which is associated with transcriptional activation (Akbarian and Huang, 2009). Similarly, enzymes that remove methyl groups from lysine residues, called histone demethylases (HDMs) also exhibit substrate specificity, with JHDM1 removing methyl groups from H3K36 and LSD1 demethylating H3K9 (Shi et al, 2004; Tsukada et al, 2006; Shi and Whetstine, 2007). Indeed, part of the promise for using HMTs and HDMs as a therapeutic treatment is that the writers and erasers of this mark are exceptionally specific.
Unlike other histone modifications, histone methylation is not distinctly an either/or modification. In fact, lysine residues can be mono-, di-, or even tri-methylated, with each distinct methyl group addition producing unique results (Scharf and Imhof, 2010). These differential states are produced by tight regulation of histone methylation machinery, as HMTs and HDMs are known to catalyze the addition or removal of different numbers of methyl groups. For example, the HDM LSD1 (also known as KDM1) requires a protonated lysine to function, and therefore cannot remove methyl groups from a trimethylated lysine (Stavropoulos et al, 2006). In addition, it is clear that proteins which contain chromodomains (and therefore bind to methylated lysine) can possess different affinities for methylation levels at a specific histone target, thereby conferring unique methylation states with unique functional consequences (Shi and Whetstine, 2007; Scharf and Imhof, 2010).
A role for histone methylation in cognition is increasingly being appreciated. Within the hippocampus, trimethylation of H3K4 and dimethylation of H3K9 are both increased immediately after contextual fear conditioning (Gupta et al, 2010). This is interesting given that, as stated above, these marks are associated with opposite transcriptional regulation. Thus, this finding likely indicates that these modifications occur at gene-specific targets, whereby they can differentially control expression of memory associated genes. Importantly, histone methylation also appears to be functionally relevant for memory formation, as mice that lack Mll (an H3K4 specific methyltransferase) exhibit impaired contextual fear conditioning (Gupta et al, 2010). Likewise, mice with neuronal deletion of the HMT G9a/GLP (an H3K9 specific methyltransferase) exhibit impaired motor behavior, decreased motivation to consume palatable sucrose, and severe learning and memory deficits (Schaefer et al, 2009). Additionally, this methyltransferase also regulates cocaine reward and cocaine-induced neuronal plasticity in the striatum (Maze et al, 2010). Together, these emerging results indicate that treatments that increase histone methylation at specific sites may be useful candidates for a range of learning and memory disorders. However, aberrant HDM signaling also appears to be relevant for human disorders, as mutations in the JARID1c gene (a lysine demethylase) have been associated with autism and X-linked mental retardation (Tzschach et al, 2006; Iwase et al, 2007; Adegbola et al, 2008). Currently, our understanding of these modifications and their role in cognitive disorders remains in its infancy. Thus, future research will be required to elucidate the complex functional consequences of altered histone methylation and determine which sites and which histone methylation enzymes are good candidates for therapeutic intervention.
Histone Phosphorylation
A third histone modification that has been shown to regulate cognitive processes is phosphorylation of serine residues on histone tails (Figure 1c). This mark, normally associated with transcriptional activation, is catalyzed by a range of protein kinases (Berger, 2007; Deng et al, 2008). This group includes the mitogen- and stress-activated protein kinase 1 (MSK1), which is activated downstream of the ERK/MAP kinase pathway. Conversely, histone phosphorylation is reversed by protein phosphatases PP1 and PP2a, which are known to be inhibited by other molecular cascades including dopamine and cyclic-AMP regulated phosphoprotein 32 (DARPP32). Perhaps the most well-characterized phosphorylation mark occurs at serine 10 on H3. This modification recruits GCN5, which contains HAT activity and therefore increases acetylation at neighboring lysine residues K9 and K14 and repressing histone methylation at H3K9 (Fischle et al, 2005). In addition to recruiting HATs, H3S10 phosphorylation enhances transcription factor binding by modifying the interaction between DNA and the histone tail (Cheung et al, 2000).
Several studies have revealed a role for histone phosphorylation at H3S10 in regulating memory formation. Mutations in the gene encoding RSK2, which has been shown to phosphorylate H3, produces Coffin–Lowry syndrome, an X-linked disorder that is associated with psychomotor retardation and physical abnormalities (Delaunoy et al, 2001; Merienne et al, 2001). In animal models, H3S10 phosphorylation and H3S10/K14 phosphoacetylation increases rapidly in the hippocampus following contextual fear conditioning, and these increases are blocked by ERK inhibition (Chwang et al, 2006). Likewise, mice lacking MSK1 exhibit impaired fear conditioning and spatial memory (Chwang et al, 2007). Interestingly, this deficit is not reversed by treatment with an HDAC inhibitor, revealing that the histone phosphorylation pathway occurs in parallel to (rather than downstream of) histone acetylation. Accordingly, inhibition of nuclear PP1, the major histone phosphatase, improves long-term object recognition memory and spatial memory without affecting short-term memory (Koshibu et al, 2009). Together, these findings suggest that enhancing histone phosphorylation via inhibition of PP1 may be a distinct and even complementary treatment for learning and memory disorders. However, this remains to be examined experimentally.
In addition to learning and memory, changes in H3S10 phosphorylation have also been implicated in behavioral responses to drugs of abuse. Thus, cocaine induces increases in H3S10 phosphorylation in the striatum and MSK1 knockout mice display impaired behavioral responses to cocaine administration (Brami-Cherrier et al, 2005). Importantly, this change occurs downstream of phosphorylation and nuclear accumulation of DARPP-32, which influences behavioral responses to both cocaine and natural rewards like sucrose (Stipanovich et al, 2008). Histone phosphorylation has also been identified as a major downstream target of anti-psychotic and anti-Parkinsonian drugs, revealing major therapeutic potential in the development of compounds that specifically target this epigenetic modification (Bertran-Gonzalez et al, 2008, 2009; Santini et al, 2009). Interestingly, the protein kinase MSK1 is found mainly in neurons and is enriched in several brain regions, including the striatum, amygdala, and hippocampus (Heffron and Mandell, 2005). This cellular and regional selectivity indicates that MSK1 may be a good candidate for therapeutic intervention in drug addiction.
