Abstract
Variation in the number and chromosomal location of nucleolar organizer regions (NORs) was studied in the house mouse, Mus musculus (2n=40). From an origin in Western Asia, this species colonized the Middle East, Europe and Asia. This expansion was accompanied by diversification into five subspecies. NOR diversity was revealed by fluorescence in situ hybridization using 18S and 28S probes on specimens spanning Asia to Western Europe. The results showed that the house mouse genome possessed a large number of NOR-bearing autosomes and a surprisingly high rate of polymorphism for the presence/absence of rRNA genes on all these chromosomes. All NOR sites were adjacent to the centromere except for two that were telomeric. Subspecific differentiation established from the NOR frequency data was concordant with the overall pattern of radiation proposed from molecular studies, but highlighted several discrepancies that need to be further addressed. NOR diversity in M. musculus consisted of a large number of polymorphic NORs that were common to at least two subspecies, and a smaller number of NORs that were unique to one subspecies. The most parsimonious scenario argues in favor of a subspecific differentiation by lineage sorting of ancestral NOR polymorphisms; only the unique NORs would have appeared by inter-chromosomal transposition, except for the two telomeric ones that may have originated by hybridization with another species. Such a scenario provides an alternative view from the one prevailing in most systematic and phylogenetic analyses that NORs have a high transposition rate due to concerted evolution of rRNA genes.
Keywords: NOR, house mouse, phylogeny, radiation, mus musculus, rDNA
Introduction
Nucleolar organizer regions (NORs) are chromosomal landmarks that consist of tandemly repeated sequences of ribosomal genes (rRNA). In eukaryotes, each unit is composed of three genes coding for 18S, 5.8S and 28S ribosomal RNA; these genes are separated by two intergenic spacers and an external transcribed spacer. Head-to-tail repeats of these units form distinct clusters on one to several chromosome pairs. The rRNA multigene family undergoes concerted evolution, whereby the coding fraction of the rDNA sequences is homogenized so that copies within a species are generally more similar than those between species (Eickbush and Eickbush, 2007). Several molecular processes are involved in sequence homogenization: unequal crossover, gene conversion and non-homologous recombination (Gonzalez and Sylvester, 2001; Parkin and Butlin, 2004). The concerted evolution of these sequences is thought to be under purifying selection to ensure the functionality of these essential translational genes. As the number and position of the rDNA clusters are often species-specific, these chromosomal characters have been widely used in systematics and phylogenetic reconstructions. Studies on NOR variation in numerous plant, insect and vertebrate groups have invariably described changes in the number and chromosomal location of NORs even in closely related species, suggesting that rDNA clusters are highly mobile components of the genome (Gallagher et al., 1999; Datson and Murray, 2006; Cabrero and Camacho, 2008; Nguyen et al., 2010; da Silva et al., 2010; Cazaux et al., 2011). This important inter-species lability is generated either by chromosomal rearrangements or transposition events (Eickbush and Eickbush, 2007). When no chromosomal repatterning is observed, several authors have ascribed the high transposition rates of NORs to the existence of transposons in the rDNA clusters; such elements have been observed in plants, invertebrates and recently in intergenic spacer (IGS) repeats of the house mouse (Grozdanov et al., 2003; Raskina et al., 2008; da Silva et al., 2010). The vast majority of these investigations are based on the silver-staining method which presents two drawbacks: it reveals only the active fraction of the rRNA genes, that is, those that were transcribed during the previous interphase, and in some cases, it may lead to non-specific staining of heterochromatic regions (Dobigny et al., 2003; Cabrero and Camacho, 2008; Carvalho et al., 2009). Access to the genes themselves by the fluorescence in situ hybridization (FISH) technique has provided a cytogenetic tool to explore the extent of NOR diversity between species irrespective of their expression status (Matsubara et al., 2004; Datson and Murray, 2006; Nguyen et al., 2008). However, many of these studies involve comparisons between species, and thus provide little information on the evolutionary processes acting on variation at the intraspecific level. To investigate the short-term evolutionary dynamics of rDNA clusters, we analyzed NOR diversity by the FISH approach in an emblematic species, the house mouse.
