Abstract
Tularemia is a severe infectious disease in humans caused by the gram-negative bacterium Francisella tularensis (Ft). Due to its low infectious dose, high mortality rate, and the threat of its large-scale dissemination in weaponized form, development of vaccines and immunotherapeutics against Ft is essential. Ft lipopolysaccharide (LPS), which contains the linear graded-length saccharide component O-antigen (OAg) attached to a core oligosaccharide, has been reported as a protective antigen. Purification of LPS saccharides of defined length and composition is necessary to reveal the epitopes targeted by protective antibodies. In this study, we purified saccharides from LPS preparations from both the Ft subspecies holarctica live vaccine strain (LVS) and the virulent Ft subspecies tularensis SchuS4 strain using liquid chromatography. We then characterized the fractions using high resolution mass spectrometry and tandem mass spectrometry. Three types of saccharides were observed in both the LVS and SchuS4 preparations: two consisting of OAg tetrasaccharide repeats attached to one of two core-oligosaccharide variants, and one consisting of tetrasaccharide repeats only (coreless). The coreless OAg oligosaccharides were shown to contain Qui4NFm (4,6-dideoxy-4-formamido-D-glucose) at the non-reducing end and QuiNAc (2-acetamido-2,6-dideoxy-O-D-glucose) at the reducing end. Purified homogeneous preparations of saccharides of each type will allow mapping of protective epitopes in Ft LPS.
Francisella tularensis (Ft) is the gram-negative bacterium that causes tularemia, an infectious disease in humans and other mammals including aquatic rodents and rabbits. Ft is classified as a category A Select Agent by the Centers for Disease Control because of its low infectious dose and high mortality rate. The infectious dose for respiratory tularemia is less than 10 colony forming units (1), with a mortality rate of 5–30% for untreated disease (2). Tularemia is normally treated with antibiotics, but the mortality rate is still 2–3% in treated patients (3–5). A strain of type B (subspecies holarctica) Ft, live vaccine strain (LVS), was developed in the former Soviet Union in 1961 (6). To date, this is the only vaccine that has partial protective effects in humans against the most virulent type A (subspecies tularensis) Ft strains that include SchuS4 (7, 8). LVS is not approved by the US Food and Drug Administration as a vaccine for the general public due to safety concerns (9, 10). Therefore development of new vaccines and immunotherapeutics for tularemia is essential.
Lipopolysaccharide (LPS) has been reported as a protective Ft antigen, with antibodies targeting primarily its saccharide components (11–14). LPS is comprised of three parts: lipid A, core-oligosaccharides, and O-antigen (OAg). Lipid A anchors in the outer membrane of Ft, and core and OAg extend from the bacterial surface (15). OAg is a linear saccharide comprised of tetrasaccharide repeats, normally attached to the non-reducing end mannose residue of the core-oligosaccharide. The structure of Ft LPS has been studied using nuclear magnetic resonance (NMR) and mass spectrometry (MS), as reviewed in (15). In 1991, Vinogradov and coworkers reported the first Ft OAg structure, determined using NMR for type B Ft strain 15 (16). The OAg was identified as containing a variable number of tetrasaccharide repeats of composition GalNAcAN2QuiNAcQui4NFm, i.e. two moles of 2-acetamido-2-deoxy-O-D-galacturonamide (GalNAcAN), one mole of 2-acetamido-2,6-dideoxy-O-D-glucose (QuiNAc), and one mole of 4,6-dideoxy-4-formamido-D-glucose (Qui4NFm) (16). In their later study, the lipid A and core oligosaccharide structures were also determined (17). There are two types of core structures: HexNAcHex3Kdo, i.e. one with one mole of N-acetyl Hexosamine (HexNAc), three moles of Hexoses (Hex), and one mole of 3-Deoxy-D-manno-oct-2-ulosonic acid (Kdo), and HexNAcHex4Kdo. The structures of OAg and core oligosaccharides in type A Ft were identical to those observed in type B Ft (18, 19). In a recent report, a Ft capsular polysaccharide (CPS) was shown to consist of the same GalNAcAN2QuiNAcQui4NFm tetrasaccharide repeat as OAg and to protect mice against LVS Ft challenge (20).
In order to develop vaccines and immunotherapeutics against Ft, it is necessary to determine the saccharide structure(s) recognized by protective antibodies. Towards this end, the compositions and linkages of saccharides from LPS preparations of both the LVS and SchuS4 strains were analyzed using liquid chromatography (LC), high resolution mass spectrometry, and tandem mass spectrometry (MSn). Contrary to earlier studies in which low resolution mass spectrometry was used and MS/MS was not employed (18), three types of saccharides were observed in LPS preparations of both strains: two consisting of OAg attached to one of two core oligosaccharide variants and one consisting of OAg only. Since all three have potential to generate an immune response toward Ft, these results represent significant new information that impact the development of immunotherapeutics based on protective antibodies.
Materials and Methods
LPS preparations
LPS from Ft LVS was purchased from Sussex Research (Ontario, Canada). LPS from Ft SchuS4 was prepared as follows: SchuS4 bacteria (NR-643, obtained from BEI Resources, Manassas, VA) were grown to confluence on chocolate agar plates (Remel, Lenexa, KS) for 3 days at 35°C. Bacteria were scraped and suspended in phosphate buffered saline, heat-killed at 82°C for 2 h, and supplemented with sodium azide to a final concentration of 0.1%. Bacteria originating from 20 plates, a total OD600 of 343, were used for LPS purification by a hot phenol-water extraction method (21, 22) with modifications. Briefly, the cell suspension was lyophilized, reconstituted with 25 mL HPLC-grade water, and the resulting suspension was heated to 68°C. An equal volume of 90% phenol solution, preheated to 68°C, was added and the mixture was incubated at 68°C for 1 h with vigorous stirring. After being kept for 1 h on ice, the mixture was centrifuged at 3,300 × g for 10 min at 4°C, and the aqueous phase was aspirated. After 25 mL water that had been heated to 68°C was added to the organic phase, the mixture was incubated at 68°C for 1 h, with vigorous stirring. After 1 h on ice, the mixture was centrifuged at 3,300 × g for 10 min at 4°C. The aqueous phase was aspirated and combined with the previous fraction. The combined aqueous solution was dialyzed extensively versus water and then concentrated using rotary evaporation at 40°C. The residual solution was lyophilized overnight. The lyophilized powder was reconstituted with 10 mL 0.3 M sodium acetate/70 mL ice-cold absolute ethanol. After cooling in a freezer at −80°C, the solution was incubated at −20°C for 2 h, centrifuged at 10,000 × g for 20 min at 4°C, and the pellet was reconstituted with 10 mL of water/70 mL ice-cold absolute ethanol. A pellet was obtained by centrifugation at 10,000 × g for 20 min at 4°C and resuspended in 1 mL of 50 mM TrisHCl pH 8.0 with 2 mM MgCl2. Benzonase (600 mU) was added and the sample was incubated at 37°C for 3 h. After the enzyme was inactivated by heating the mixture at 99°C for 5 min, the buffer was adjusted to 1 mM CaCl2. Pronase (1 mg) was added and the sample was incubated at 55°C for 24 h. After overnight lyophilization, 20 mg of material was obtained.
