Abstract
As important as the events that influence selection for specific chromosome types in the derivation of novel karyotypes, are the events that initiate the changes in chromosome number and structure between species, and likewise polymorphisms, variants and disease states within species. Although once thought of as transcriptional ‘noise', noncoding RNAs (ncRNAs) are now recognized as important mediators of epigenetic regulation and chromosome stability. Here we highlight recent work that illustrates the influence short and long ncRNAs have as participants in the function and stability of chromosome regions such as centromeres, telomeres, evolutionary breakpoints and fragile sites. We summarize recent evidence that ncRNAs can facilitate chromosome change and present mechanisms by which ncRNAs create DNA breaks. Finally, we present hypotheses on how they may create novel karyotypes and thus affect chromosome evolution.
Keywords: non-coding, ncRNA, small RNA, chromosome, evolution, karyotype
‘Although the genome of a species as a whole is important, chromosomes are the basic units subjected to genetic events that coin evolution to a large extent' (Li et al., 2011).
Introduction: Non-coding does not mean non-essential
The identification of tens of thousands of non-protein coding genes in the human genome (Birney et al., 2007) has challenged the classical definition of a gene and the dogma that DNA is transcribed into RNA, which is subsequently translated into functional proteins. In toto, these noncoding RNAs (ncRNAs) are not translated into proteins, yet are highly abundant, responsible for fundamental biological processes in many species and contribute significantly to disease and evolution. Genes were originally viewed as ‘elements of heredity', then as a location responsible for a phenotypic trait and later as the information that codes for a protein. More recently, we have seen a shift in the definition of genes to reflect the addition of ncRNAs and pseudogenes to their ranks. There are a number of different classes of ncRNAs, each categorized by their size, biogenesis and/or function, although these categories grow and shift constantly (Brosnan and Voinnet, 2009). On a gross scale, ncRNAs are divided into small (<200 nt) or long (>200 nt, lncRNAs) representing a variety of different classes. Their diversity is most obvious with respect to their functional importance in cellular processes, including translation, RNA splicing, cell cycle control, gene regulation, genome defense and chromosome structure (Table 1). The mechanisms through which ncRNAs, both large and small, are derived are the focus of recent, extensive reviews (Kim, 2005; Brosnan and Voinnet, 2009; Ponting et al., 2009). Rather, in this review we will focus on the role ncRNAs have in structural features of the genome and chromosome change.
Table 1. List of major classes of small ncRNAs, including their abbreviations, size range and function.
| Name | Abbreviation | Size | Function(s) |
|---|---|---|---|
| transfer RNA | tRNA | 73–93 nt | Transfers amino acids to the ribosome during translation of mRNA to protein (Hoagland et al., 1958) |
| ribosomal RNA | rRNA | 120–5035 nt | Translation of mRNA to protein (Watson, 1963) |
| small nuclear RNA | snRNA | 90–220 nt | RNA splicing (Lerner et al., 1980) |
| small nucleolar RNA | snoRNA | C/D box-70–120 nt H/ACA-100–200 nt | Guide the modification and maturation of other ncRNAs (Crouch et al., 1983) |
| long non-coding RNA/long intergenic non-coding RNA | lncRNA/lincRNA | >200 nt | Epigenetic gene regulation (Sleutels et al., 2002) |
| microRNA | miRNA | 21–22 nt | Translation inhibition; binds mRNA to block translation (Lee et al., 1993) |
| short/small interfering RNA | siRNA | 21–25 nt | Translation inhibition; binds mRNA to signal cleavage (Hamilton and Baulcombe, 1999) |
| endoribonuclease-prepared short interfering RNA | esiRNA | 20–25 nt | RNA interference (Yang et al., 2002) |
| repeat-associated small interfering RNA *previous name for piRNAs | rasiRNA | 24–29 nt | Silencing of retrotransposons in the germline (Aravin et al., 2003) |
| piwi-interacting RNA | piRNA | 26–31 nt | Silencing of retrotransposons in the germline (Brennecke et al., 2007) |
| centromere repeat-associated small interacting RNA | crasiRNA | 35–42 nt | Centromere identity and function (Carone et al., 2008) |
| small RNAs derived from snoRNAs | sdRNA | 17–19, >27 nt | Gene regulation (Ender et al., 2008) and alternative splicing (Kishore et al., 2010) |
| transcription initiation RNA | tiRNA | 18 nt | Unclear; may function in transcription regulation (Taft et al., 2009) |
| splice site RNA | spliRNA | 17–18 nt | Unclear; generated from the donor splice site in animal internal exons (Taft et al., 2010) |
| tRNA-derived RNA fragments | tRFs | 18–22 nt, 30–35 nt | miRNA-like functions; gene regulation (Lee et al., 2009) |
Envisioning chromosome change involves thinking of the chromosome as an entity with defined subregions. These subregions can be categorized not only by their specific cellular function (for example, telomeres and centromeres) but also by their presentation as a visible structure on the normal chromosome (for example, fragile sites, centromeres and telomeres). These chromosome locations are inherently susceptible to breakage and thus are major contributors to chromosome change. For example, centromeres are the target of myriad chromosome rearrangements, including Robertsonian translocations, neocentromerization, fissions and fusions. These same types of rearrangements typify those that define species-specific karyotypes in many lineages and those that accompany many disease states. The many and recent studies highlighted in this review show a striking coincidence of ncRNA with both the occurrence and recurrence of rearrangements at these chromosome subregions, and suggest that perhaps this coincidence may be the result of the function and/or the structural restraints imposed by ncRNA on chromosomes.