The house mouse, Mus musculus, is one of the 14 species comprising the subgenus Mus, which is remarkable for its highly conserved karyotype (2n=40). A previous phylogenetic analysis of NOR variation within the subgenus indicated that the number and location of clusters between species differed, each taxon showing its own chromosomal distribution (Cazaux et al., 2011). This extensive NOR diversity indicated that high rates of inter-chromosomal transposition existed within the Mus genome in the absence of cytogenetically visible rearrangements. The house mouse is an Eurasian polytypic species with five currently recognized subspecies: M. m. domesticus, the Western European house mouse, occupies western Europe and the Mediterranean Basin to southwestern Iran; M. m. musculus, the eastern European house mouse, extends in fact from northern Europe to China; M. m. castaneus, the Asian house mouse, is present from central to southeast Asia (Bonhomme et al., 2007; Geraldes et al., 2008; Rajabi-Maham et al., 2008); M. m. molossinus, the Japanese house mouse, is restricted to Japan and neighboring countries and originated by hybridization between M. m. musculus and M. m. castaneus (Yonekawa et al., 2012); finally M. m. gentilulus, the Arabian house mouse, is the most recently rehabilitated subspecies occupying the southern Arabian peninsula (Prager et al., 1998; Duplantier et al., 2002). Two extant hybrid zones exist involving M. m. musculus, one with M. m. domesticus in Europe and the other with M. m. castaneus in China (Bonhomme et al., 2007). From its native Eurasian distribution, the house mouse subsequently expanded its range worldwide through passive transport with humans. The evolutionary history of the subspecific radiation of M. musculus has been extensively documented and most of the molecular analyses concur with the paleontological data in identifying the Indian subcontinent as the cradle from which the species expanded (Bonhomme et al., 2007; Geraldes et al., 2008; Duvaux et al., in press). This region is currently occupied by M. m. castaneus and represents the ancestral range of the species. Analyses based on mitochondrial or sex-linked sequences generally define monophyletic subspecific lineages, particularly in the peripheral populations of the subspecies, that is, Europe, Arabia, Japan and Southeast Asia (Prager et al., 1998; Duplantier et al., 2002; Geraldes et al., 2008; Rajabi-Maham et al., 2008). In contrast, the fewer studies using autosomal nuclear markers have highlighted the genetic interrelatedness of the subspecies gene pools due to an ancient episode of gene flow or to lineage sorting (Bonhomme et al., 2007; Geraldes et al., 2008; Duvaux et al., in press).
The present study investigates micro-evolutionary patterns of genomic change by determining the dynamics of NOR diversity within M. musculus. The performance of this nuclear chromosomal marker in tracking subspecific differentiation is assessed and compared with previous radiation scenarios. In addition, the processes generating the intraspecific distribution of NOR diversity within the house mouse are evaluated.
Materials and methods
Mouse samples
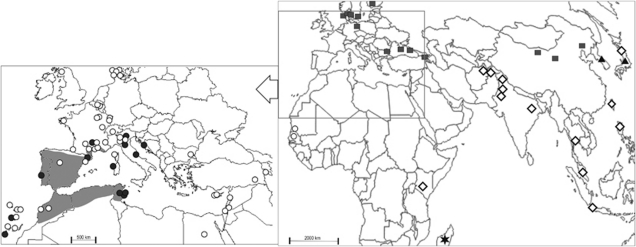
A total of 74 wild house mice were analyzed from 68 localities (see Table 1, Figure 1 and Supplementary Table S1) distributed in Europe (Armenia, Belgium, Bulgaria, Finland, France, Italy, Portugal, Scotland, Spain, Switzerland), the Mediterranean Basin (Cyprus, Egypt, Israel, Lebanon, Mauritania, Morocco, Tunisia, Turkey), Africa (Madagascar, Senegal, Kenya) and Asia (India, Pakistan, Thailand). In most cases, only one individual per locality was studied. Four of the five subspecies of M. musculus are represented in this survey: M. m. domesticus (DOM), M. m. musculus (MUS), M. m. castaneus (CAS), and M. m. gentilulus (GEN). The data pertaining to the fifth subspecies, M. musculus molossinus (MOL) were sampled from the literature (Suzuki et al., 1990; Ito et al., 2007, 2008). DOM individuals carrying Robertsonian fusions (see Piálek et al., 2005) were included in the analyses to increase the geographical coverage of this subspecies (see Supplementary Table S1). The NOR distribution between the samples carrying a standard karyotype (2n=40) and the Robertsonian mice (2n<40) was not significantly different (FST=0.0015; P=0.41). Subsequently, DOM specimens were pooled irrespective of their karyotype. The subspecific assignation of the studied specimens was determined from published molecular analyses of these individuals (mitochondrial and nuclear sequences; Duplantier et al., 2002; Auffray et al., 2003; Bonhomme et al., 2007). These studies revealed that the mouse from Madagascar had a mosaic genome: while it clearly harbored a distinct mitochondrial genome attributed to GEN (Prager et al., 1998; Duplantier et al., 2002), nuclear markers indicated an affinity with CAS (Bonhomme et al., 2007). Where possible, the samples were pooled by geographic origin; given the extensive data for DOM, the specimens were grouped into three regional clusters: the Mediterranean Basin, including southern Europe (S), northern Europe (N) and the Atlantic Coast (A).