Release and derivatization of saccharides purified from LPS
Saccharides were released from LPS by incubation in 1% acetic acid at 100°C for 2.5 h, and the resulting oligosaccharides were separated from lipid A by aspirating the supernatant after centrifugation at 11,000 × g for 1 h. Reduction and permethylation were performed using published procedures (18, 23, 24).
Size exclusion chromatography (SEC)
Saccharides from SchuS4 and LVS LPS were fractionated using SEC (Superdex Peptide 10/300 GL, GE Healthcare) using 100 mM ammonium acetate/10% acetonitrile as the mobile phase. The flow rate was 0.2 mL/min with a 2-h isocratic gradient and UV detection at 225 nm was used for monitoring sample elution. An automatic fraction collector was used with an interval of two min per eluted fraction.
Mass spectrometry and data analysis
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS
A Reflex IV MALDI-TOF mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with a nitrogen laser (337 nm, 3 nsec pulse width) was used. The matrix solution was 10 mg/mL 2,5-dihydroxybenzoic acid (DHB) in 0.1% trifluoroacetic acid (TFA)/ 10 μM sodium bicarbonate/ 50% acetonitrile. The saccharide samples (0.5 μL) were allowed to co-crystallize with 0.5 μL of matrix solution on a stainless steel target. Signals from 400 laser shots at 25–40% laser power were accumulated in positive ion mode over the range m/z 500 – 8000. To optimize detection of high molecular weight saccharides in the derivatized sample, the detection range was set to m/z 2500 – 8000. External calibration was achieved using peptide calibration standard II (Bruker Daltonics, Billerica, MA). DataAnalysis 4.0 (Bruker Daltonics) was used for data analysis.
Chip-based liquid chromatography mass spectrometry
Saccharides from LVS and SchuS4 strains without SEC fractionation were analyzed by hydrophilic interaction chromatography (HILIC)/MS using a makeup flow (MUF)-chip-based mass spectrometer interface (Agilent Technologies, Santa Clara, CA) and a published procedure (25). Formic acid (50 mM, pH 4.4) with ammonia containing 10% acetonitrile was used as mobile phase A. Mobile phase B contained 95% acetonitrile and 5% mobile phase A. A gradient composed of 90% B was used to load oligosaccharide samples on the enrichment column at 4 μL/min flow rate for 30 μL volume. A gradient from 90% B to 0% B was delivered over 39 min using a nanoflow pump at 200 nL/min. Pure acetonitrile was used as MUF at 200 nL/min. An Agilent 6520 quadrupole TOF mass spectrometer was used for detection in negative ionization mode.
High resolution and high accuracy mass spectrometric analysis
SEC fractionated SchuS4 OAg saccharides were analyzed with an LTQ-Orbitrap XL (ThermoFisher Scientific, San Jose, CA) and a hybrid qQh FT-ICR mass spectrometer (SolariX 12T, Bruker Daltonics, Billerica, MA) to achieve high resolution and high accuracy measurements. The samples were diluted in 50% methanol for negative ion mode detection and in 20 μM sodium bicarbonate/50% acetonitrile for positive ion mode. The FT-ICR mass spectrometer was calibrated with ESI tuning mix (part number G2432A, Agilent Technologies, Santa Clara, CA). The capillary voltage was set to 1200 V and nitrogen was used as the drying gas at a flow rate of 2.5 L min−1. The mass spectra were acquired over the range m/z 400 –3000. The collision voltage was set to 5 V for fragmentation of [4,2,2][0,0,0] and 10 V for [6,3,3][0,0,0], and argon was used as the collision gas. DataAnalysis 4.0 software (Bruker Daltonics) was used for data analysis.
The LTQ-Orbitrap XL is equipped with a NanoMate TriVersa ion source (Advion, Ithaca, NY). The capillary voltage was set to 1.5 kV and the gas pressure was set to 0.4 psi. The mass spectra were acquired over the range m/z 200 – 2000 at a resolution of 100,000. After external calibration, the mass accuracy remained within 5 ppm. The normalized collision energy for collision-induced dissociation (CID) was set to 20% and helium was used as the collision gas. All spectra were recorded in profile mode. Xcalibur 2.0.7 (ThermoFisher Scientific) was used for data analysis.