Other Histone Modifications: Ubiquitination and Poly-ADP Ribosylation
Although the three modifications reviewed above (acetylation, methylation, and phosphorylation) represent the bulk of ongoing research in the area of epigenetic control in cognitive processes, a number of additional histone modifications can occur in vivo and regulate many aspects of cellular signaling and function. Among these are histone ubiquitination, poly-ADP ribosylation, sumoylation, and O-GLCNAcylation. Below we will consider two specific modifications (ubiquitination and poly-ADP ribosylation), which through recent discoveries have been linked to cognition.
Histone ubiquitination occurs at all four histone proteins, but is most well characterized at the carboxy terminus of H2A and H2B. It is regulated by a three-step enzymatic process in which the enzyme E1 first activates ubiquitin, and the ubiquitin-carrier enzyme E2 directs ubiquitin to the substrate. Finally, ubiquitin is linked to a lysine residue by the E3 ubiquitin ligase family of enzymes (Wang et al, 2004; Higashi et al, 2010). Likewise, a wide family of deubiquitinases (DUBs) remove ubiquitin from lysine residues (Atanassov et al, 2010; Higashi et al, 2010). Although histone ubiquitination is known to have a wide range of cellular functions, most notably controlling transcriptional initiation and elongation (Zhu et al, 2007). In fact, it is known that ubiquitination/deubiquitination enzymes interact with other epigenetic machinery, and thus correlate (or are even prerequisites for) other histone marks, specifically histone methylation (Jason et al, 2002; Lee et al, 2007; Sadri-Vakili et al, 2007; Zhu et al, 2007). However, perhaps the clearest link between histone ubiquitination and neurodegenerative impairments comes from Huntington's disease, which is characterized by a mutation in the gene-encoding huntingtin protein and aberrant transcriptional regulation. Interestingly, huntingtin interacts with the ubiquitin ligase hPRC1L, suggesting a role for histone ubiquitination in Huntington's disease (Kim et al, 2008). Moreover, ubiquitinated H2A is increased in a mouse model of Huntington's disease, whereas ubiquitinated H2B is decreased, resulting in modified histone methylation patterns and transcriptional dysregulation (Kim et al, 2008). Accordingly, targeting ubiquitin ligases may be a potential therapeutic avenue for patients with Huntington's disease.
Poly-ADP ribosylation of histones occurs at all histone proteins as well (Quenet et al, 2009; Messner et al, 2010), and is catalyzed by Poly (ADP-ribose) polymerases such as PARP1 and PARP2, with NAD+ acting as a substrate. Likewise, poly-ADP ribosylation is reversed by poly (ADP-ribose)glycohydrolase (Quenet et al, 2009). Interestingly, although poly-ADP ribosylation and PARP1 activity are most well known for their role in DNA repair following damage, PARP1 activation has been shown to increase in cortical neurons following depolarization (Homburg et al, 2000). Likewise, PARP1 is required for long-term synaptic facilitation and long-term memory in aplysia, suggesting a functional role in the nervous system (Cohen-Armon et al, 2004; Hernandez et al, 2009). Likewise, in mouse models, poly-ADP ribosylation is increased at the histone linker protein H1 following novel object exposure, and PARP1 is required for hippocampal LTP and novel object recognition memory (Fontan-Lozano et al, 2010). These findings indicate that, in addition to the mechanisms reviewed above, histone poly-ADP ribosylation may be an interesting target for novel therapeutics for learning and memory disorders.
DIRECT COVALENT MODIFICATION OF DNA
Methylation and Demethylation
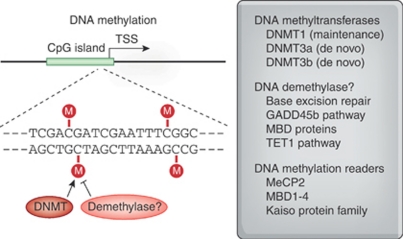
Methylation of cytosine nucleotides on DNA itself is the so-called ‘fifth-base' of DNA and the ‘prima donna' of the epigenetic landscape (Santos et al, 2005). This modification is catalyzed by DNA methyltransferases (DNMTs), which are divided into two groups (see Figure 2). DNMT1 is a maintenance methyltransferase, meaning that it recognizes hemi-methylated DNA and initiates methylation of the complementary strand. In contrast, DNMT3a and 3b are de novo methyltransferases, which can methylate both unmethylated and hemimethylated DNA (Klose and Bird, 2006). The methyl donor for this reaction is S-adenosyl methionine. Methylated cytosine residues serve as a binding site for a variety of methyl-binding domain (MBD) proteins (Ho et al, 2008), with MeCP2 being the prototypical MBD (Nan et al, 1997). When bound, MeCP2 can direct both transcriptional repression and transcriptional activation in different contexts (Chahrour et al, 2008).
Figure 2.
DNA methylation and demethylation. A majority of mammalian gene promoters contain dense clusters of cytosine-guanine dinucleotides called CpG islands, at which methylation can occur to dramatically influence gene transcription. In this example, the CpG island (green bar) overlaps the transcription start site. At CpG dinucleotides, methylation is catalyzed by DNA methyltransferases (DNMTs). De novo DNMTs direct the methylation of unmethylated CpGs, whereas maintenance DNMTs recognize hemi-methylated DNA and methylate the complementary strand. The existence of a direct demethylase is controversial, but a number of different mechanisms have been proposed to regulate removal of the methyl moiety, including excision and replacement of the entire base pair. DNA methylation marks are read by a family of proteins with methyl binding domains (MBD proteins), which includes MeCP2. Each of these targets may represent candidates for therapeutic treatments of disorders characterized by aberrant DNA methylation. See text for additional details.