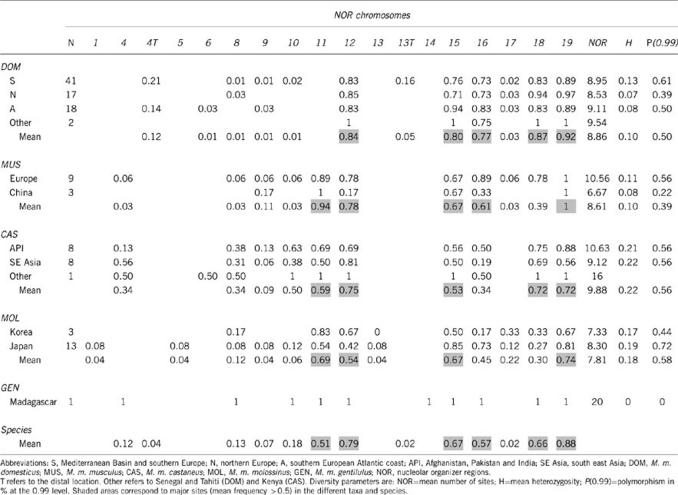
Table 1. NOR frequency distribution in the different geographic regions and subspecies.
Figure 1.
Distribution maps of the 68 localities sampled in the present analysis (74 mice) and those taken from the literature (40 localities and 50 mice). See Supplementary Table S1 for the geographical coordinates and references. Four subspecies are presented in the worldwide map: M. m. musculus (square; MUS), M. m. castaneus (diamond; CAS), M. m. molossinus (triangle, MOL), M. m. gentilulus (star, GEN). The M. m. domesticus samples (circle: DOM) are presented in the insert map; the distribution of mice with (dark circle) or without (white circle) NORs 4T and/or 13T are indicated. The shaded area refers to the distribution range of M. spretus.
Methods
Mitotic metaphases were obtained by the air-drying method from bone marrow cells after yeast stimulation. The number and location of the rRNA genes were detected by FISH using 18S (SalC-pSP64; 2 kb) and 28S (BE-2-pSP64; 1.5 kb) probes, following the procedure detailed in Cazaux et al., 2011. Identification of chromosomes was performed by DAPI (4,6-diamidino-2-phenylindole)-Banding following the standard nomenclature. A minimum of 10 metaphases was observed per individual. All observations were made with a Zeiss Axiophot fluorescence microscope (Oberkochen, Germany) equipped with an image analyzer (Cytovision 3.93.2, Genetix, New Milton, UK).
Analyses
rDNA genotypes
The rDNA genotypes were scored directly from the metaphase FISH signals, considering each chromosomal location as a locus with two allelic states (presence and absence) and three possible genotypes (homozygous with or without an rDNA cluster, heterozygous). Thus, rRNA genes were considered as present on a chromosome when a signal was detected regardless of its intensity, and as absent when no signal was visible. Given the sensitivity of the FISH method (lower limit: 3.1 kb; Wang et al., 2006), the size of the probes (3.5 kb) and the protocol (that is, checking signals over several metaphases), the procedure used is at the detection limit for a single copy of the rDNA unit. The following nomenclatures were adopted: NOR followed by a number designates the NOR-bearing chromosome; telomeric NORs are indicated by a T after the chromosome number; a NOR site refers to the location of the rRNA genes on a single chromosome. The data were completed with those available in the literature for wild individuals of DOM, MUS, CAS and MOL, using either the FISH approach (Fel-Clair et al., 1998; Ito et al., 2007, 2008; Cazaux et al., 2011), or the Ag-NOR staining method when genotype scores could be extracted (Winking et al., 1980; Suzuki et al., 1990; Ramalhinho et al., 2005). Details on the procedure used for genotype scoring are indicated in Supplementary Table S1.
Diversity analyses
All diversity indices were computed from the complete genotypic dataset (see Supplementary Table S1). Variability parameters included: frequency, mean heterozygosity, mean rate of polymorphism and linkage disequilibrium. An additional diversity measure, that is, the mean number of sites, was determined. Genotypic differentiation between samples and subspecies was assessed by Wright's FST index. Relationships between samples were investigated by constructing a distance tree using the FST-based Reynold's distance index and the Fitch program available in the PHYLIP v3.5c package (Felsenstein, 1985). The tree topology was drawn with the Treeview v1.6.6 software distributed by Page (1996). All parameters were calculated using GENETIX v4.03 (Belkhir et al., 1996–2004).
Results
Differentiation between subspecies
The cytogenetic analysis identified a total of 16 NOR-bearing chromosomes and 18 NOR sites in M. musculus (Table 1; Figure 2). All of them were autosomal and centromerically located except in DOM where rRNA genes were located at the distal ends of chromosomes 4 and 13 (hereafter 4T, 13T). From the frequency data, two types of NORs were determined in the species as well as in each subspecies: major (>0.5) and minor (⩽0.5). This distribution identified six chromosomes (11, 12, 15, 16, 18 and 19) as carrying major NORs in M. musculus, although they differed in frequency between subspecies. The remaining 10 chromosomes bore minor NORs, some of which were common to all (8, 9, 10 and 17) or several subspecies (4, 6), whereas others appeared as diagnostic subspecific characters (1, 4T, 5, 13, 13T, 14). The number of NOR sites per individual ranged from a minimum of 6 in DOM to a maximum of 16 in CAS and even 20 in GEN (Supplementary Table S1). With the exception of the African samples, the lowest value was present in the MUS sample from China.