Results
MALDI-TOF MS of LVS LPS
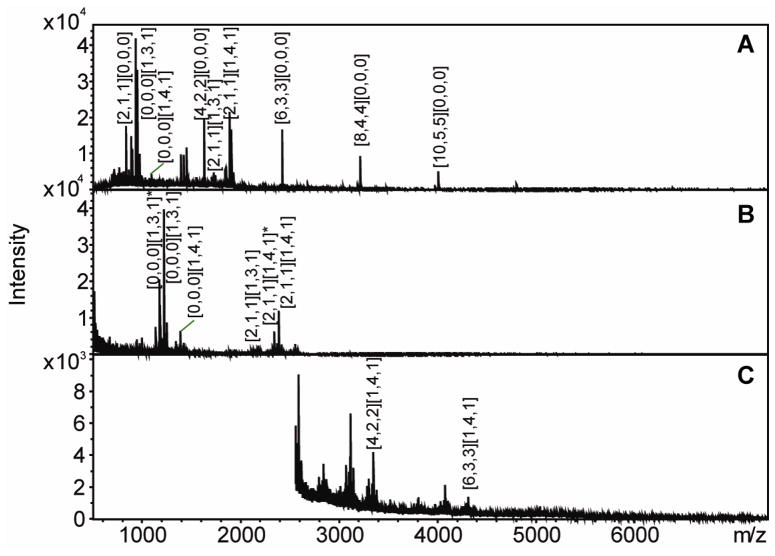
To confirm published reports and establish separation conditions, native saccharides derived from a commercial preparation of LVS LPS and their derivatives that had been subjected to reduction and permethylation were analyzed using MALDI-TOF MS in positive ion mode. Peaks corresponding to the oligosaccharide compositions HexNAcHex3Kdo and HexNAcHex4Kdo, which are the two types of core structures of LPS (18), were observed in both the native and permethylated samples (Figure 1 and Table 1). Glycans resulting from sequential addition of a tetrasaccharide unit consisting of GalNAcAN2QuiNAcQui4NFm to the HexNAcHex4Kdo core structure were also observed in both samples. However, peaks corresponding to the saccharides composed of one to nine tetrasaccharide repeats without an attached core, which had compositions identical to that of Ft CPS (20), were observed only in the mass spectrum generated from the native sample. The mass spectra of the saccharides after permethylation (Figure 1B and 1C) are largely consistent with the previously published work of Prior et al (18), and the observed ions correspond to OAg with the HexNAcHex4Kdo core. It is noteworthy that OAg attached to the HexNAcHex3Kdo core, [2,1,1][1,3,1] (the saccharide composition is given as [GalNAcAN, QuiNAc, Qui4NFm] [HexNAc, Hex, Kdo]), has been detected in both the native and permethylated samples. These data represent the first time that [2,1,1][1,3,1] has been reported. The mass assignments for the MALDI-TOF MS results are listed in Table 1. The detection of native saccharides in the LPS preparation demonstrates that the material previously thought to consist only of OAg attached to HexNAcHex4Kdo core, is actually comprised of three types of chains: OAg attached to HexNAcHex4Kdo core, OAg attached to HexNAcHex3Kdo core, and OAg with no attached core (coreless saccharides).
Figure 1. MALDI-TOF mass spectra of native (A) and reduced and permethylated LVS saccharide chains in low (B) and high (C) molecular weight range.
Saccharide compositions are given as [GalNAcAN, QuiNAc, Qui4NFm] [HexNAc, Hex, Kdo].
Table 1. Observed ions for native and reduced and permethylated LVS saccharides in positive ion reflectron mode on a MALDI-TOF mass spectrometer.
Saccharide compositions are given as [GalNAcAN, QuiNAc, Qui4NFm] [HexNAc, Hex, Kdo].
| Saccharide Composition | m/z Observed (native) | m/z Observed (Permethylated) |
|---|---|---|
| [2,1,1][0,0,0] | 833.8 | |
| [0,0,0][1,3,1]* | 1172.6 | |
| [0,0,0][1,3,1] | 932.7 | 1218.6 |
| [0,0,0][1,4,1] | 1094.7 | 1421.2 |
| [4,2,2][0,0,0] | 1626.2 | |
| [2,1,1][1,3,1] | 1725.2 | 2179.2 |
| [2,1,1][1,4,1]* | 2338.3 | |
| [2,1,1][1,4,1] | 1887.3 | 2384.3 |
| [4,2,2][1,4,1] | 3345.8 | |
| [6,3,3][1,4,1] | 4307.5 | |
| [6,3,3][0,0,0] | 2419.8 | |
| [8,4,4][0,0,0] | 3212.3 | |
| [10,5,5][0,0,0] | 4005.7 |
represents loss of formic acid.
Relative abundances of saccharides from LVS and SchuS4 LPS preparations
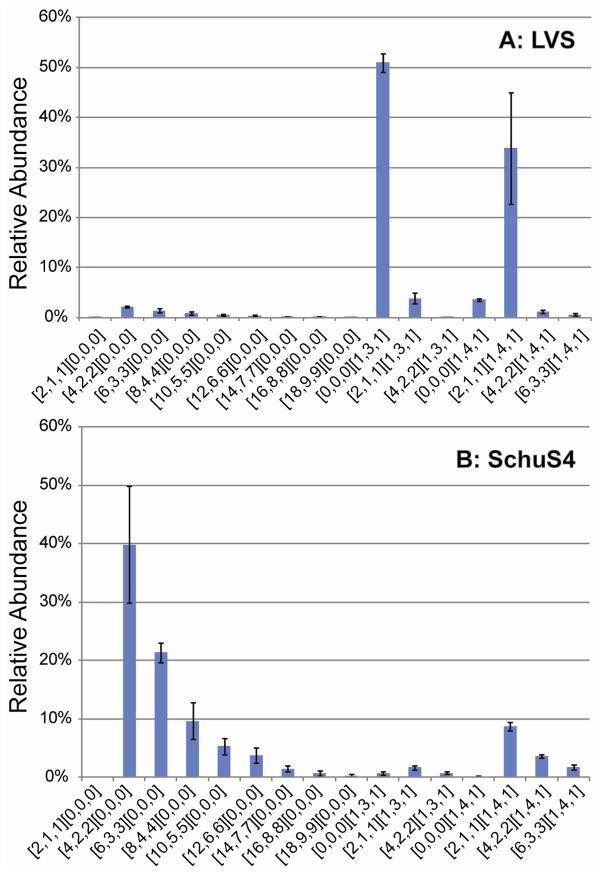
To determine whether the three types of saccharide chains observed in the LVS LPS preparation are also present in an LPS preparation derived from the virulent SchuS4 strain, we analyzed saccharides from in-house purified SchuS4 LPS. The relative abundances of the saccharide chains in the LPS preparations from both the LVS and SchuS4 strains were determined using a chip-based LC/MS system. All saccharides found in LVS were also found in SchuS4 and both strains were found to have higher abundance of OAg-core saccharides containing the HexNAcHex4Kdo core ([1,4,1]) than the corresponding saccharides containing the HexNAcHex3Kdo core ([1,3,1]) (Figure 2). Coreless saccharides [2,1,1]1–9[0,0,0] were found in considerably higher abundances in SchuS4 than LVS. In the LVS preparation, [0,0,0][1,3,1] core-oligosaccharide was the most abundant component and LVS saccharide [2,1,1][1,4,1] was also detected in high abundance. The results show dramatically different abundances of saccharides in the OAg fractions from the LVS and SchuS4 preparations, but these differences may not be explained solely by the strain differences because growth conditions can affect LPS saccharide composition in Ft (26).
Figure 2. Relative abundances of saccharide chains in LPS preparations from LVS (A) and SchuS4 (B) Ft strains.