Links between ncRNA and chromosomal domains
Centromeres
Centromeres are the site of kinetochore assembly and spindle attachment during meiosis and mitosis. Thus, the proper functioning of centromeres is a prerequisite for faithful segregation of chromosomes, the failure of which is fundamental in cancer, infertility and genomic instability. Once thought of as a transcriptionally inert region of the eukaryotic genome, it has become clear that centromeres are in fact transcriptionally active and that ncRNAs are fundamental to centromere function (Wong et al., 2007; Bergmann et al., 2011) and consequently to the process of chromosome evolution (O'Neill and Carone, 2009; Brown and O'Neill, 2010).
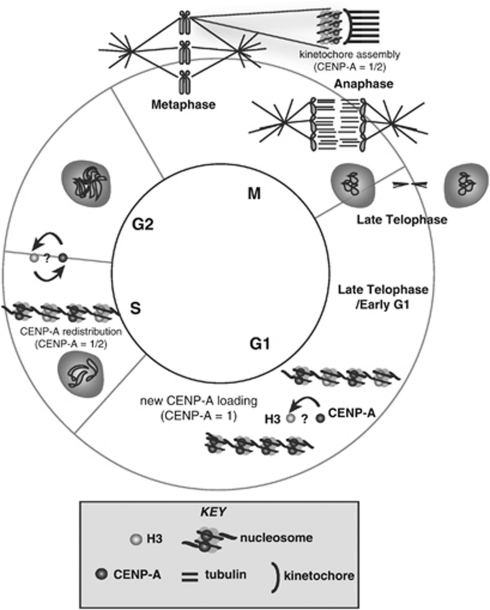
Kinetochore formation during mitosis is the culmination of the cycle comprising centromere function; the pivotal event is the ‘loading', or deposition, of newly synthesized CENP-A, the histone H3 variant specific to centromeres that replaces conventional H3 in centromeric nucleosomes (Figure 1). Failure to load CENP-A into centromeric chromatin in late telophase/early G1 of the cell cycle in humans leads to malsegregation and cell-division defects in subsequent cell cycles. Although only a few key players involved in the epigenetic cascade that leads to CENP-A loading during centromere assembly have been identified (reviewed in Mellone et al., 2009), mounting evidence supports the hypothesis that a ncRNA component is a crucial part of this epigenetic cascade.
Figure 1.
During telophase/early G1, newly synthesized CENP-A is loaded onto centromeric nucleosomes. During S phase and DNA replication, CENP-A is redistributed to sister chromosomes through either a replacement or a dilution process (indicated by ‘?') before entering G2. After mitosis, new CENP-A loading is likely facilitated by a priming mechanism, which may prepare the centromeric nucleosome for the replacement of H3 with CENP-A, although the actual mechanism of CENP-A/H3 replacement is currently unknown (indicated by a ‘?'). Key is shown in inset at bottom.
Studies have shown that CentO satellites and centromeric retroposons reside within the kinetochore-binding region of rice centromeres (Nagaki et al., 2004). As might be expected for a retroposon, centromeric retroposon of rice elements are actively transcribed, but transcripts derived from centromere satellites have also been identified in Arabidopsis (May et al., 2005), maize (Topp et al., 2004), mouse, human and many other eukaryotic species (Ugarkovic, 2005). Although prevalent in complex eukaryotic centromeres, the role of these satellite-derived transcripts in centromere function is only recently becoming apparent. Chromosome missegregation has been associated with aberrant satellite transcription in animals (Valgardsdottir et al., 2005) and satellite RNA has been implicated in the assembly of centromere components, such as CENP-A and CENP-C, in humans (Wong et al., 2007; Bergmann et al., 2011). An elegant study using an artificial chromosome in human cells recently showed that although another modified histone, H3K4me2, is necessary for long-term kinetochore maintenance, the active transcription of centromeric satellite DNA is also required for targeting CENP-A loading into active centromeric chromatin (Bergmann et al., 2011).
It was recently shown that mouse major satellite transcripts at the periphery of the pericentric heterochromatin produce lncRNAs that associate in the forward orientation with SUMO-modified HP1 (heterochromatin protein 1) (Maison et al., 2011). The association between SUMO-HP1 and satellite lncRNA at the pericentric heterochromatin domain may serve as a primary ‘seeding' step in the establishment of pericentric heterochromatin that may subsequently trigger the cascade to further recruit HP1alpha and stabilize heterochromatin domains, in a manner analogous to the action of another ncRNA, Xist (reviewed in Augui et al., 2011). Although the relationship of this lncRNA–HP1 association to centromere function is currently unknown, the correlation of bursts of pericentric satellite RNA transcription immediately before the formation of HP1 domains on the paternal genome in early mouse development suggests that this type of lncRNA-facilitated heterochromatin seeding may be developmentally regulated (Probst and Almouzni, 2008; Probst et al., 2010; Maison et al., 2011). Maison et al. (2011) further postulate that heterochromatin seeding that demarcates specific chromosome regions may contribute to developmental and cell-specific genome rearrangements (for example, differentiation) in mouse.
Studies of the centromeres of budding yeast, Shizosaccharomyces pombe, have shown that dsRNAs are actively transcribed from the pericentric repeats dh and dg, and are subsequently processed into small interfering RNAs. These small interfering RNAs are bound to a complex of proteins (the RNA-induced initiation of transcriptional gene silencing, RITS) and result in targeted H3 lysine-9 methylation through RNA interference (Volpe et al., 2002, 2003). Moreover, the disruption of RNA interference components in fungi compromises heterochromatin assembly (Volpe et al., 2002) and CENP-A deposition (Folco et al., 2008), linking a small RNA component to centromere function.