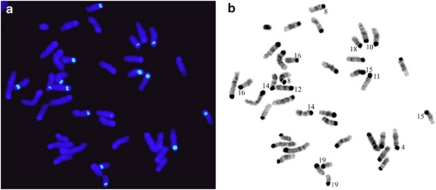
Figure 2.
Detection by FISH (a) and identification of the NOR-bearing chromosomes by DAPI banding (b) of the M. m. gentilulus (GEN) mouse from Madagascar.
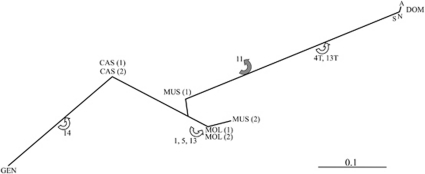
NOR differentiation levels were measured by the FST index (Table 2) and phylogenetic relationships were established from FST-derived distance values using NOR genotypes as genetic markers (Figure 3). The samples outside of the native range of DOM and CAS were omitted as only one individual was studied in each case. The tree depicted several notable patterns. First, two subspecies were highly differentiated (DOM and GEN), whereas the other samples were grouped in the central part of the tree and exhibited lower levels of divergence. Second, MUS did not appear monophyletic, as the European sample did not cluster together with the Chinese one, which in fact was more closely related to both MOL samples. Thirdly, MOL held an intermediate position between the Chinese MUS and CAS.
Table 2. FST differentiation values within and between subspecies.
| Subspecies | Region | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DOM | 1 | S | |||||||||
| 2 | N | 0.015 | |||||||||
| 3 | A | 0.008 | 0.034 | ||||||||
| MUS | 4 | Europe | 0.261 | 0.320 | 0.314 | ||||||
| 5 | China | 0.479 | 0.623 | 0.583 | 0.325 | ||||||
| CAS | 6 | API | 0.287 | 0.316 | 0.335 | 0.123 | 0.210 | ||||
| 7 | SE Asia | 0.313 | 0.347 | 0.368 | 0.246 | 0.258 | 0.035 | ||||
| MOL | 8 | Korea | 0.348 | 0.462 | 0.450 | 0.190 | −0.027 | 0.064 | 0.038 | ||
| 9 | Japan | 0.235 | 0.282 | 0.239 | 0.131 | 0.058 | 0.139 | 0.274 | 0.580 | ||
| GEN | 10 | Madagascar | 0.655 | 0.777 | 0.742 | 0.605 | 0.729 | 0.272 | 0.355 | 0.459 | 0.513 |
Abbreviations: S, Mediterranean Basin and southern Europe; N, northern Europe; A, southern European Atlantic coast; API, Afghanistan, Pakistan and India; SE Asia, south east Asia; DOM, M. m. domesticus; MUS, M. m. musculus; CAS, M. m. castaneus; MOL, M. m. molossinus; GEN, M. m. gentilulus.
Figure 3.
Fitch-Margoliash tree depicting the phylogenetic relationships between regional samples of M. musculus. DOM=M. m. domesticus (S=Mediterranean Basin and southern Europe; N=northern Europe; A=southern European Atlantic coast), MUS=M. m. musculus (1: Europe, 2: China), CAS=M. m. castaneus (1: Afghanistan, Pakistan, India, 2: Southeast Asia), MOL=M. m. molossinus (1: Korea, 2: Japan), GEN=M. m. gentilulus. Losses (grey arrow) and gains (white arrow) of unique NORs are indicated along the branches. Sum of squares=9.91; average standard deviation=33.6%.
Diversity patterns within subspecies
One of the major results of this cytogenetic study was the high degree of polymorphism observed within subspecies. With the exception of the Chinese MUS, rates of polymorphism fluctuated around 50%, meaning that on average, variation was detected in at least one individual in half of the NOR-bearing chromosomes. This large variability was also evident in the mean heterozygosity values, which reached a particularly high level in CAS (Table 1). This was, for a large part, due to the high variability of the minor NOR-bearing chromosomes present in this subspecies (NORs 4, 8, 10). Within MUS, the two geographic samples differed considerably in diversity (mean number of sites, mean heterozygosity, mean rate of polymorphism; see Table 1), the Chinese individuals showing much lower levels than the European MUS. Among the geographic groups within DOM, the sample from the Southern region exhibited the highest level of diversity, most likely due to the relatively large frequency of the distal NORs (4T and 13T). As for MOL, a difference between the FISH (2 mice) and the Ag-staining data (14 mice) was apparent in the Japanese sample, as the three unique NORs were only revealed using FISH; this suggests that these NORs were either locally distributed or were present but never active.