The relative abundances were obtained from integration of the areas under the peaks in the extracted ion chromatograms for each compound from HILIC Chip-based LC/MS. Error bars were calculated as standard deviation from the means of triplicates. Saccharide compositions are given as [GalNAcAN, QuiNAc, Qui4NFm] [HexNAc, Hex, Kdo].
Tandem MS of LPS coreless saccharides
[4,2,2][0,0,0]
Tandem MS was performed on the saccharides with two and three tetrasaccharide repeats. The oligosaccharide containing two tetrasaccharide repeats, [4,2,2][0,0,0], was enriched using SEC. The peak at m/z 824.2961 corresponds to the [M + 2Na]2+ of [4,2,2][0,0,0] with -1.1 ppm mass error. This ion was subjected to tandem MS using CID in positive ion mode on an LTQ-Orbitrap XL mass spectrometer (Supplemental Figure 1A). To further characterize the topology of this saccharide, the product ion at m/z 833.3009 from the MS2 spectrum, which corresponds to the sodium adducted fragment containing the tetrasaccharide [2,1,1][0,0,0], was isolated and subjected to further CID fragmentation. In the resulting MS3 spectrum (Supplemental Figure 1B), the two ions with the highest signal intensities corresponding to [2,0,1][0,0,0] and [2,1,0][0,0,0] were each isolated and fragmented with another stage of CID. The resulting MS4 spectra for fragmentation of ions at m/z 646.2158 ([2,0,1][0,0,0]) and 660.2314 ([2,1,0][0,0,0]) are shown in Supplemental Figure 1C and 1D. The detailed interpretations of the multistage tandem MS results are listed in Supplemental Table 1. The fragment ions were reported using the Domon and Costello nomenclature (27). Fragments corresponding to [2,0,0][0,0,0] and [1,0,1][0,0,0] were observed in the MS4 spectrum of [2,0,1][0,0,0], and these indicate the monosaccharide sequence of Qui4NFm-GalNAcAN-GalNAcAN. In the MS4 spectrum of [2,1,0][0,0,0], the observation of [2,0,0][0,0,0] and [1,1,0][0,0,0] fragments suggests the monosaccharide sequence of GalNAcAN-GalNAcAN-QuiNAc. The existence of [2,0,1][0,0,0] and [2,1,0][0,0,0] fragments in the MS3 spectrum, together with the monosaccharide sequences deduced from the MS4 spectra, show that Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc is the monosaccharide sequence of [2,1,1][0,0,0]. Analysis of the fragments in the MS2 spectrum which resulted from glycosidic bond cleavages, led to the conclusion that the precursor ion at m/z 824.2961, assigned to [4,2,2][0,0,0] is composed of two tetrasaccharide repeats, Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc, in series.
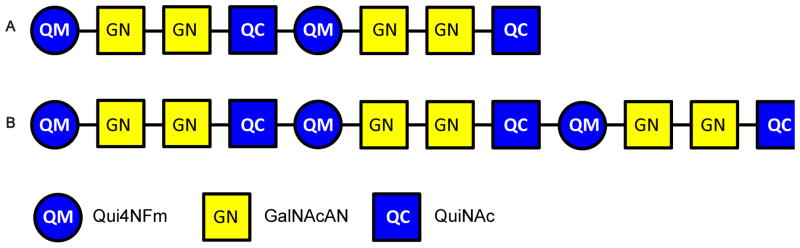
Although the monosaccharide sequence [4,2,2][0,0,0] was assigned on the basis of MSn analysis of the native glycan, tandem MS of the 18O-labeled sample was necessary to unambiguously assign the residue at the reducing end (Supplemental Figure 2). The ions observed at m/z 825.2987 correspond in composition to [M + 2Na]2+ for [4,2,2][0,0,0], with one 16O being replaced by one 18O with −0.5 ppm mass error. After isolation of the ions in the quadrupole ion trap, CID resulted in the formation of the fragments at m/z 835.3053 in the MS2 spectrum, which correspond to the sodium adduct of [2,1,1][0,0,0] with one 18O atom replacing one 16O atom (Supplemental Figure 2A). After isolation of the peak at m/z 835.3053, MS3 was performed (Supplemental Figure 2B). The detailed interpretations of the multi-stage tandem MS results are listed in Supplemental Table 2. The two most abundant MS3 product ions were observed at m/z 646.2163 and 662.2362 and were consistent with trisaccharide compositions of [2,0,1][0,0,0] and one 18O atom replacing one 16O atom of [2,1,0][0,0,0], respectively. From this result, a QuiNAc residue was determined to be located at the reducing-end of [4,2,2][0,0,0]. The [4,2,2][0,0,0] glycan is therefore composed of two tetrasaccharide repeats, Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc, in series, with a Qui4NFm residue at the non-reducing end and a QuiNAc residue at the reducing end (Figure 3A).
Figure 3.
The structures of coreless saccharides [4,2,2][0,0,0] (A) and [6,3,3][0,0,0] (B) deduced from tandem MS in both positive and negative ion modes.