Despite the fact that there is an established link between RNA processing and centromere function, small RNAs derived from the centromere core have been difficult to discern in vertebrate species. Until recently, no sequence containing a promoter has been identified within vertebrate centromeres that may facilitate transcription of native centromere sequences (but see Carone et al., 2009), complicating models that evoke satellite transcription as a requirement for centromere function. The combined observation that retrotransposons harbor strong promoters and are resident within centromeres of many plants is suggestive of a role for these cognate elements in centromere transcription of lncRNAs in vertebrates. Recent work uncovering a link between LINE transcription and CENP-A deposition within a neocentromere in human supports this association (Chueh et al., 2009). However, the role of retrotransposons and RNA in endogenous mammalian centromeres remains poorly understood (but see Dawe, 2003; O'Neill and Carone, 2009).
A novel class of small RNA (35–42nt, crasi RNAs, see Table 1) has been uncovered in mammals that is derived from retroelements (Carone et al., 2009) found within CENP-A nucleosomes (Renfree et al., 2011). Small RNAs of ∼40 nt were also identified for CentO satellites in rice centromeres (Cheng et al., 2002), indicating that RNAs 35–42 nt may be a conserved functional feature of eukaryotic centromeres. Moreover, work in maize and human have shown that ncRNAs bind to, and stabilize, CENP-C protein complexes and may serve as the epigenetic mark preparing the centromere for final assembly with CENP-A-RNA-bound nucleosomes (Du et al., 2010). Illustrating the complexity of the protein–ncRNA network involved in centromere function, WDHD1 is a recently identified HMG-1 (high mobility group) protein that participates in the localization of HP1alpha through an interaction with centromere satellite ncRNAs in a cell-cycle dependent manner (Hsieh et al., 2011). Moreover, abrogation of WDHD1 results in chromosome segregation defects and disruption of satellite ncRNA processing.
Satellite repeats in heterochromatin-rich pericentric regions are transcribed into ncRNAs that accumulate in both mouse and human cell lines in response to DNA demethylation and cellular stress (Valgardsdottir et al., 2005; Bouzinba-Segard et al., 2006). As both demethylation and cellular stress are participants in tumorigenesis, not surprisingly, ncRNA satellite transcripts are overexpressed in both mouse and human epithelial cancers (Ting et al., 2011). The derepression of satellite transcripts correlates with an overexpression of a LINE-1 retrotransposon and aberrant expression of genes proximal to the LINE-1 insertions (Ting et al., 2011). The mechanism and consequences of aberrant satellite ncRNA transcripts in cancer tissues is yet unknown, but hypothesized to result from dysregulation of epigenetic silencing (methylation and histone H3 lysine 9 trimethylation) (Ting et al., 2011).
It has recently been shown that loss of the tumor suppressor BRCA1 protein is coincident with a decrease in ubiquinated H2A (ub-H2A) in pericentric heterochromatin and subsequently an increase in the production of satellite ncRNAs (Zhu et al., 2011). This marked increase in satellite ncRNAs is also associated with cell-cycle defects associated with BRCA1's role in tumorigenesis, such as centrosome amplification, increased DNA double strand breaks (DSBs), lagging and bridging chromosomes. Thus, satellite transcripts may contribute to large-scale genome instability and consequently to chromosome evolution in the cancer phenotype (Zhu et al., 2011). Given the association between aberrant ncRNA expression, and chromosome instability and centromere dysfunction it stands to reason that such ncRNAs may also contribute to the karyotypic rearrangements that typify species differences. However, the circumstances and mechanisms by which these ncRNAs become destabilized in a non-disease situation and thus may facilitate chromosome change are currently unknown.
Telomeres
Telomeres protect the DNA ends of a chromosome from being recognized as sites of DNA damage, but progressively shorten because of problems replicating this highly repetitive region, which are in turn overcome by the reverse transcribing enzyme telomerase. Telomere shortening is linked both to chromosomal stability and tumorigenesis. In fact, telomere length abnormalities are proposed to be one of the earliest chromosome changes in malignancy, with telomere abnormalities resulting in aggregation, breakage-bridge-fusion cycles and chromosome instability (reviewed in Knecht and Mai, 2011). Telomeres are heterochromatic, composed of tandem arrays of TTAGGG repeats and yet recent studies have shown that eukaryotic telomeres are transcribed into TElomeric Repeat containing RNA (TERRA) (reviewed in Feuerhahn et al., 2010). These lncRNAs function in the nucleus, and in telomeric and subtelomeric heterochromatin structures. Some of these telomeric transcripts in Arabidopsis thaliana are processed into small interfering RNAs which promote methylation of the cytosines in the telomeric CCCTAAA repeats (Vrbsky et al., 2010). Confounding this, however, is the observation that while some of the TERRA in Arabidopsis and in S. cerevisiae are derived from telomeres, most are in fact derived from centromeres that contain simply remnants of telomeric DNA (Feuerhahn et al., 2010).
In mammals, only UUAGGG telomeric transcripts are detected (for example, transcripts from only the leading DNA strand); however, in Arabidopsis both telomeric strands appear to be transcribed, suggesting that under certain circumstances the telomere strand may act as a promoter (Vrbsky et al., 2010). RNA chromatin immunoprecipitation for TERRA using antibodies against HP1alpha and H3K9me3, protein marks for constitutive heterochromatin, further suggests that TERRA has a role in heterochromatization (Deng et al., 2009). Along with the satellite lncRNA derived from pericentric domains (Maison et al., 2011), TERRA is the only other lncRNA shown to specifically bind this heterochromatin protein.