Discussion
The present investigation provides an extensive survey of NOR variation within a species. These data extend previous analyses that used mostly Ag-staining methods in M. musculus (Winking et al., 1980; Suzuki et al., 1990; Fel-Clair et al., 1998; Ramalhinho et al., 2005). A total of 16 NOR-bearing chromosomes were identified comprising 16 centromeric and 2 telomeric locations on these chromosome pairs. All of these NOR sites (except NOR 14) have previously been identified in the species (Winking et al., 1980; Suzuki et al., 1990; Ramalhinho et al., 2005; Ito et al., 2007, 2008), but their subspecific distribution is here refined (for example, NORs 8, 9, 10 and 17 were previously unknown in DOM). The frequency distribution of NORs showed considerable variation between chromosome pairs within and between subspecies. Of the six major NORs in the species, three were present at a relatively high frequency in all subspecies (NORs 12, 15 and 19), whereas three showed a mixed pattern: NORs 11, 16 and 18. The former was absent in DOM, whereas the frequency of the latter varied considerably among subspecies. Six minor NORs were unique to one subspecies and might be considered as diagnostic: NOR 14 in GEN, NORs 1, 5 and 13 in MOL, and NORs 4T and 13T in DOM. However, except for DOM, these results need to be confirmed by additional sampling particularly in GEN as only one individual was analyzed.
Do NORs retrace the evolutionary history of the M. musculus radiation?
The NOR-based phylogeny is essentially compatible with the currently established relationships for the three main subspecies: DOM is the most differentiated subspecies whereas MUS and CAS are more closely related (Bonhomme et al., 2007), although all sister group associations have been found depending on the marker used (see Bonhomme et al., 2007; Geraldes et al., 2008). MOL, which carries a MUS/CAS composite genome has, as expected, an intermediate position in the tree. However, the NOR tree highlights a discrepancy with previous studies in the position of GEN by way of considering it either as a sister taxon of DOM (Duplantier et al., 2002; Rajabi-Maham et al., 2008), or as closely related to CAS (Bonhomme et al., 2007). This apparent conflict with published data may stem from the genomic compartments analyzed (mitochondrial vs nuclear markers), and requires further analyses with increased sample sizes to be resolved. The NOR data also retrieved an unexpected differentiation within MUS, with Chinese populations sharing stronger affinities with MOL rather than European MUS. This result agrees with increasing evidence of a southeast Asian lineage within MUS, as well as current views of the geographic origin of the MUS genomic contribution to MOL (Yonekawa et al., 2012). NOR variability levels are in agreement with colonization patterns that predict higher diversity in populations from the area of origin compared with those present in peripheral regions (Geraldes et al., 2008; Rajabi-Maham et al., 2008). Thus, CAS from the Indo-Pakistani region has the highest mean heterozygosity value among the subspecies (Table 1), although MOL is the one that shows the highest diversity in number of NOR-bearing chromosomes (13 vs 11 in the other subspecies). Within DOM, the variability levels observed between regions also match those predicted by the colonization of Northern Europe from the Mediterranean Basin (Auffray et al., 1990) with loss of diversity by genetic drift and/or founder effects. In conclusion, the study of NOR variation is concordant with the overall pattern of divergence proposed from molecular data with a subspecific radiation following an expansion from a west–central Asian cradle. In addition, subspecific differentiation of NORs provides several diagnostic genetic markers for several subspecies, and supports an intra-subspecific divergence within MUS.