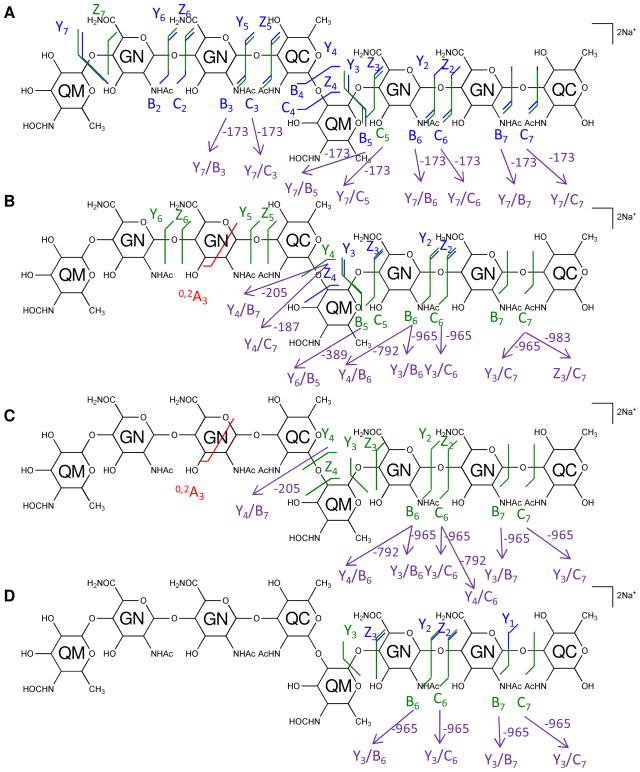
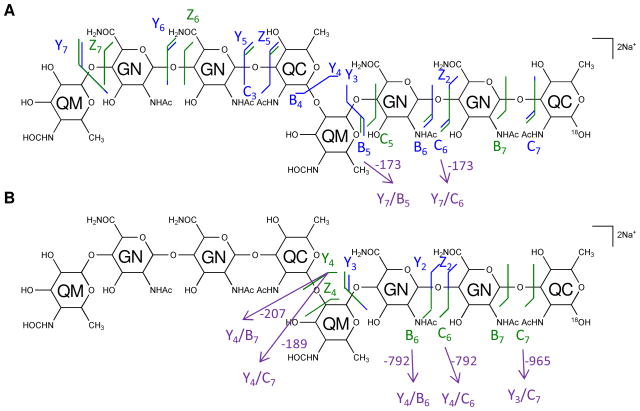
The multi-stage tandem MS fragments of native and 18O-labeled [4,2,2][0,0,0] are displayed on their chemical structures in Figures 4 and 5, with glycosidic bond cleavages labeled in blue, internal cleavages in green, and cross-ring cleavages in red. Observed glycosidic bond cleavages provided monosaccharide sequence information and the presence of the 0,2A3 cleavage in Figure 4B and 4C verified the non-reducing end saccharide mass.
Figure 4. Multi-stage tandem MS reveals the structural linkage for [4,2,2][0,0,0].
Tandem MS of the saccharide [4,2,2][0,0,0], [M + 2Na]2+ m/z 824.2961, and its product ions was performed on an LTQ-Oribtrap XL mass spectrometer in positive ion mode. CID fragmentations of the precursor ion m/z 824.2961 (A), MS3 of the ion at m/z 833.3009 (B), and MS4 of the ions at m/z 646.2158 (C) and 660.2314 (D) are displayed on the chemical structures as glycosidic bond cleavages labeled in blue, cross-ring cleavages in red, and internal cleavages in green. Monosaccharide residues abbreviations QM (Qui4NFm), GN (GalNAcAN), and QC (QuiNAc) are shown inside the six-member rings for clarity. One pair of internal cleavage is shown in purple to represent the alternative assignments. All fragments contain Na.
Figure 5. Multi-stage tandem mass spectra of the 18O labeled saccharide [4,2,2][0,0,0] reveals the reducing-end residue.
Tandem MS of the 18O labeled saccharide [4,2,2][0,0,0] was performed on an LTQ-Oribtrap XL mass spectrometer in positive ion mode. CID fragmentations of the precursor [M + 2Na]2+ m/z 825.2987 (A) and MS3 of the secondary product ion at m/z 835.3053 (B) are displayed on the chemical structures. Monosaccharide residues abbreviations QM (Qui4NFm), GN (GalNAcAN), and QC (QuiNAc) are shown inside the six-member rings for clarity. All fragments contain Na.
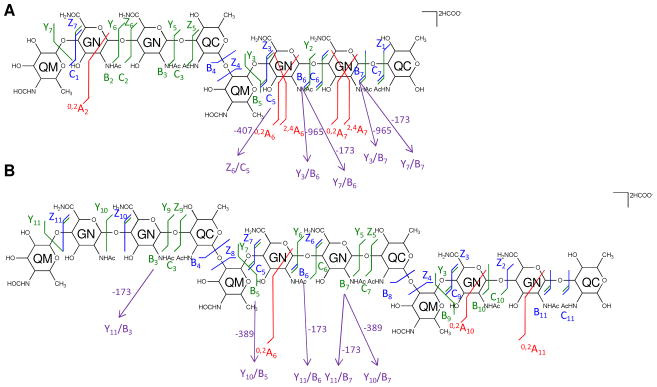
To confirm these results, tandem MS of native [4,2,2][0,0,0] was carried out in negative ion mode using a Fourier transform-ion cyclotron resonance (FT-ICR) mass spectrometer (Supplemental Figure 3A). The mass spectral data were more accurate than those acquired with the LTQ-Orbitrap XL mass spectrometer and thus provided the advantage of greater certainty of mass assignments. The detailed interpretations of the tandem MS result are presented in Figure 6A and Supplemental Table 3. Fragmentation patterns similar to those found in the negative ion MS/MS spectra were observed in the MS/MS spectra acquired in the positive ion mode using the LTQ-Orbitrap XL mass spectrometer. The observed glycosidic bond cleavages confirmed that [4,2,2][0,0,0] is composed of two tetrasaccharide repeats, Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc, in series. Cross-ring cleavages were observed in higher abundances in negative ion mode using the FT-ICR mass spectrometer than positive ion mode using the LTQ-Orbitrap mass spectrometer, but did not allow unambiguous assignment of linkages.
Figure 6. Similar fragment ions are observed for saccharides in negative ion mode.
CID MS2 fragmentations of the [M + 2HCOO]2− of the saccharide chains [4,2,2][0,0,0] at m/z 846.3049 (A) and [6,3,3][0,0,0] at m/z 1242.4535 (B) were obtained in negative ion mode using a Solarix FT-ICR mass spectrometer and are displayed on the chemical structures. Monosaccharide residues abbreviations QM (Qui4NFm), GN (GalNAcAN), and QC (QuiNAc) are shown inside the six-member rings for clarity.