The general chemical reaction of DNA methylation suggests that it should be static and long lasting. Direct removal of the methyl moiety from cytosine is extremely difficult given the strength of the carbon-carbon bond. In cases where a methyl group is passively removed from one strand, maintenance DNMT activity will recognize hemi-methylated DNA, leading again to double-stranded methylation (Ma et al, 2009a). Therefore, this modification is seemingly ideal for the long-term perpetuation of cellular phenotype, gene imprinting, and silencing of repetitive DNA elements (Dulac, 2010). However, despite their clear roles in embryonic development and cellular proliferation and differentiation (Wu and Zhang, 2010), DNMTs and MBD proteins are also found in relatively high levels in postmitotic, non-proliferating neurons (Goto et al, 1994; Dulac, 2010). This fact alone suggests that DNA methylation may have an active function beyond the static role in cellular phenotype that had long been assumed.
DNA Methylation in Learning and Memory
Interestingly, the general form of the chemical reaction, which maintains DNA methylation is precisely what memory biochemists hypothesized would be necessary for the perpetuation of long-term memory traces (Razin and Friedman, 1981; Crick, 1984; Lisman, 1985). Thus, the ability of maintenance DNMTs to recognize hemi-methylated DNA and re-methylate the complementary strand ensures that this modification could remain stable over time despite ongoing molecular turnover within a cell, even in the absence of the initiating agent (Day and Sweatt, 2010). Consistent with this idea, a number of reports over the past several years have indicated that DNA methylation does indeed have a key role in memory formation and other types of long-term behavioral change. Initial studies revealed that contextual fear conditioning increases DNMT3a expression and induces changes in DNA methylation at the promoters of several key plasticity genes in the hippocampus (Miller and Sweatt, 2007; Lubin et al, 2008). These changes correspond with gene expression profiles, as increases in DNA methylation at the promoter for the memory suppressor gene PP1 was associated with decreased gene expression, whereas decreased DNA methylation at promoters for memory-enhancing genes such as reelin and BDNF were associated with increases in the expression of these genes. Moreover, blocking DNMT activity in the hippocampus with distinct DNMT inhibitors zebularine, 5-aza-deoxycytidine, or RG-108 immediately after learning impaired the consolidation of conditioned freezing (Miller and Sweatt, 2007; Lubin et al, 2008).
It is important to note that the use of DNMT inhibitors comes with several caveats. First, both zebularine and 5-aza-deoxycytidine are nucleoside analogs that operate by first incorporating into DNA, a process that is thought to require cell division (Szyf, 2009). Thus, the mechanism of these drugs in non-dividing neurons is unclear. However, RG-108, which blocks the active site on DNMTs, has a well-understood mechanism and is therefore the preferred compound. A second caveat is that there is no evidence that the activity of these drugs is limited to neurons, and in fact this is likely not the case. Nevertheless, additional studies using genetic tools have revealed that neuron and forebrain-specific deletion of both DNMT isoforms impairs contextual fear conditioning, spatial memory, and hippocampal LTP (Feng et al, 2010). Together, these results reveal that changes in DNA methylation in adult neurons are critical transcriptional components of the memory consolidation process. However, these studies also reveal that very few of these learning-induced changes in DNA methylation in the hippocampus are enduring, suggesting that these changes alone do not form the basis of a molecular memory switch.
In contrast, DNA methylation changes within the cortex and in response to other types of behavioral experience have been found to be long lasting. For example, contextual fear conditioning induces sustained increases in promoter methylation and a corresponding downregulation of the calcineurin gene within the anterior cingulate cortex. Additionally, inhibition of DNA methylation via three distinct compounds (RG-108, zebularine, and 5-aza-deoxycytidine) prevents the remote maintenance of a previously formed memory, suggesting that long-term DNA methylation changes are necessary for the persistence of behavioral memory (Miller et al, 2010). Likewise, differences in maternal care in infants produces enduring changes in DNA methylation at several gene targets and in several brain regions (including the hippocampus) that are maintained well into adulthood (Weaver et al, 2004, 2005; Roth et al, 2009). Thus, changes in DNA methylation are characterized by regional differences as well as temporally distinct responses to different environmental events.
One interesting finding from these studies is that in addition to increases in DNA methylation at one set of genes, learning experiences just as often result in decreases in methylation at another set of genes. Furthermore, very rapid increases in methylation often return back to baseline levels. In combination, these results appear to conflict with previous assertions that DNA methylation is always a stable and self-perpetuating epigenetic modification. However, several recent reports indicate that DNA methylation is much more cyclical than previously assumed (Kangaspeska et al, 2008; Metivier et al, 2008), leading to the hypothesis that a separate mechanism was governing rapid demethylation of DNA (Ma et al, 2009a; Wu and Zhang, 2010). A number of distinct mechanisms have now been proposed for this demethylation activity, most notably control of DNA demethylation by the growth arrest and DNA damage inducible protein 45 (GADD45) protein family (Ma et al, 2009a, 2009b; Wu and Zhang, 2010). Importantly, these possible mechanisms indicate that DNA methylation is a two-way street, and reveal a number of novel candidates for potential therapeutic regulation of DNA methylation (see Figure 2).