Interestingly, studies of telomeric ncRNAs in single-celled protists have uncovered more complex roles for telomere-heterochromatin modulation through ncRNAs. The parasite Plasmodium falciparum, causative agent of malaria, lacks RNA interference and methylation, but does have conserved histone-modifying enzymes, a high number of RNA-binding proteins, and requires chromatin remodeling and epigenetic regulations for blood stage-specific expression and antigenic variation of virulence genes (reviewed in Broadbent et al., 2011). Recent transcriptional profiling and analysis of the parasite also revealed lncRNAs under developmental regulation during the parasite's pathogenic human blood stage, 15 of which are long telomere-associated ncRNA genes (long ncRNA telomere-associated repetitive element transcripts (lncRNA-TARE) located on 22 of 28 P. falciparum telomeres (Broadbent et al., 2011)). UpsB-type var genes are located near these lncRNA-TARE, subtelomerically, and are subject to coordinated silencing and activity as part of antigenic variation in defense against the human immune response during malaria infection (reviewed in Cui and Miao, 2010). Gene regulation of the multigene var family is cell-cyle dependent, and is facilitated by epigenetic marks and nuclear repositioning (for example, Petter et al., 2011). LncRNA-TARE transcription is in sync with the chromatin-remodeling events that regulate var expression and thus, may facilitate the change in nucleosome architecture (Broadbent et al., 2011) or entire chromosome compartmentalization within the nucleus, pointing to a complex role for lncRNAs in both heterochromatin and nuclear architecture.
Fragile sites
Fragile sites, heritable loci expressed as chromosome breaks or gaps, are inherently unstable in their DNA structure and provide one mechanistic explanation for chromosome rearrangements. Fragile sites are highly conserved; for example, orthologous fragile site loci are found between mouse and rat (Elder and Robinson, 1989), and among primates (Ruiz-Herrera et al., 2004). Fragile sites are also linked to evolutionary chromosome rearrangements in human and other mammalian species (Ruiz-Herrera et al., 2006). Moreover, fragile sites are hotspots for constitutional and cancer translocations, gene amplifications and integration of foreign DNA. For the chromosome regions containing sequences or genes producing ncRNAs, complex, large or even small chromosome rearrangements could explain how non-transcribed regions of the genome have gained transcriptional activity and proliferated throughout the karyotype. Under such a scenario, fragile sites could be viewed as hotspots for both the origin of novel ncRNA genes as well as a major contributor to the transcriptional catastrophe that often accompanies carcinogenesis.
In support of this proposition, Calin et al. (2004) mapped miRNA precursor genes to both common and rare chromosome fragile sites, and reported a nine-fold higher occurrence of miRNA genes at fragile sites versus non-fragile chromosomal loci. In fact, >50% of miRNA genes are in cancer-associated breakpoints or in fragile sites (Calin et al., 2004). In addition, miRNA gene copy number changes, gains and losses, as well as copy number changes of miRNA regulatory genes (Dicer1 and Argonaute2) are shared among cancer types, with some changes unique to specific tumors (Zhang et al., 2006). Thus, fragile sites represent a major contributor to the cancer phenotype not only through structural changes but also through changes to gene regulatory networks by ncRNAs.
In another example, upregulation of the oncogene SKI in acute myeloid leukemia patients has been demonstrated to be resultant from loss of miRNAs on chromosome 7q (whole chromosome or long arm of 7 deletions) (Teichler et al., 2011). A direct role for MIR29A, an miRNA found at the fragile site in 7q32.3, as a tumor suppressor is demonstrated in that there is a functional binding site for MIR29A in the 3′UTR of SKI and overexpression of MIR29A reduces the expression of the SKI oncogene (Teichler et al., 2011). This chromosome fragile site (7q32.3) also coincides with FLJ43663, a presumed ncRNA. Downregulation of the miRNA gene MIR29B1 occurs concomitantly with chromosome breaks at FLJ43663 in B-cell lymphomas, yet both MIR29A and MIR29B1 are overexpressed in anaplastic large cell lymphomas (Feldman et al., 2011). Thus, MIR29A has tumor suppressor activity for an oncogene present in B-cell lymphomas, but an oncogenic function in anaplastic large cell lymphomas in which the SKI oncogene is not expressed (Feldman et al., 2011). A fine balance must be achieved in transcriptional regulatory networks when disruption of ncRNAs (such as miRNA genes) is associated with chromosome breaks. A fundamental question, however, is whether this disruption precedes the rearrangement or is a consequence thereof.
Pevzner and Tesler (2003) observed that breakpoints involved in chromosome rearrangements in the evolution of species are non-random and clustered, with certain chromosome regions repeatedly used in evolution (Murphy et al., 2005). These recurring, specific chromosome breakpoints, often associated with fragile sites, are known as Evolutionary Breakpoints (EB) and are regions reused for karyotype reorganization (Murphy et al., 2005). EBs comprise only about 3% of the mammalian genome (Robinson et al., 2006) and yet are three times more likely to co-localize with cancer-associated breakpoints than with breakpoints found less commonly in cancer (Ruiz-Herrera et al., 2005). As in the observation of miRNA genes clustered within fragile sites, the reuse of EBs during karyotypic change may facilitate the ‘shuffling' of transcriptional modules, thereby creating lineage-specific miRNA gene families (or any other ncRNA gene family). In other words, a simple translocation event at a fragile site, or EB, could reorganize ncRNA modules, thereby affecting large cascades of gene networks in a lineage-specific fashion.