[6,3,3][0,0,0]
The oligosaccharide corresponding to the coreless chain [6,3,3][0,0,0] was also analyzed using multi-stage tandem MS in positive ion mode on an LTQ-Orbitrap XL mass spectrometer. The [M + 2Na]2+ peak at m/z 1220.4479 was isolated and subjected to CID (Supplemental Figure 4A). The tetrasaccharide product ion observed at m/z 833.3003 in the MS2 spectrum, which corresponds to [2,1,1][0,0,0], was isolated and fragmented using CID (Supplemental Figure 4B). The detailed interpretations of the multi-stage tandem MS results are listed in Supplemental Table 4. As was the case for the MS3 spectrum of [4,2,2][0,0,0], singly-charged product ions at m/z 646.2155 and 660.2312, corresponding to the compositions [2,0,1][0,0,0] and [2,1,0][0,0,0], were observed in the MS3 spectrum of [6,3,3][0,0,0], suggesting the Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc sequence. The presence of the ions at m/z 444.1574 and 473.1473, corresponding to the compositions [1,1,0][0,0,0] and [2,0,0][0,0,0], also confirmed the monosaccharide sequence. The base peak in the MS2 spectrum, m/z 1126.9037, corresponding to the doubly-charged [6,2,3][0,0,0] species, implies the position of a QuiNAc residue at either the non-reducing end or the reducing end of the sequence. In the tandem MS spectrum of the [M + 2Na]2+ of the 18O-labeled [6,3,3][0,0,0] (Supplemental Figure 5 and supplemental Table 5), the base peak was observed at m/z 1126.9044, corresponding to [6,2,3][0,0,0]. This result indicates that a QuiNAc residue is located at the reducing end of the linear oligosaccharide chain. As was observed in the MS2 spectrum of 18O-labeled [4,2,2][0,0,0], the appearance of the signals at m/z 662.2365 and 646.2166, which correspond to [2,1,0][0,0,0] with one 18O atom replacing one 16O atom and [2,0,1][0,0,0], supports the conclusion that a QuiNAc residue serves as the reducing-end of this oligosaccharide. Therefore, [6,3,3][0,0,0] is composed of three tetrasaccharide repeats, Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc, in series, with a Qui4NFm residue at the non-reducing end and a QuiNAc residue at the reducing end (Figure 3B).
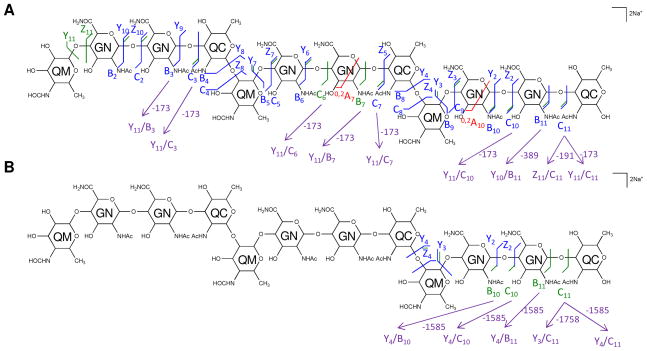
The multi-stage tandem MS fragments of native and 18O labeled [6,3,3][0,0,0] are displayed on their chemical structures in Figures 7 and 8. A few cross-ring cleavages were observed on the GalNAcAN residues in the MS2 spectrum of [6,3,3][0,0,0], and no linkage information can be deduced from them.
Figure 7. Multi-stage tandem MS reveals the structural linkage for [6,3,3][0,0,0].
Tandem MS of the saccharide [6,3,3][0,0,0], [M + 2Na]2+ m/z 1220.4479 was performed on an LTQ-Oribtrap XL mass spectrometer in positive ion mode. CID fragmentations of the precursor ion m/z 1220.4479 (A) and MS3 of the product ion at m/z 833.3003 (B) are displayed on the chemical structures. Monosaccharide residues abbreviations QM (Qui4NFm), GN (GalNAcAN), and QC (QuiNAc) are shown inside the six-member rings for clarity. All fragments contain sodium.
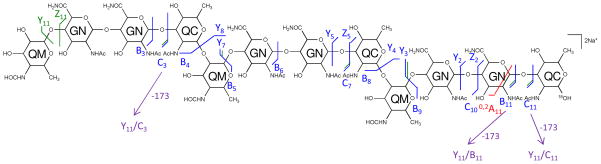
Figure 8. Tandem MS of the 18O labeled saccharide [6,3,3][0,0,0] reveals the reducing-end residue.
Tandem MS of the 18O labeled saccharide [6,3,3][0,0,0], [M + 2Na]2+ m/z 1221.4487 was performed on an LTQ-Oribtrap XL mass spectrometer in positive ion mode. CID fragmentations of the precursor ion are indicated on the chemical structure. Monosaccharide residues abbreviations QM (Qui4NFm), GN (GalNAcAN), and QC (QuiNAc) are shown inside the six-member rings for clarity. All fragments contain sodium.
Tandem MS of native [6,3,3][0,0,0] was also carried out in negative ion mode using an FT-ICR mass spectrometer (Supplemental Figure 3B). The detailed interpretations of the tandem MS result are presented in Figure 6B and Supplemental Table 3. From the observed products that arose via glycosidic bond cleavages, [6,3,3][0,0,0] is confirmed to be composed of three tetrasaccharide repeats, Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc, in series. The cross-ring cleavages observed in tandem mass spectra acquired in the negative ion mode did not allow unambiguous assignment of linkages.
Discussion
OAg-containing saccharides obtained after hydrolysis of the components in preparations of Ft LPS have been assumed to consist of OAg chains attached to the HexNAcHex4Kdo core (17). In the present work, OAg saccharides attached to HexNAcHex3Kdo core, HexNAcHex4Kdo core as well as OAg saccharides without an attached core were identified in hydrolysates of LPS preparations from both the LVS and SchuS4 strains. It is likely that the published studies using NMR failed to detect the coreless saccharides. In the present study and in previous work (18), coreless saccharides were not detected in the permethylated samples, possibly due to their poor solubility in organic solvents during the derivatization procedure. Similarly, previous studies may not have detected OAg-core saccharides containing the HexNAcHex3Kdo core because of the higher abundance of OAg-HexNAcHex4Kdo saccharides in both LVS and SchuS4.
It was necessary to use high resolution and high mass accuracy MS in order to reach a definitive interpretation of the compositions of saccharides derived from Ft LPS preparations. For example, the monoisotopic peak at m/z 846.3090 (2+) could have been interpreted as [2,0,1][1,4,1] with a ~30 ppm mass error that is within the tolerable range for low resolution mass spectrometers (Table 2). By contrast, the assignment of the same peak as [4,2,2][0,0,0] results in a mass error of 3.5 ppm, which is within the mass error tolerance for the high resolution and high accuracy mass spectrometers used in this study. Thus, the application of high resolution and high accuracy MS enabled the identification of previously unidentified coreless saccharides (17, 18) in fractions released from Ft LPS preparations.
Table 2. High resolution and high mass accuracy MS is necessary for interpretation of saccharide composition.