Evolutionary dynamics of NORs
The present study has brought to light two remarkable traits in M. musculus: the high number of NOR-bearing chromosomes, as well as the high level of polymorphism within and between subspecies. When no chromosomal repatterning is evident, evolution of the number and location of NORs is thought to proceed by gains and losses of sites. The major driving forces are transposition of rDNA sequences from one chromosome pair to a new one, or unequal crossover exchanges between non-homologous chromosomes with subsequent in situ expansion or contraction of rDNA copies (Eickbush and Eickbush, 2007). Indications from restriction fragment length polymorphism and sequence data of rRNA genes in M. musculus suggest that intra- and inter-chromosomal homogenization through concerted evolution is occurring at a relatively high rate in this species (Suzuki et al., 1986), the latter being apparently less frequent than the former (Sasaki et al., 1987). The chromosomal distribution and variability pattern of NORs between subspecies of M. musculus suggest the following scenario of evolutionary change. The six major NOR-bearing chromosomes common to all subspecies are most likely ancestral in the species and subspecific differentiation took place by the independent loss of NOR 11 in the DOM lineage as previously noted by Suzuki et al., 1990. Minor NORs, however, may have two origins: independent transposition events in each lineage with subsequent gene flow, or lineage sorting of ancestral polymorphisms. These two processes may in fact be involved depending on the distribution of these NORs among subspecies. The existence of minor NOR-bearing chromosomes that are common to several subspecies (NORs 4, 6, 8, 9, 10, 17) suggests that they either correspond to low frequency ancestral polymorphisms or, if they originated by de novo transposition, this event occurred sufficiently early during the differentiation process for them to spread to the different sublineages. That a large number of ancestral NORs may be present in this species is supported by the high number of NORs recorded in the subgenus Mus (Cazaux et al., 2011). In fact, in each of the three main clades of the subgenus, one to several species carry NORs on all (19) or almost all (16) autosome pairs. From this, it may be inferred that all autosomes may potentially bear NORs in species of the subgenus Mus. Further support for such a view stems from a previous study that identified rDNA-related sequences (that is, without coding sequences) on chromosomes 5, 6 and 17 in the DOM-derived laboratory strain C57Bl/6J, which are undetected by the FISH technique (Rowe et al., 1996). These NOR traces may represent the signatures of previous rDNA locations in the subgenus. As for the minor NORs that are rare or unique to one subspecies (NORs 1, 4T, 5, 13, 13T, 14), an independent transposition event in each subspecies or group of subspecies may be more likely.
What does this scenario tell us of the rates of NOR evolution in M. musculus? The most parsimonious scenario developed above posits that the majority of NORs in this species existed as ancestral (major NORs) or near ancestral (shared minor ones) sites. Three processes would have led to the subspecific NOR distribution detected: subsequent complete or incomplete lineage sorting, sporadic gene flow during divergence and/or unequal crossover events within homologous chromosomes. In other words, few or no mutation events (that is, transposition) are required to account for the subspecific patterns observed, which would be mainly due to stochastic processes that accompanied the expansion of the different sublineages. If rare or unique sites are the only ones to have been generated by de novo transposition, this would imply an inter-chromosomal transposition rate of six events in 0.5 MY at the most (Geraldes et al., 2008; Duvaux et al., in press).
Where do the 4T and 13T NORs in DOM come from?
The existence of telomeric NORS on two chromosome pairs (4T, 13T) in DOM is exceptional in the species and even the genus Mus. Among the 41 species belonging to the genus, NORs are always centromerically located except in one species, Mus spretus, in which three telomeric NORs have been described (4T, 13T, 19T; Winking et al., 1980; Cazaux et al., 2011). The coincidence in location in the two taxa is intriguing. Two explanations come to mind. The first considers that ancestral sequences that act as receptors for the integration of ribosomal gene units exist at these locations (Bodega et al., 2006) and persisted in the genome of M. musculus after the split with M. spretus (Rowe et al., 1996). Their occurrence in DOM would then be ascribed to two transposition events resulting in convergence between the two species. The second possibility posits that the telomeric NORs introgressed into the DOM genome by hybridization with M. spretus. That these species do hybridize in nature, albeit exceptionally, has been confirmed by molecular analyses (Orth et al., 2002). To investigate this further, the geographical distribution of the DOM samples carrying the telomeric NORs was mapped onto that of M. spretus (Figure 3). The comparison indicates that the two distributions overlap, suggesting the potential introgressive origin of the telomeric NORs in DOM. This interpretation is further sustained by the presence of mice in Tunisia carrying both 4T and 13T NORs that are in linkage disequilibrium (P=0.0001; see Supplementary Table S1). However, several DOM samples lie outside the zone of sympatry (Italy, Croatia and France). As there is no fossil evidence that the distribution area of M. spretus may have been larger at the time the house mouse arrived (Auffray et al., 1990), the outlier samples may be the signature of rare instances of long-distance migration within Southern Europe from a western Mediterranean source.
Consequences for phylogenetic reconstructions
NORs have been widely and successfully used as cytogenetic markers to assess patterns of chromosomal evolution particularly in non-mammalian organisms for which banding data are less tractable (Gallagher et al., 1999; Cabrero and Camacho, 2008; Raskina et al., 2008; Carvalho et al., 2009; Nguyen et al., 2010; da Silva et al., 2010). However, these investigations often included only one or a few individuals per species, so population surveys of NOR variability using FISH remain scarce (Veltsos et al., 2009). One indication of intraspecific NOR variability stems from a study in humans in which 54.2% exhibited a polymorphism for the absence/presence of rRNA genes on the NOR-bearing chromosomes (Zavala Guillén et al., 2004). Even though additional intraspecific estimates of NOR variation are required to assess the extent of intrapopulation variability, polymorphism may be more widespread than previously thought. If so, this may provide an explanation for the apparent high rates of transposition between taxa that are often inferred from inter-species comparisons. In phylogenetic reconstructions, such results have sometimes been interpreted as a signature of homoplasy, when in fact they may correspond to hemiplasy or lineage sorting (Robinson et al., 2008). An illustration of this may be found in a recent phylogenetic analysis of Bovidae using chromosomal characters, 17 of which were NORs (Gallagher et al., 1999; Nguyen et al., 2008). Several of the individuals analyzed were heterozygous at seven NOR-bearing chromosomes, four of which were homoplasic in the reconstructed tree. An alternative explanation worthy of consideration would involve a trans-specific polymorphism for these NORs with subsequent lineage sorting between species.