For example, a deletion of chromosome 13 at the EB band q14.3 is found in 55% of chronic lymphocytic leukemia cases with chromosome abnormalities (Mitelman et al., 2011). Deletion at this chromosomal breakpoint encompasses a non-transcribed gene and two miRNA genes leading researchers to first propose that miRNA gene disruption is linked to cancer (Calin et al., 2002). The last 10 years have witnessed the discovery of the myriad roles that miRNAs have in cancer as a result of their oncogenic and tumor-suppressive functions through the interference with genes regulating metastasis and apoptosis (Munker and Calin, 2011). What is noteworthy is the observation that chromosome rearrangements are often linked to ncRNA disruption. Chromosome rearrangements at 19q13.4, another EB, are the most frequent non-random chromosome translocations in human epithelial tumors (Mitelman et al., 2011). Two miRNA clusters (C19MC and miR-371-3), located close to this breakpoint on chromosome 19, are overexpressed in thyroid adenoma cells carrying 19q13.4 abnormalities, and may be causally associated with tumorigenesis, with large RNA Pol (RNAP)-II mRNA fragments as the most likely source of upregulation of the miRNA cluster (Rippe et al., 2010). Of note, rearrangements of 19q13.4 are found in other human cancers, suggesting that activation of these miRNA clusters might be a more general characteristic of human tumors.
ncRNA and structural changes to the karyotype
Recent studies provide tantalizing new clues into how ncRNAs may actually facilitate chromosome change, in addition to their post-rearrangement transcriptional effects. A genome-wide scan in yeast showed that meiotic DSBs are directed preferentially to loci that express poly-adenylated ncRNAs (Wahls et al., 2008), leading to the proposal that these ncRNAs direct recombination hotspots; how they do so is unknown. But, there are several mechanisms by which ncRNAs could orchestrate chromosome rearrangement.
One hypothetical mechanism by which ncRNAs could orchestrate chromosome rearrangement is that replication timing may be mediated by some ncRNAs in the eukaryotic genome. Chromosomes with a delay in replication timing, a delay in initiation and completion of DNA synthesis along the entire chromosome, display a delay in mitotic chromosome condensation. Delayed replication timing (DRT) and delayed mitotic chromosome condensation (DMC) are commonly detected in tumors and in cells exposed to ionizing radiation and are associated with persistent chromosome instability (Breger et al., 2005). Recent work (see below) links DRT/DMC resulting from chromosome translocations with ncRNA disruption, providing evidence that ncRNAs are a molecular mechanism accounting for secondary chromosome changes associated with genome instability.
An elegant chromosome engineering strategy showed that rearrangements of chromosome 6q16.1, an EB and a locus carrying mono-allelic expression of an intergenic lncRNA called asynchronous replication and autosomal RNA on chromosome 6 (ASAR6), results in delayed DNA replication of the entire chromosome 6 (Stoffregen et al., 2011). Disruption of this locus resulted in activation of previously silenced mono-allelically expressed genes linked to ASAR6, and none of the nine other Cre/loxp-mediated rearranged chromosomes displayed DRT, thus indicating that inactivation of the lncRNA at this site is responsible for the abnormalities observed (Stoffregen et al., 2011). Remarkably, cells containing chromosomes with DRT also have a 30–80-fold increase in the occurrence of gross chromosome rearrangements (Stoffregen et al., 2011), linking stability of specific lncRNA transcription to subsequent chromosome change. The mechanism by which ASAR6 lncRNA protects a chromosome from DRT and rearrangement is currently unknown; unlike XIST, ASAR6 does not coat its native chromosome (Stoffregen et al., 2011). Perhaps ASAR6 functions as a recruiter for chromatin-modifying enzymes, or polycomb group proteins, that protects the locus from DSBs and/or remodeling.
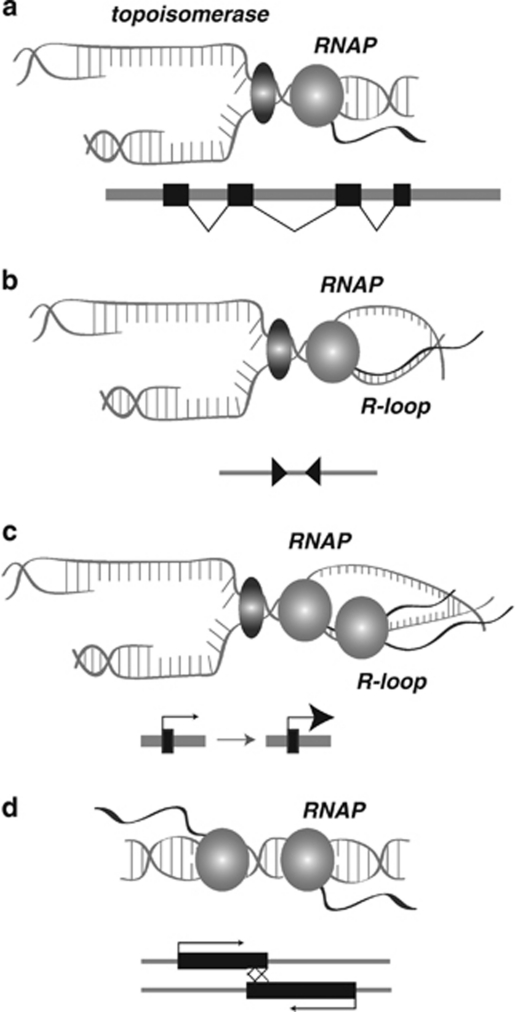
A common theme for ncRNAs is the ability of these transcripts to recruit proteins to a locus, region or entire chromosome. Letessier et al., 2011 propose that common fragile sites are, in fact, epigenetically defined loci, such that chromatin organization at these regions changes in order to delay replication initiation. However, active transcription of ncRNAs solely may contribute to the instability that precedes a DSB. Typically, DNA replication and transcription proceed independently (Figure 2a); however, factors that stall replication or increase transcription aberrantly can potentially cause replication fork collapse and collision between the fork and transcriptional machinery (Figures 2b and c). Likewise, bidirectional transcription can also lead to collision, but in this case between two RNAP complexes traveling towards one another (Figure 2d). One consequence of collision is the generation of large, stable or even double R-loops (Sikdar et al., 2008; Reddy et al., 2011; Figures 2b and c), RNA–DNA hybrids that are normally small and transient during transcription. These R-loops are then susceptible to targeted deamination, which may trigger single or DSBs (Ruiz et al., 2011), resulting in gross chromosome rearrangement (Sikdar et al., 2008). ncRNA transcripts are also potential facilitators of RNA–DNA hybrid formation in the subtelomeric and telomeric regions of chromosomes. TERRA ncRNA transcripts accumulate at telomeres in patients with reduced levels of CpG methylation (Yehezkel et al., 2008). Telomere dysfunction in these patients may be due to an increased frequency of replication fork collapse with the potential for an exacerbated phenotype because TERRA also inhibits telomerase activity, thus preventing healing at the collapsed fork (Yehezkel et al., 2008). An interesting caveat to the fork stalling theory is that FRA3B appears to rely not on slowing or stalling, but on the combination of late replication completion and scarcity of initiation events (Letessier et al., 2011).