The mass errors from two different potential assignments for the observed monoisotopic masses for saccharides composed of two to five tetrasaccharide repeats are listed in the table. Saccharide compositions are given as [GalNAcAN, QuiNAc, Qui4NFm] [HexNAc, Hex, Kdo].
| Observed Peaks (m/z) | Charge | Interpretation #1 | Mass error for assignment #1 (ppm) | Interpretation #2 | Mass error for assignment #2 (ppm) |
|---|---|---|---|---|---|
| 846.3090 | 2 | [2,0,1][1,4,1] | 32.4 | [4,2,2][0,0,0] | 3.5 |
| 1242.4604 | 2 | [4,1,2][1,4,1] | 22.2 | [6,3,3][0,0,0] | 2.5 |
| 1107.4107 | 3 | [6,2,3][1,4,1] | 19.8 | [8,4,4][0,0,0] | 5.1 |
| 1371.5132 | 3 | [8,3,4][1,4,1] | 17.2 | [10,5,5][0,0,0] | 5.3 |
CID was used to achieve fragmentation of the coreless saccharides with two and three tetrasaccharide repeats in this study. Extensive glycosidic bond cleavages were observed in both positive and negative ionization mode tandem MS experiments. The information enabled a consistent interpretation regarding the monosaccharide sequences of the saccharides. Other fragmentation techniques, such as electron-transfer dissociation (ETD) (28) and electron-capture dissociation (ECD) (29), which generate mainly cross-ring cleavages for carbohydrates (30, 31), and/or other chemical derivatization of the native saccharides, such as peracetylation, might be helpful to provide additional detail concerning the glycosidic linkage positions in these coreless saccharides.
Although mass spectrometry has the advantage of accurately detecting minor components comparing to other detection methods such as NMR, it is not sufficient to distinguish isomers of the monosaccharide residues because it only provides m/z values of the analytes. It is insufficient to determine the identity of the monosaccharide residues, and other complimentary detection techniques, for example, NMR and glycosyl composition, are needed to identify the actual monosaccharide residues.
Ft OAg had been reported by Vinogradov and coworkers, based on NMR analysis (16, 17), and by Prior et al., based on MS analysis (18), to have repeats of the structure: →4)-GalNAcAN-(α1→4)-GalNAcAN-(α1→3)-QuiNAc-(β1→2)-Qui4NFm-(β1→ or to be composed of (GalNAcAN)2QuiNAcQui4NFm, respectively. Both groups proposed that the QuiNAc residue is attached to the core based on similarities in the sugar composition of the OAg repeat-unit of Ft compared to other Gram negative bacteria (16, 17) and the deduced putative functions of the proteins encoded in the OAg biosynthetic gene-cluster of Ft (18). The present study confirmed the previously reported Ft OAg repeat structure and showed that QuiNAc is the reducing end residue and that a Qui4NFm is the residue at the non-reducing end of coreless OAg saccharides.
The present high resolution and mass accuracy tandem MS study demonstrated that the coreless saccharides have the monosaccharide sequence of Qui4NFm-GalNAcAN-GalNAcAN-QuiNAc attached in series. This is the same as the recently reported Ft CPS tetrasaccharide repeat sequence (20). The saccharides observed in the present study may derive from LPS chains in the process of synthesis, before or during transfer from the carrier oligosaccharide to core-lipid A in the Ft inner membrane, a process documented for other Gram negative bacteria (32–34). The considerably higher abundance of coreless saccharides in the SchuS4 vs. the LVS strain observed in the present study is consistent with the presence of thicker capsules in virulent Ft strains compared with LVS (35). Differences in abundances of saccharides may also arise from the culture conditions of the bacteria used for the commercial LVS versus that for the in-house SchuS4 LPS preparations (26). Other methods of separating LPS and CPS (36)(20) might be helpful in revealing the biological role of the coreless OAg saccharides observed in this study..
This study identified three structurally distinct types of OAg-containing saccharides present in hydrolysates of Ft LPS preparations: those with HexNAcHex4Kdo core, those with HexNAcHex3Kdo core, and those with no core. Each of these has the potential to present unique epitopes for antibody reactivity. Preparative chromatographic separation of homogeneous OAg-containing and core saccharides is therefore necessary in order to map epitopes targeted by protective anti-Ft LPS antibodies towards development of vaccines and immunotherapeutics for tularemia.
Supplementary Material
Acknowledgments
The authors thank Mathew Walsh for preliminary results, Yu Huang for help with data analysis, and Xiang Yu and Liang Han for technical help in oligosaccharide derivatization.
Abbreviations
- Ft
Francisella tularensis
- LPS
lipopolysaccharide
- OAg
O-antigen
- LVS
live vaccine strain
- CPS
capsular polysaccharide
- Qui4NFm
4,6-dideoxy-4-formamido-D-glucose
- QuiNAc
2-acetamido-2,6-dideoxy-O-D-glucose
- GalNAcAN
2-acetamido-2-deoxy-O-D-galacturonamide
- Hex
Hexoses
- HexNAc
N-acetyl Hexosamine
- Kdo
3-Deoxy-D-manno-oct-2-ulosonic acid
- NMR
nuclear magnetic resonance
- MS
mass spectrometry
- LC
liquid chromatography
- MSn
tandem mass spectrometry
- SEC
size exclusion chromatography
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- DHB
2,5-dihydroxybenzoic acid
- TFA
trifluoroacetic acid
- HILIC
hydrophilic interaction chromatography
- MUF
makeup flow
- CID
collision-induced dissociation
- FT-ICR
Fourier transform-ion cyclotron resonance
- ETD
electron-transfer dissociation
- ECD
electron-capture dissociation
Footnotes
This research is supported by NIH contract HHSN272200900054C and NIH grants P41 RR010888, S10 RR015942, S10 RR020946 and S10 RR025082.