DNA Methylation in Rett Syndrome
Aberrant DNA methylation and changes in DNA methylation machinery or binding proteins have been implicated in a number of cognitive disorders. The prototypical example of this is Rett syndrome, which is an X-linked disorder associated with progressive mental retardation in females (Amir et al, 1999; Wan et al, 1999). The vast majority of Rett syndrome cases are caused by either sporadic or germline mutations in the MeCP2 gene, and the specific nature of these mutations correlates highly with phenotypic outcomes (Amir et al, 1999; Amir and Zoghbi, 2000). Given that MeCP2 binds selectively to methylated DNA, this indicates that aberrant readout of DNA methylation is a critical component of the cognitive impairments observed in this syndrome.
Various aspects of Rett syndrome impairments have now been recapitulated in animal models. For example, a range of studies using mice with selective mutations in the MeCP2 gene have revealed that loss of MeCP2 function produces deficits in spatial memory, contextual and cued fear conditioning memory, and deficits on social interaction tests (Shahbazian et al, 2002; Moretti et al, 2005, 2006; Pelka et al, 2006; Stearns et al, 2007). Likewise, MeCP2 mutant mice exhibit deficits in hippocampal LTP and LTD (Asaka et al, 2006; Moretti et al, 2006), revealing a physiological correlate for impaired learning and memory in Rett syndrome. Importantly, deficits observed in MeCP2 mutant mice may not completely represent developmental problems alone, but also disruption of neuronal function in the mature nervous system. Thus, selective deletion of MeCP2 in the adult basolateral amygdala induces impaired anxiety and learning and memory deficits (Adachi et al, 2009).
The most straightforward treatment for MeCP2 mutations in Rett syndrome would presumably be to overexpress or enhance the function of MeCP2. Along these lines, overexpression of MeCP2 results in enhanced fear conditioning and hippocampal LTP (Collins et al, 2004), revealing some therapeutic potential. However, given that MeCP2 function is in part linked to interaction with transcriptionally repressive HDACs (Nan et al, 1998; Chahrour et al, 2008), another possibility is to target HDAC function. Thus, deficits in learning and memory and hippocampal LTP induced by DNMT inhibitors can be rescued by pre-treatment with HDAC inhibitors (Miller et al, 2008). However, treatment with HDAC inhibitors in other brain regions like the basolateral amygdala can mimic the behavioral deficits associated with MeCP2 deletion in this area (Adachi et al, 2009), indicating that caution must be used when generalizing results from hippocampus-dependent memory tasks to memory functions supported by other brain structures.
DNA Methylation in Schizophrenia
A second psychiatric condition in which altered DNA methylation has long been implicated is schizophrenia (Pollin et al, 1961; Bredy et al, 2010). The first evidence for a role of DNA methylation in schizophrenia came from the observation that treatment with an S-adenosyl-methionine precursor -methionine, actually intensifies a number of schizophrenic symptoms (Pollin et al, 1961). Given that S-adenosyl-methionine is the methyl donor for the DNA methylation reaction, this result suggests that hypermethylation of DNA may contribute to schizophrenia. Consistent with this hypothesis, patients with schizophrenia exhibit hypermethylation at the synaptic plasticity gene reelin in the cortex, which is associated with decreased reelin expression (Grayson et al, 2005). Additionally, post-mortem samples from schizophrenic patients have revealed an overexpression of DNMT1 and DNMT3a in GABAergic neurons (Zhubi et al, 2009), which is consistent with the observation that DNMT1 regulates reelin methylation in vivo (Noh et al, 2005).
A number of drug treatments appear to be capable of reversing DNA hypermethylation (Szyf, 2009). Interestingly, the anti-psychotic drugs clozapine and sulpiride, which are both used for schizophrenia treatment in clinical settings, have been found to demethylate the reelin gene in vivo (Dong et al, 2008). Likewise, a number of HDAC inhibitors are capable of inducing demethylation in the brain, and co-application of the HDAC inhibitor valproate along with clozapine and sulpiride dramatically enhances the ability of these drugs to reverse reelin methylation (Dong et al, 2008, 2010; Szyf, 2009). Thus, treatment with HDAC inhibitors may provide a new avenue of treatment for schizophrenia, whether alone or in combination with existing pharmacotherapies. Finally, another obvious option for decreasing aberrant DNA methylation in schizophrenia is the DNMT inhibitors, such as zebularine, 5-aza-deoxycytidine, and RG108. However, the success of HDAC inhibitors and DNMT inhibitors in alleviation of schizophrenic symptoms remains to be investigated in human populations.
ADDITIONAL TARGETS
Epigenetic mechanisms are traditionally defined as mitotically and/or meiotically heritable changes in gene expression that do not involve changes in DNA sequence (Russo et al, 1996). This definition has undergone revision in recent years, in large part to include marks that occur at chromosomes in non-dividing cells such as neurons. Thus, a more inclusive definition considers epigenetic mechanisms to be as follows: ‘the structural adaptation of chromosomal regions so as to register, signal, or perpetuate altered activity states' (Bird, 2007). Although this definition includes the mechanisms discussed above, it excludes a number of additional molecular mechanisms that, while not considered ‘epigenetic' in the strict sense, are capable of exerting powerful control over gene expression or protein function, and therefore are of potential interest when considering therapies for cognitive disorders. Additionally, these mechanisms have considerable interplay with traditionally defined epigenetic mechanisms (Rouhi et al, 2008), such as DNA methylation and it is therefore important to address these mechanisms in the context of epigenetics.
MicroRNA
The first of these mechanisms derive from the activity of small RNA molecules, which include microRNAs, small interfering RNA, and small nuclear RNA. Small RNAs have multiple functions within a cell, including activation, repression, or interference with gene expression (He and Hannon, 2004), and have been implicated in a number of cognitive disorders. For example, microRNA binds to 3′ untranslated regions of messenger RNA and thereby either cleaves and degrades the messenger RNA or controls its expression through translational mechanisms (Bartel, 2004; Vasudevan et al, 2007). MicroRNA thereby control expression of the majority of genes within the genome, and represent a critical component of normal physiology and function in the developing and adult nervous system (Bartel, 2004; Miranda et al, 2006).