Figure 2.
Mechanisms of ncRNA facilitation of chromosome breaks. (a) Normal transcription and DNA replication proceed independently with no collisions. Topoisomerase (green) and RNAPII (light purple) run independently on the same segment of DNA. The gene structure is shown below, with exons as boxes and spliced introns indicated by lines. (b) An inverted duplication of two retroelements (or other ncRNA gene), shown at bottom as red triangles, causes recruitment of specific proteins (for example, Sap1) that cause a replication stall between the RNAP machinery and replication fork. This leads to the formation of an R-loop. (c) Increased transcription of a ncRNA (purple box with large arrowhead) leads to a buildup of RNAP, resulting in a replication stall and an R-loop. (d) Bidirectional ncRNA transcripts (purple) can also lead to a collision between two RNAP complexes. A full color version of this figure is available at the Heredity journal online.
With relevance to this review is how ncRNAs could lead to the stall, collapse or collision. One intriguing observation is that tDNA, long tandem repeats (LTRs), Ty elements and inverted repeats in yeast cause replication fork stalling, resulting in DSBs (Lemoine et al., 2005; Zaratiegui et al., 2011). Coordination of lagging strand synthesis and prevention of single-strand annealing between direct repeated LTRs was proposed for fission yeast in which LTRs recruit a DNA-binding factor, switch-activating protein 1 (Sap1), to increase its persistence in the genome through replication fork stalling (Zaratiegui et al., 2011). Sap1 mutants not only show direct progression of the replication fork through the LTR, but also chromosome rearrangements detrimental to the retrotransposon (Zaratiegui et al., 2011). Sap1 also works with rDNA in which the protein blocks replication-fork progress through rDNA repeats, resulting in directional replication and prevention of mitotic recombination between these repeats (Noguchi and Noguchi, 2007). A striking similarity among tDNA, LTRs, Ty elements and rDNA is their production of ncRNAs. Moreover, inverted repeats, as well as many types of ncRNAs (for example, cis-NATs, cis-acting natural antisense transcripts), are often capable of producing bidirectional transcripts. Perhaps the ncRNAs produced from these sites either recruit proteins, such as Sap1, or are involved in a collision with RNAPII complexes that stall the replication fork (Figure 2b). Although it would appear that locations of rDNAs and LTRs (or other retroelements) would cause inherent instability on chromosomes, there are genome defenses in place that act in conflict with these selfish elements. For example, Zaratiegui et al., 2011 showed that the centromere protein CENP-B promotes replication through Sap1-stalled forks at LTRs. While LTRs and LTR-derived satellites are most often found in direct repeat orientation at centromere/pericentromere regions, it stands to reason that CENP-B would persist in such chromosome locations and thus reduce the frequency of DSB therein (Zaratiegui et al., 2011). In contrast, oppositely oriented LTRs, tRNAs and inverted repeats are a persistent feature that demarcate fragile sites in mammals and yeast through potentially unprotected, slow-progressing forks that facilitate a high rate of DSBs (Szilard et al., 2010; Figures 2b and d).
A question yet unanswered is why fragile sites (and their inclusive ncRNA genes) perpetuate in the genome. A recent proposal shows that these fragile loci are sequestered in nuclear compartments marked by p53-binding protein 1 (53BP1) and other chromatin-associated proteins (Lukas et al., 2011). Although some DNA lesions might be resolved by DNA repair mechanisms, the ‘shielding' in nuclear bodies may reduce the risk of further genome instability and potential loss (Hung et al., 2011). Another mechanism may be their ability to introduce new regulatory networks through the shuffling of ncRNA gene regions, promoters and so on. Thus, a single rearrangement can lead to significant shifts in gene networks that may provide selective advantage to the organism (or population), leading to the preservation of the rearrangement.
Fragile sites and EBs also appear to be ‘sinks' for the accumulation of endogenous retroelements (Darai-Ramqvist et al., 2008). Such an association for retroelement accumulation at fragile sites and EB (Longo et al., 2009) begs the question: are these chromosomal regions in and of themselves selfish? Perhaps the same mechanisms that aide in the propagation of retroelements, namely replication fork stall and collapse (Zaratiegui et al., 2011), are also the mechanisms that perpetuate fragile sites and EB through genome ‘shuffling' events.