SUPPORTING INFORMATION AVAILABLE
Supplemental tables and figures are included in the Supporting Information section. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. Tularemia vaccine study. II. Respiratory challenge. Arch Intern Med. 1961;107:702–714. doi: 10.1001/archinte.1961.03620050068007. [DOI] [PubMed] [Google Scholar]
- 2.Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 3.Sjostedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 4.Tarnvik A, Chu MC. New approaches to diagnosis and therapy of tularemia. Ann N Y Acad Sci. 2007;1105:378–404. doi: 10.1196/annals.1409.017. [DOI] [PubMed] [Google Scholar]
- 5.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Tonat K. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 6.Eigelsbach HT, Downs CM. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J Immunol. 1961;87:415–425. [PubMed] [Google Scholar]
- 7.Burke DS. Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J Infect Dis. 1977;135:55–60. doi: 10.1093/infdis/135.1.55. [DOI] [PubMed] [Google Scholar]
- 8.Sandstrom G. The tularaemia vaccine. J Chem Technol Biotechnol. 1994;59:315–320. doi: 10.1002/jctb.280590402. [DOI] [PubMed] [Google Scholar]
- 9.Conlan JW. Tularemia vaccines: recent developments and remaining hurdles. Future Microbiol. 2011;6:391–405. doi: 10.2217/fmb.11.22. [DOI] [PubMed] [Google Scholar]
- 10.Oyston PC. Francisella tularensis vaccines. Vaccine. 2009;27(Suppl 4):D48–51. doi: 10.1016/j.vaccine.2009.07.090. [DOI] [PubMed] [Google Scholar]
- 11.Fulop MJ, Webber T, Manchee RJ, Kelly DC. Production and characterization of monoclonal antibodies directed against the lipopolysaccharide of Francisella tularensis. J Clin Microbiol. 1991;29:1407–1412. doi: 10.1128/jcm.29.7.1407-1412.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulop M, Mastroeni P, Green M, Titball RW. Role of antibody to lipopolysaccharide in protection against low- and high-virulence strains of Francisella tularensis. Vaccine. 2001;19:4465–4472. doi: 10.1016/s0264-410x(01)00189-x. [DOI] [PubMed] [Google Scholar]
- 13.Lu Z, Roche MI, Hui JH, Unal B, Felgner PL, Gulati S, Madico G, Sharon J. Generation and characterization of hybridoma antibodies for immunotherapy of tularemia. Immunol Lett. 2007;112:92–103. doi: 10.1016/j.imlet.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roche MI, Lu Z, Hui JH, Sharon J. Characterization of monoclonal antibodies to terminal and internal O-antigen epitopes of Francisella tularensis lipopolysaccharide. Hybridoma (Larchmt) 2011;30:19–28. doi: 10.1089/hyb.2010.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunn JS, Ernst RK. The structure and function of Francisella lipopolysaccharide. Ann N Y Acad Sci. 2007;1105:202–218. doi: 10.1196/annals.1409.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinogradov EV, Shashkov AS, Knirel YA, Kochetkov NK, Tochtamysheva NV, Averin SF, Goncharova OV, Khlebnikov VS. Structure of the O-antigen of Francisella tularensis strain 15. Carbohydr Res. 1991;214:289–297. doi: 10.1016/0008-6215(91)80036-m. [DOI] [PubMed] [Google Scholar]
- 17.Vinogradov E, Perry MB, Conlan JW. Structural analysis of Francisella tularensis lipopolysaccharide. Eur J Biochem. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- 18.Prior JL, Prior RG, Hitchen PG, Diaper H, Griffin KF, Morris HR, Dell A, Titball RW. Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J Med Microbiol. 2003;52:845–851. doi: 10.1099/jmm.0.05184-0. [DOI] [PubMed] [Google Scholar]
- 19.Thirumalapura NR, Goad DW, Mort A, Morton RJ, Clarke J, Malayer J. Structural analysis of the O-antigen of Francisella tularensis subspecies tularensis strain OSU 10. J Med Microbiol. 2005;54:693–695. doi: 10.1099/jmm.0.45931-0. [DOI] [PubMed] [Google Scholar]
- 20.Apicella MA, Post DM, Fowler AC, Jones BD, Rasmussen JA, Hunt JR, Imagawa S, Choudhury B, Inzana TJ, Maier TM, Frank DW, Zahrt TC, Chaloner K, Jennings MP, McLendon MK, Gibson BW. Identification, characterization and immunogenicity of an O-antigen capsular polysaccharide of Francisella tularensis. PLoS ONE. 2010;5:e11060. doi: 10.1371/journal.pone.0011060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westphal O, Jann K. Bacterial Lipopolysaccharides Extraction with Phenol-Water and Further Applications of the procedure. Methods in Carbohydrate Chemistry. 1965:83–91. [Google Scholar]
- 22.Apicella MA, Griffiss JM, Schneider H. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol. 1994;235:242–252. doi: 10.1016/0076-6879(94)35145-7. [DOI] [PubMed] [Google Scholar]
- 23.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 24.Ciucanu I, Costello CE. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J Am Chem Soc. 2003;125:16213–16219. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- 25.Staples GO, Naimy H, Yin H, Kileen K, Kraiczek K, Costello CE, Zaia J. Improved hydrophilic interaction chromatography LC/MS of heparinoids using a chip with postcolumn makeup flow. Anal Chem. 2010;82:516–522. doi: 10.1021/ac901706f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherwonogrodzky JW, Knodel MH, Spence MR. Increased encapsulation and virulence of Francisella tularensis live vaccine strain (LVS) by subculturing on synthetic medium. Vaccine. 1994;12:773–775. doi: 10.1016/0264-410x(94)90284-4. [DOI] [PubMed] [Google Scholar]
- 27.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
- 28.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 30.Han L, Costello CE. Electron Transfer Dissociation of milk oligosaccharides. J Am Soc Mass Spectrom. 2011;22:997–1013. doi: 10.1007/s13361-011-0117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamson JT, Hakansson K. Electron capture dissociation of oligosaccharides ionized with alkali, alkaline earth, and transition metals. Anal Chem. 2007;79:2901–2910. doi: 10.1021/ac0621423. [DOI] [PubMed] [Google Scholar]
- 32.Lengeler J, Drews G, Schlegel H. Biology of the Prokaryotes. Blackwell Science Ltd; Oxford, UK: 2009. Assembly of cellular surface structures; pp. 555–570. [Google Scholar]
- 33.Metzler D. Biochemistry: the chemical reactions of living cells. Academic Press; 2003. Biosynthesis of bacterial cell walls; pp. 1160–1169. [Google Scholar]
- 34.Zhao J, Raetz CR. A two-component Kdo hydrolase in the inner membrane of Francisella novicida. Mol Microbiol. 2010;78:820–836. doi: 10.1111/j.1365-2958.2010.07305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hood AM. Virulence factors of Francisella tularensis. J Hyg (Lond) 1977;79:47–60. doi: 10.1017/s0022172400052840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adam O, Vercellone A, Paul F, Monsan PF, Puzo G. A nondegradative route for the removal of endotoxin from exopolysaccharides. Anal Biochem. 1995;225:321–327. doi: 10.1006/abio.1995.1161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.