An example of the importance of microRNA regulation of gene expression comes from Fragile X syndrome, a commonly inherited disorder characterized by mental retardation and autism-like behavioral abnormalities. Fragile X is caused by a mutation in the FMR1 gene, which causes gene hypermethylation and resulting absence of its product, Fragile X mental retardation protein (FMRP) (Penagarikano et al, 2007). Interestingly, FMR1 is a RNA-binding protein that interacts with several microRNAs to regulate gene expression that is critical for neural development (Jin et al, 2004). In fact, FMR1's association with a specific microRNA, miR125b, has been shown to underlie NMDA receptor expression in mouse models, providing a specific mechanism for behavioral impairments in Fragile X syndrome (Edbauer et al, 2010).
Further implicating a role for microRNAs in psychiatric diseases, recent evidence has revealed that Alzheimer's Disease is associated with a altered expression of several microRNAs, some of which interact with the amyloid precursor protein's cleavage machinery, which has long been associated with this disease (Provost, 2010). Similarly, in an animal model of drug addiction, prolonged experience with cocaine self-administration dramatically increased expression of miR-212 in the striatum (Hollander et al, 2010). However, in this case, this overexpression was associated with a dampening of the motivational properties of the drug by modulating CREB pathways linked to the stimulatory effects of cocaine. These data, together with the substrate specificity of microRNAs, suggest that microRNA targets may be excellent therapeutic candidates for cognitive disorders.
Prion-Like Proteins
A second set of mechanisms, prion-like proteins, involves proteins capable of two conformational states. Interestingly, prion proteins are able to not only undergo a conformational change, but are also then capable of converting normal isoforms into the conformationally altered version. Prion proteins have recently been shown to have functional roles in memory maintenance, and therefore represent a potential therapeutic candidate for treatment of memory disorders. For example, cytoplasmic polyadenylation element binding protein (CPEB), which exhibits prion characteristics, is essential for synaptic facilitation in aplysia as well as normal memory function in both mice and drosophila (Si et al, 2003a, 2003b, 2010; Berger-Sweeney et al, 2006; Keleman et al, 2007; Miniaci et al, 2008), indicating that prion function may be a necessary and common component of synaptic plasticity. Likewise, another prion protein, PrPC, is critical for inhibitory avoidance memory in rats (Coitinho et al, 2006), and mice that overexpress PrPC fail to show age-related declines in social recognition memory (Rial et al, 2009). Together, these results reveal that prion proteins may have a normal functional role in the establishment of long-term memory.
FUTURE RESEARCH DIRECTIONS AND CONCLUSIONS
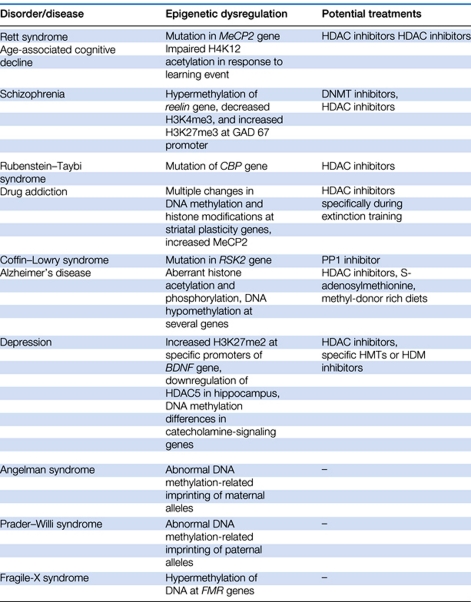
Taken together, the wide variety of findings reviewed above suggest that a range of cognitive disorders are characterized by aberrant epigenetic changes and that epigenetic drug targets may be useful for treatments for these disorders (see Table 1). However, interpreting these results comes with a number of caveats. For example, for a number of disorders discussed above, it remains unclear if epigenetic changes represent the root cause of the behavioral symptoms or instead simply reflective of other neuronal changes. As epigenetic mechanisms like histone acetylation and DNA methylation represent downstream integration templates for a number of signal transduction and second messenger cascades, it is possible that epigenetic alterations are only one of many functional outcomes resulting from altered intracellular signaling. An example of this is the specific impairment in learning-induced H4K12 acetylation in aged animals (Peleg et al, 2010). Conceptually, this deleterious lack of epigenetic function could result from dysregulation in many components of neuronal physiology, including neurotransmitter release, receptor function, and upstream molecular signaling. Therefore, continued investigation of whether epigenetic changes are the cause or outcome of specific cognitive disorders will enhance our ability to treat or in some cases prevent those disorders. However, even when altered epigenetic function is known to be the cause of a disorder, such as Rett syndrome, it is still unclear in many cases how this change plays out across thousands of gene targets and across functionally diverse brain structures. Thus, in addition to examining the epigenetic endpoints of a disorder, it remains a clear necessity to continue to evaluate how these endpoints affect gene expression and function of entire neural circuits.
Table 1. Selected List of Psychiatric Disorders and Syndromes with Epigenetic Origins or Treatments.
The use of drugs that alter epigenetic mechanisms also comes with a number of challenges. A major difficulty is that available epigenetic compounds such as HDAC inhibitors and DNMT inhibitors are not selective for specific brain regions, neuronal subtypes, or specific genes. This lack of selectivity becomes a key issue for treatment of disorders that are characterized by changes in epigenetic state at a small group of gene sites or impaired generation of a specific histone modification (eg, H3S10). For example, as mentioned above, schizophrenia is associated with hypermethylation of the reelin gene (Grayson et al, 2005). However, applying DNMT inhibitors systemically or even in a region-specific manner would presumably result not only in decreased reelin methylation, but also in decreased methylation of multitudes of other genes. Likewise, treatment with HDAC inhibitors, which are also capable of reversing reelin hypermethylation, would result in increased histone acetylation of other unrelated genes. A related challenge is that as histone modifications and DNA methylation work in concert to regulate transcription, altering one mechanism will consequently affect other mechanisms in a complex manner. In terms of epigenetic treatments, each of the challenges will likely produce a number of side effects that may be deleterious and prohibit widespread applications. Thus, future research will be required to develop more specific regulators of epigenetic machinery.