Concluding remarks
Certain genomic regions are conserved phylogenetically, others, EB and fragile sites are frequently involved in chromosome rearrangements. Conservation and reuse result from functional constraints at the chromosome breakpoints or the presence of fragile sites. For example, in macropodid marsupials, centromeres (both active and latent) are sites of breakpoint reuse (O'Neill et al., 2004; Bulazel et al., 2007), yet retain function as centromere-viable regions of the genome. In contrast, fragile sites have been the main constraint on the evolution of Drosophila chromosomes within nine species, especially for the gene order patterns observed on the X chromosome (von Grotthuss et al., 2010). Interestingly, orthologous fragile sites among these species that are under constraint have internal functional heterogeneity and lack common functional themes, except for the presence of highly conserved non-coding elements (for example, miRNA genes) (von Grotthuss et al., 2010).
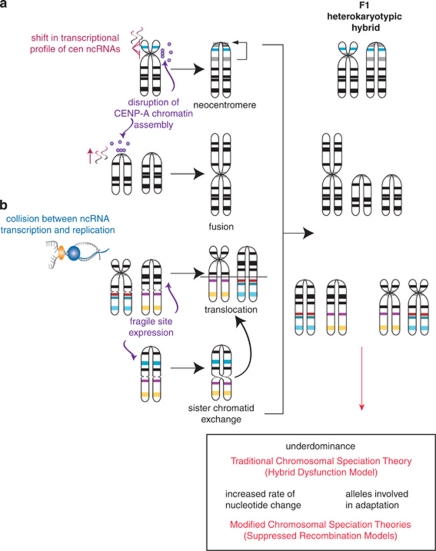
Thus, functionally constrained regions of the genome such as EB, centromeres (as well as telomeres) and fragile sites share a commonality in the density of repeats and ncRNAs, and inferentially in their potential to create new rearrangements through the mechanisms described above (Figure 3). Furthermore, the genome adapts and evolves through chromosome change, potentially a result of the function of ncRNA genes themselves or the structural restraints that ncRNA transcription places on specific chromosome regions. Although the exact mechanisms of chromosome change mediated by ncRNAs are still an area of active research, it is clear that their activity along the length of the entire chromosome can contribute to evolutionary novelty through the introduction of neocentromeres, translocations, fusions and sister chromatid exchange (Figure 3)—chromosome features that delineate lineage-specific karyotypes. Following establishment of a rearrangement in the germ line, the formation of an F1 heterokaryotypic hybrid can then lead to chromosomal speciation through a variety of possible evolutionary processes (Figure 3), including underdominance, an increased rate of nucleotide change within rearranged regions, selection for new alleles formed within rearrangement and genetic conflict (reviewed in Brown and O'Neill, 2010). In essence, ncRNAs not only provide both fragility, preserving chromosome breaks, but also strength and flexibility of chromosomes that can provide the fodder for new speciation events.
Figure 3.
ncRNA facilitates chromosome breaks that can lead to karyotypic speciation. (a) A shift in centromeric (cen) ncRNAs (pink) from the native centromere to another location in the same chromosome results in the disruption of CENP-A nucleosomes (purple) and a change in active centromere location (neocentromere) (top). An increase in cen ncRNAs causes a disruption of CENP-A nucleosomes, resulting in a chromosome fusion event (bottom). (b) Collision of replication forks (yellow) and transcriptional machinery (blue) (as in Figure 3) results in R-loop formation and subsequent expression of fragile sites. These sites then lead to events such as translocations and sister chromatid exchange, which can also lead to translocations. All of the rearrangements shown in A and B lead to F1 heterokaryotypic hybrids after a carrier and normal individual reproduce. This hybrid event can lead to speciation through various means (box). Note: not all possible mechanisms or chromosome derivatives are represented.
They always say its the quiet ones who make the most noise.
Acknowledgments
We apologize to colleagues whose work could not be cited because of space limitations. We are grateful to Julianna Crivello for assistance during the drafting of this manuscript and Michael O'Neill for insightful conversations over speciation theory. JB, SM and RO are supported by a grant from the NSF to RO.
The authors declare no conflict of interest.
References
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breger KS, Smith L, Thayer MJ. Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Hum Mol Genet. 2005;14:2813–2827. doi: 10.1093/hmg/ddi314. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Broadbent K, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, et al. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 2011;12:R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Brown JD, O'Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genomics Hum Genet. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- Bulazel K, Ferreri GC, Eldridge MD, O'Neill RJ. Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol. 2007;8:R170. doi: 10.1186/gb-2007-8-8-r170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2008;118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell. 2002;14:1691–1704. doi: 10.1105/tpc.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch RJ, Kanaya S, Earl PL. A model for the involvement of the small nucleolar RNA (U3) in processing eukaryotic ribosomal RNA. Mol Biol Rep. 1983;9:75–78. doi: 10.1007/BF00777476. [DOI] [PubMed] [Google Scholar]
- Cui L, Miao J. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryot Cell. 2010;9:1138–1149. doi: 10.1128/EC.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darai-Ramqvist E, Sandlund A, Muller S, Klein G, Imreh S, Kost-Alimova M. Segmental duplications and evolutionary plasticity at tumor chromosome break-prone regions. Genome Res. 2008;18:370–379. doi: 10.1101/gr.7010208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RK. RNA interference, transposons, and the centromere. Plant Cell. 2003;15:297–301. doi: 10.1105/tpc.150230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder FF, Robinson TJ. Rodent common fragile sites: are they conserved? Evidence from mouse and rat. Chromosoma. 1989;97:459–464. doi: 10.1007/BF00295030. [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, et al. A Human snoRNA with MicroRNA-Like Functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Feldman AL, Dogan A, Smith DI, Law ME, Ansell SM, Johnson SH, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. 2011;117:915–919. doi: 10.1182/blood-2010-08-303305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958;231:241–257. [PubMed] [Google Scholar]
- Hsieh CL, Lin CL, Liu H, Chang YJ, Shih CJ, Zhong CZ, et al. WDHD1 modulates the post-transcriptional step of the centromeric silencing pathway. Nucleic Acids Res. 2011;39:4048–4062. doi: 10.1093/nar/gkq1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cell. 2005;19:1–15. [PubMed] [Google Scholar]
- Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum mol genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht H, Mai S. 3D imaging of telomeres and nuclear architecture: An emerging tool of 3D nano-morphology-based diagnosis. J Cell Physiol. 2011;226:859–867. doi: 10.1002/jcp.22425. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing. Nature. 1980;283:220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu C, Lin Z, Wu Y, Zhang D, Bai G, et al. Chromosome size in diploid eukaryotic species centers on the average length with a conserved boundary. Mol Biol Evol. 2011;28:1901–1911. doi: 10.1093/molbev/msr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo MS, Carone DM, Green ED, O'Neill MJ, O'Neill RJ. Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genomics. 2009;10:334. doi: 10.1186/1471-2164-10-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, de Oca RM, Probst AV, Vassias I, et al. SUMOylation promotes de novo targeting of HP1á to pericentric heterochromatin. Nat Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1:e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Zhang W, Karpen GH. Frodos found: Behold the CENP-a ‘Ring' bearers. Cell. 2009;137:409–412. doi: 10.1016/j.cell.2009.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens FE.(eds). (2011Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer ( http://cgap.nci.nih.gov/Chromosomes/Mitelman ).
- Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci. 2011;121:141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Noguchi C, Noguchi E. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics. 2007;175:553–566. doi: 10.1534/genetics.106.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RJ, Carone DM. The role of ncRNA in centromeres: a lesson from marsupials. Prog Mol Subcell Biol. 2009;48:77–101. doi: 10.1007/978-3-642-00182-6_4. [DOI] [PubMed] [Google Scholar]
- O'Neill RJ, Eldridge MD, Metcalfe CJ. Centromere dynamics and chromosome evolution in marsupials. J Hered. 2004;95:375–381. doi: 10.1093/jhered/esh063. [DOI] [PubMed] [Google Scholar]
- Petter M, Lee CC, Byrne TJ, Boysen KE, Volz J, Ralph SA, et al. Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter. PLoS Pathog. 2011;7:e1001292. doi: 10.1371/journal.ppat.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci USA. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Probst A, Almouzni G. Pericentric heterochromatin: dynamic organization during early development in mammals. Differentiation. 2008;76:15–23. doi: 10.1111/j.1432-0436.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Nichol Edamura K, et al. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39:1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biology. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe V, Dittberner L, Lorenz VN, Drieschner N, Nimzyk R, Sendt W, et al. The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS ONE. 2010;5:e9485. doi: 10.1371/journal.pone.0009485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TJ, Ruiz-Herrera A, Froenicke L. Dissecting the mammalian genome--new insights into chromosomal evolution. Trends Genet. 2006;22:297–301. doi: 10.1016/j.tig.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Ruiz JF, Gomez-Gonzalez B, Aguilera A. AID induces double-strand breaks at immunoglobulin switch regions and c-MYC causing chromosomal translocations in yeast THO mutants. PLoS Genet. 2011;7:e1002009. doi: 10.1371/journal.pgen.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Castresana J, Robinson TJ. Is mammalian chromosomal evolution driven by regions of genome fragility. Genome Biol. 2006;7:R115. doi: 10.1186/gb-2006-7-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Garcia F, Fronicke L, Ponsa M, Egozcue J, Caldes MG, et al. Conservation of aphidicolin-induced fragile sites in Papionini (Primates) species and humans. Chromosome Res. 2004;12:683–690. doi: 10.1023/B:CHRO.0000045753.88789.ea. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Garcia F, Mora L, Egozcue J, Ponsa M, Garcia M. Evolutionary conserved chromosomal segments in the human karyotype are bounded by unstable chromosome bands. Cytogenet Genome Res. 2005;108:161–174. doi: 10.1159/000080812. [DOI] [PubMed] [Google Scholar]
- Sikdar N, Banerjee S, Zhang H, Smith S, Myung K. Spt2p defines a new transcription-dependent gross chromosomal rearrangement pathway. PLoS Genet. 2008;4:e1000290. doi: 10.1371/journal.pgen.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Stoffregen EP, Donley N, Stauffer D, Smith L, Thayer MJ. An autosomal locus that controls chromosome-wide replication timing and mono-allelic expression. Hum Mol Genet. 2011;20:2366–2378. doi: 10.1093/hmg/ddr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, Jacques PE, Laramee L, Cheng B, Galicia S, Bataille AR, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Str Mol Biol. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Str Mol Biol. 2010;17:1030–1034. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- Teichler S, Illmer T, Roemhild J, Ovcharenko D, Stiewe T, Neubauer A. MicroRNA29a regulates the expression of the nuclear oncogene Ski. Blood. 2011;118:1899–1902. doi: 10.1182/blood-2010-09-306258. [DOI] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic D. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6:1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- von Grotthuss M, Ashburner M, Ranz JM. Fragile regions and not functional constraints predominate in shaping gene organization in the genus Drosophila. Genome Res. 2010;20:1084–1096. doi: 10.1101/gr.103713.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, et al. siRNA-mediated methylation of Arabidopsis telomeres. PLoS genetics. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Siegel ER, Davidson MK. Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PLoS One. 2008;3:e2887. doi: 10.1371/journal.pone.0002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. Involvement of RNA in the synthesis of proteins. Science. 1963;140:17–26. doi: 10.1126/science.140.3562.17. [DOI] [PubMed] [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen C-Y, Brodsky FM, et al. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Aca Sci U S A. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Vaughn MW, Irvine DV, Goto D, Watt S, Bahler J, et al. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]