Conclusion
This survey of NOR diversity in the house mouse has highlighted the extensive polymorphism of all rDNA clusters within and between subspecies. NOR clusters thus behave as neutral genetic markers tracking evolutionary variation in the nuclear genome among subspecies of M. musculus (see Veltsos et al., 2009). The processes regulating this variability remain, however, poorly understood. Is the extent of polymorphism related to the number of NORs in the species: is it higher in species with many NOR-bearing chromosomes (such as mice) than in those with few? How does this polymorphism relate to regulation of the activity of the rRNA gene copies? The presence of shared polymorphic NORs among subspecies in M. musculus argues in favor of subspecific divergence by lineage sorting of ancestral polymorphisms. In addition, if the two unique telomeric NORs in DOM have an introgressive origin, only four NORs would have appeared by transposition to a new location in the species. Thus, if NOR polymorphisms are in fact widespread among organisms, then rates of transposition may be lower than previously thought. Definite resolution of the mode of NOR evolution in M. musculus awaits comparative sequence analyses of the flanking regions of NORs as well as of the rDNA arrays (Bodega et al., 2006). Such studies will shed light on the extent of transposition rates and its relationship with concerted evolution.
Data archiving
There was no data to deposit.
Acknowledgments
We acknowledge R Moussa, M Karam, R Kabalan and the Davidians' for their field support, and G Lefranc as well as A Mégarbané with all his team for access to the cytogenetics laboratory at the Université St Joseph in Beyrouth. The rDNA 18S and 28S probes were gifts from J-P Bachellerie. We wish to thank P Boursot for insightful discussions and F Veyrunes for useful comments on the MS. All FISH analyses were performed on the Evolutionary Cytogenomics Platform of the IFR119. Funding was provided by recurrent grants from the CNRS and the UM2 to the ISEM.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Auffray JC, Orth A, Catalan J, Gonzalez JP, Desmarais E, Bonhomme F. Phylogenetic position and description of a new species of subgenus Mus (Rodentia, Mammalia) from Thailand. Zool Scripta. 2003;32:119–127. [Google Scholar]
- Auffray JC, Vanlerberghe F, Britton-Davidian J. The house mouse progression in Eurasia: a palaeontological and archaeozoological approach. Biol J Linn Soc. 1990;41:13–25. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, Logiciel Sous Windows TM Pour la Génétique Des Populations. Université de Montpellier II: Montpellier, France; 1996–2004. [Google Scholar]
- Bodega B, Cardone MF, Rocchi M, Neusser M, Orzan F, Rossi E, et al. The boundary of macaque rDNA is constituted by low-copy sequences conserved during evolution. Genomics. 2006;88:564–571. doi: 10.1016/j.ygeno.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Bonhomme F, Rivals E, Orth A, Grant GR, Jeffreys AJ, Bois PRJ. Species-wide distribution of highly polymorphic minisatellite markers suggests past and present genetic exchanges among house mouse subspecies. Genome Biol. 2007;8:R80. doi: 10.1186/gb-2007-8-5-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrero J, Camacho JPM. Location and expression of ribosomal RNA genes in grasshoppers: abundance of silent and cryptic loci. Chrom Res. 2008;16:595–607. doi: 10.1007/s10577-008-1214-x. [DOI] [PubMed] [Google Scholar]
- Carvalho KA, Garcia PCA, Recco-Pimentel SM. NOR dispersion, telomeric sequence detection in centromeric regions and meiotic multivalent configurations in species of the Aplastodiscus albofrenatus group (Anura, Hylidae) Cytogenet Genome Res. 2009;126:359–367. doi: 10.1159/000264179. [DOI] [PubMed] [Google Scholar]
- Cazaux B, Catalan J, Veyrunes F, Douzery EJP, Britton-Davidian J. Are ribosomal DNA clusters rearrangement hotspots? A case study in the genus Mus (Rodentia, Muridae) BMC Evol Biol. 2011;11:124. doi: 10.1186/1471-2148-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva CRM, Quintas CC, Vanzela ALL. Distribution of 45S and 5S rDNA sites in 23 species of Eleocharis (Cyperaceae) Genetica. 2010;138:951–957. doi: 10.1007/s10709-010-9477-5. [DOI] [PubMed] [Google Scholar]