One promising avenue for the development of more specific epigenetic treatments is the HDACs. As these enzymes exist in many different isoforms (see Figure 1a) that differ in their expression across brain structures as well as their molecular functions, it may be possible to enhance spatial and genetic specificity by targeting only one HDAC isoform. For example, given that HDAC2 and HDAC3 have been found to negatively regulate learning and memory, it may be useful to create a compound, which targets these isoforms for learning and memory disorders (Guan et al, 2009; McQuown et al, 2011). Likewise, given the diverse range of specific HDM and methyltransferase enzymes, it may be worthwhile to investigate the manipulation of these enzymes in disorders characterized by aberrant histone methylation. However, in cases where this type of targeting may not be a possibility, such as the DNMT inhibitors, another strategy would be to focus on the specific factors and proteins that target histone and DNA-modifying enzymes to specific gene targets. This approach would ensure that resulting epigenetic changes are restricted to a smaller group of genes than global interference with epigenetic machinery.
Although the majority of epigenetic changes discussed in this review center upon individual modifications and their contribution to disease in a single lifetime, a final consideration for future research is the transgenerational inheritance of epigenetic alterations (Richards, 2006). Indeed, it has recently been appreciated that complex environmentally induced epigenetic changes may influence epigenetic state in offspring (Roth et al, 2009), providing a mechanism for non-genetic behavioral traits to be inherited. In addition, it is now clear that a large number of genes are imprinted in the brain (Gregg et al, 2010a, 2010b), resulting in favored expression of either the maternal or paternal allele. Likewise, it has long been understood that epigenetic mechanisms (in particular DNA methylation) regulate this imprinting process (Li et al, 1993). In our view, transcriptional inheritance of epigenetic states has the potential to revolutionize our understanding of disease vulnerability, specifically for behavioral disorders such as drug addiction, depression, and anxiety. However, future research will be required to tease apart the complex effects of environmental experience, parental history, and genetic makeup on epigenetic patterns. Nevertheless, along with the development of more specific epigenetic drugs, understanding how epigenetic patterns develop and how they affect diverse brain systems will greatly enhance our ability to treat and possibly even prevent cognitive disorders.
Acknowledgments
Research in the our laboratory is supported by funds from the NINDS, NIMH, NIA, NIDA, the Rett Syndrome Foundation, the Ellison Medical Foundation, and the Evelyn F McKnight Brain Research Foundation.
The authors declare no conflict of interest.
References
- Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi M, Autry AE, Covington HE, III, Monteggia LM. MeCP2-mediated transcription repression in the basolateral amygdala may underlie heightened anxiety in a mouse model of Rett syndrome. J Neurosci. 2009;29:4218–4227. doi: 10.1523/JNEUROSCI.4225-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A:505–511. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Huang HS. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol Psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Alberini CM. The role of protein synthesis during the labile phases of memory: revisiting the skepticism. Neurobiol Learn Mem. 2008;89:234–246. doi: 10.1016/j.nlm.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY.1999Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2 Nat Genet 23185–188.This important finding revealed that MeCP2 mutations were the underlying cause of Rett Syndrome, leading to a groundswell of interest in the epigenetic regulation of cognitive processes. [DOI] [PubMed] [Google Scholar]
- Amir RE, Zoghbi HY. Rett syndrome: methyl-CpG-binding protein 2 mutations and phenotype-genotype correlations. Am J Med Genet. 2000;97:147–152. doi: 10.1002/1096-8628(200022)97:2<147::aid-ajmg6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21:217–227. doi: 10.1016/j.nbd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Atanassov BS, Koutelou E, Dent SY.2010The role of deubiquitinating enzymes in chromatin regulation FEBS Lettdoi: 10.1016/j.febslet.2010.10.042 [DOI] [PMC free article] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- Bertran-Gonzalez J, Bosch C, Maroteaux M, Matamales M, Herve D, Valjent E, et al. Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. J Neurosci. 2008;28:5671–5685. doi: 10.1523/JNEUROSCI.1039-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran-Gonzalez J, Hakansson K, Borgkvist A, Irinopoulou T, Brami-Cherrier K, Usiello A, et al. Histone H3 phosphorylation is under the opposite tonic control of dopamine D2 and adenosine A2A receptors in striatopallidal neurons. Neuropsychopharmacology. 2009;34:1710–1720. doi: 10.1038/npp.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.2007Perceptions of epigenetics Nature 447396–398.Clear and concise outline of what makes something ‘epigenetic', this review argues for a more inclusive definition that encompasses chromatin modifications that lack heritability, such as those that occur in mature neurons.17522671 [Google Scholar]
- Bonda DJ, Lee HG, Camins A, Pallas M, Casadesus G, Smith MA, et al. The sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerations. Lancet Neurol. 2011;10:275–279. doi: 10.1016/S1474-4422(11)70013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brami-Cherrier K, Valjent E, Herve D, Darragh J, Corvol JC, Pages C, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Sun YE, Kobor MS. How the epigenome contributes to the development of psychiatric disorders. Dev Psychobiol. 2010;52:331–342. doi: 10.1002/dev.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruserud O, Stapnes C, Ersvaer E, Gjertsen BT, Ryningen A. Histone deacetylase inhibitors in cancer treatment: a review of the clinical toxicity and the modulation of gene expression in cancer cell. Curr Pharm Biotechnol. 2007;8:388–400. doi: 10.2174/138920107783018417. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27:12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Katzoff A, Levitan D, Susswein AJ, Klein R, et al. Long-term memory requires polyADP-ribosylation. Science. 2004;304:1820–1822. doi: 10.1126/science.1096775. [DOI] [PubMed] [Google Scholar]
- Coitinho AS, Freitas AR, Lopes MH, Hajj GN, Roesler R, Walz R, et al. The interaction between prion protein and laminin modulates memory consolidation. Eur J Neurosci. 2006;24:3255–3264. doi: 10.1111/j.1460-9568.2006.05156.x. [DOI] [PubMed] [Google Scholar]
- Collins A, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunoy J, Abidi F, Zeniou M, Jacquot S, Merienne K, Pannetier S, et al. Mutations in the X-linked RSK2 gene (RPS6KA3) in patients with Coffin-Lowry syndrome. Hum Mutat. 2001;17:103–116. doi: 10.1002/1098-1004(200102)17:2<103::AID-HUMU2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Deng H, Bao X, Cai W, Blacketer MJ, Belmont AS, Girton J, et al. Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development. 2008;135:699–705. doi: 10.1242/dev.015362. [DOI] [PubMed] [Google Scholar]
- Dong E, Chen Y, Gavin DP, Grayson DR, Guidotti A. Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics. 2010;5:730–735. doi: 10.4161/epi.5.8.13053. [DOI] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci USA. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DJ, Waters MF, Williams CA, Zori RT, Glenn CC, Avidano KM, et al. A DNA methylation imprint, determined by the sex of the parent, distinguishes the Angelman and Prader-Willi syndromes. Genomics. 1992;13:917–924. doi: 10.1016/0888-7543(92)90001-9. [DOI] [PubMed] [Google Scholar]
- Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson MH, Rose S, Mahadevan LC. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front Biosci. 2001;6:D853–D865. doi: 10.2741/dyson. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. 2010Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons Nat Neurosci 13423–430.Using genetic tools, these authors extend insights from pharmacological studies and reveal that neuron and forebrain-specific deletion of two DNMT isoforms leads to impairments in memory and synaptic plasticity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fontan-Lozano A, Suarez-Pereira I, Horrillo A, del-Pozo-Martin Y, Hmadcha A, Carrion AM. Histone H1 poly[ADP]-ribosylation regulates the chromatin alterations required for learning consolidation. J Neurosci. 2010;30:13305–13313. doi: 10.1523/JNEUROSCI.3010-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490 (Pt 3:703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988;452:57–65. doi: 10.1016/0006-8993(88)90008-x. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, et al. 2010A novel pathway regulates memory and plasticity via SIRT1 and miR-134 Nature 4661105–1109.Whereas other HDACs have been shown to negatively regulate learning and memory, this report suggests that SIRT1, a NAD+-dependent deacetylase, positively regulates plasticity and memory by limiting expression of miR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- Graff J, Mansuy IM. Epigenetic dysregulation in cognitive disorders. Eur J Neurosci. 2009;30:1–8. doi: 10.1111/j.1460-9568.2009.06787.x. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, et al. 2010bHigh-resolution analysis of parent-of-origin allelic expression in the mouse brain Science 329643–648.First study to characterize genome-wide imprinting in the brain, this report reveals that parental expression bias is much more common than previously believed, especially in the neural circuits controlling motivated behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. 2009HDAC2 negatively regulates memory formation and synaptic plasticity Nature 45955–60.Intriguing report revealing isoform specificity in the contribution of HDACs to hippocampal plasticity, learning, and memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Heffron D, Mandell JW. Differential localization of MAPK-activated protein kinases RSK1 and MSK1 in mouse brain. Brain Res Mol Brain Res. 2005;136:134–141. doi: 10.1016/j.molbrainres.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Hernandez AI, Wolk J, Hu JY, Liu J, Kurosu T, Schwartz JH, et al. Poly-(ADP-ribose) polymerase-1 is necessary for long-term facilitation in Aplysia. J Neurosci. 2009;29:9553–9562. doi: 10.1523/JNEUROSCI.1512-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi M, Inoue S, Ito T. Core histone H2A ubiquitylation and transcriptional regulation. Exp Cell Res. 2010;316:2707–2712. doi: 10.1016/j.yexcr.2010.05.028. [DOI] [PubMed] [Google Scholar]
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homburg S, Visochek L, Moran N, Dantzer F, Priel E, Asculai E, et al. A fast signal-induced activation of Poly(ADP-ribose) polymerase: a novel downstream target of phospholipase c. J Cell Biol. 2000;150:293–307. doi: 10.1083/jcb.150.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jason LJ, Moore SC, Lewis JD, Lindsey G, Ausio J. Histone ubiquitination: a tagging tail unfolds. Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MO, Chawla P, Overland RP, Xia E, Sadri-Vakili G, Cha JH. Altered histone monoubiquitylation mediated by mutant huntingtin induces transcriptional dysregulation. J Neurosci. 2008;28:3947–3957. doi: 10.1523/JNEUROSCI.5667-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Koshibu K, Graff J, Beullens M, Heitz FD, Berchtold D, Russig H, et al. Protein phosphatase 1 regulates the histone code for long-term memory. J Neurosci. 2009;29:13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Lisman JE. A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proc Natl Acad Sci USA. 1985;82:3055–3057. doi: 10.1073/pnas.82.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009a;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009b;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA.2010Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference Biol Psychiatry 6736–43.Important result suggesting that treament with HDAC inhibitors facilitates extinction of drug-associated memories, revealing therapeutic potential for treatment of addiction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, III, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. Quantitative analysis of CBP- and P300-induced histone acetylations in vivo using native chromatin. Mol Cell Biol. 2003;23:7611–7627. doi: 10.1128/MCB.23.21.7611-7627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Pannetier S, Harel-Bellan A, Sassone-Corsi P. Mitogen-regulated RSK2-CBP interaction controls their kinase and acetylase activities. Mol Cell Biol. 2001;21:7089–7096. doi: 10.1128/MCB.21.20.7089-7096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu H, Kandel ER, et al. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Moretti P, Bouwknecht JA, Teague R, Paylor R, Zoghbi HY. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Noh JS, Sharma RP, Veldic M, Salvacion AA, Jia X, Chen Y, et al. DNA methyltransferase 1 regulates reelin mRNA expression in mouse primary cortical cultures. Proc Natl Acad Sci USA. 2005;102:1749–1754. doi: 10.1073/pnas.0409648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, et al. 2010Altered histone acetylation is associated with age-dependent memory impairment in mice Science 328753–756.Reveals that age-associated memory decline in a mouse model is linked to inability to generate a specific histone acetylation mark, and that treatment with HDAC inhibitors reverses this deficit and improves learning and memory. [DOI] [PubMed] [Google Scholar]
- Pelka GJ, Watson CM, Radziewic T, Hayward M, Lahooti H, Christodoulou J, et al. Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain. 2006;129:887–898. doi: 10.1093/brain/awl022. [DOI] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Barnes CA, Sweatt JD. An epigenetic hypothesis of aging-related cognitive dysfunction. Front Aging Neurosci. 2010a;2:9. doi: 10.3389/fnagi.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, et al. 2010bAge-related changes in Arc transcription and DNA methylation within the hippocampus Neurobiol Agingdoi: 10.1016/j.neurobiolaging.2010.01.009 [DOI] [PMC free article] [PubMed]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Pollin W, Cardon PV, Jr, Kety SS. Effects of amino acid feedings in schizophrenic patients treated with iproniazid. Science. 1961;133:104–105. doi: 10.1126/science.133.3446.104. [DOI] [PubMed] [Google Scholar]
- Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer's and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Razin A, Friedman J. DNA methylation and its possible biological roles. Prog Nucleic Acid Res Mol Biol. 1981;25:33–52. doi: 10.1016/s0079-6603(08)60482-1. [DOI] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, III, Maze I, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, III, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial D, Duarte FS, Xikota JC, Schmitz AE, Dafre AL, Figueiredo CP, et al. Cellular prion protein modulates age-related behavioral and neurochemical alterations in mice. Neuroscience. 2009;164:896–907. doi: 10.1016/j.neuroscience.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhi A, Mager DL, Humphries RK, Kuchenbauer F. MiRNAs, epigenetics, and cancer. Mamm Genome. 2008;19:517–525. doi: 10.1007/s00335-008-9133-x. [DOI] [PubMed] [Google Scholar]
- Russo VEA, Martienssen RA, Riggs AD.eds (1996Epigenetic Mechanisms of Gene Regulation Cold Spring Harbor Laboratory Press: Woodbury [Google Scholar]
- Sadri-Vakili G, Bouzou B, Benn CL, Kim MO, Chawla P, Overland RP, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington's disease models. Hum Mol Genet. 2007;16:1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34:2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- Santini E, Alcacer C, Cacciatore S, Heiman M, Herve D, Greengard P, et al. L-DOPA activates ERK signaling and phosphorylates histone H3 in the striatonigral medium spiny neurons of hemiparkinsonian mice. J Neurochem. 2009;108:621–633. doi: 10.1111/j.1471-4159.2008.05831.x. [DOI] [PubMed] [Google Scholar]
- Santos KF, Mazzola TN, Carvalho HF. The prima donna of epigenetics: the regulation of gene expression by DNA methylation. Braz J Med Biol Res. 2005;38:1531–1541. doi: 10.1590/s0100-879x2005001000010. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, et al. 2009Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex Neuron 64678–691.Among first evidence that histone methylation in adult neurons contributes to normal adaptive behavior, in part by suppressing the expression of non-neuronal genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf AN, Imhof A.2010Every methyl counts - epigenetic calculus FEBS Lettdoi: 10.1016/j.febslet.2010.11.029 [DOI] [PubMed]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003a;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003b;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- Stearns NA, Schaevitz LR, Bowling H, Nag N, Berger UV, Berger-Sweeney J. Behavioral and anatomical abnormalities in Mecp2 mutant mice: a model for Rett syndrome. Neuroscience. 2007;146:907–921. doi: 10.1016/j.neuroscience.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proc Natl Acad Sci USA. 2009;106:9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanovich A, Valjent E, Matamales M, Nishi A, Ahn JH, Maroteaux M, et al. 2008A phosphatase cascade by which rewarding stimuli control nucleosomal response Nature 453879–884.Critical finding linking striatal cyto-nuclear shuttling of DARPP-32 to phosphorylation at H3S10 in response to cocaine exposure and motivated behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J Neurosci. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tzschach A, Lenzner S, Moser B, Reinhardt R, Chelly J, Fryns JP, et al. Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum Mutat. 2006;27:389. doi: 10.1002/humu.9420. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Lee SS, Zhang X, Houwink-Manville I, Song HR, Amir RE, et al. Rett syndrome and beyond: recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am J Hum Genet. 1999;65:1520–1529. doi: 10.1086/302690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. 2004Epigenetic programming by maternal behavior Nat Neurosci 7847–854.First report to reveal that maternal behavior produces long-lasting changes in DNA methylation in the brain, and that these changes are reversible. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y.2010Active DNA demethylation: many roads lead to Rome Nat Rev Mol Cell Biol 11607–620.Thorough review considering the different mechanisms that underlie demethylation of DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, et al. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res. 2009;111:115–122. doi: 10.1016/j.schres.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]