- Datson PM, Murray BG. Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism. Chrom Res. 2006;14:845–857. doi: 10.1007/s10577-006-1092-z. [DOI] [PubMed] [Google Scholar]
- Dobigny G, Ozouf-Costaz C, Bonillo C, Volobouev V. ‘Ag-NORs' are not always true NORs: new evidence in mammals. Cytogenet Genome Res. 2003;98:75–77. doi: 10.1159/000068541. [DOI] [PubMed] [Google Scholar]
- Duplantier J-M, Orth A, Catalan J, Bonhomme F. Evidence for mitochondrial lineage originating from the Arabian peninsula in the Madagascar house mouse (Mus musculus) Heredity. 2002;89:154–158. doi: 10.1038/sj.hdy.6800122. [DOI] [PubMed] [Google Scholar]
- Duvaux L, Boulesteix M, Belkhir K, Boursot P.(in press)Isolation and gene flow: inferring the speciation history of European house mice Mol Ecol [DOI] [PubMed]
- Eickbush TH, Eickbush DG. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fel-Clair F, Catalan J, Lenormand T, Britton-Davidian J. Centromeric incompatibilities in the hybrid zone between house mouse subspecies from Denmark: evidence from patterns of NOR activity. Evolution. 1998;52:592–603. doi: 10.1111/j.1558-5646.1998.tb01657.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Gallagher DSJ, Davis SK, De Donato M, Burzlaff JD, Womack JE, Taylor JF, et al. A molecular cytogenetic analysis of the tribe Bovini (Artiodactyla: Bovidae: Bovinae) with an emphasis on sex chromosome morphology and NOR distribution. Chrom Res. 1999;7:481–492. doi: 10.1023/a:1009254014526. [DOI] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, et al. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 2008;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE. Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics. 2001;73:255–263. doi: 10.1006/geno.2001.6540. [DOI] [PubMed] [Google Scholar]
- Grozdanov P, Geogiev O, Karagyozov L. Complete sequence of the 45-kb mouse ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 2003;82:637–643. doi: 10.1016/s0888-7543(03)00199-x. [DOI] [PubMed] [Google Scholar]
- Ito T, Osawa S, Shibata H, Kanda N. Identification of rRNA gene loci in the wild mouse (Mus musculus molossinus) captured at Hachioji, Tokyo. J Vet Med Sci. 2007;69:1277–1279. doi: 10.1292/jvms.69.1277. [DOI] [PubMed] [Google Scholar]
- Ito T, Tsuchiya K, Osawa S, Shibata H, Kanda N. Mapping of rRNA gene loci in the mice, Mus musculus molossinus (Japan) captured and Mus musculus musculus (Russia) by double color FISH. J Vet Med Sci. 2008;70:997–1000. doi: 10.1292/jvms.70.997. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Nishida-Umehara C, Tsuchiya K, Nukaya D, Matsuda Y. Karyotypic evolution of Apodemus (Muridae, Rodentia) inferred from comparative FISH analyses. Chrom Res. 2004;12:383–395. doi: 10.1023/B:CHRO.0000034103.05528.83. [DOI] [PubMed] [Google Scholar]
- Nguyen P, Sahara K, Yoshido A, Marec F. Evolutionary dynamics of rDNA clusters on chromosomes of moths and butterflies (Lepidoptera) Genetica. 2010;138:343–354. doi: 10.1007/s10709-009-9424-5. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Aniskin VM, Gerbault-Seureau M, Planton H, Renard JP, Nguyen BX, et al. Phylogenetic position of the saola (Pseudoryx nghetinhensis) inferred from cytogenetic analysis of eleven species of Bovidae. Cytogenet Genome Res. 2008;122:41–54. doi: 10.1159/000151315. [DOI] [PubMed] [Google Scholar]
- Orth A, Belkhir K, Britton-Davidian J, Boursot P, Benazzou T, Bonhomme F. Natural hybridization between two sympatric species of mice Mus musculus domesticus L and Mus spretus Lataste. C R Acad Sci. 2002;325:89–97. doi: 10.1016/s1631-0691(02)01413-0. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Parkin EJ, Butlin RK. Within- and between-individual sequence variation among ITS1 copies in the meadow grasshopper Chorthippus parallelus indicates frequent intrachromosomal gen conversion. Mol Biol Evol. 2004;21:1595–1601. doi: 10.1093/molbev/msh163. [DOI] [PubMed] [Google Scholar]
- Piálek J, Hauffe HC, Searle JB. Chromosomal variation in the house mouse. Biol J Linn Soc. 2005;84:535–563. [Google Scholar]
- Prager EM, Orrego C, Sage RD. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics. 1998;150:835–861. doi: 10.1093/genetics/150.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabi-Maham H, Orth A, Bonhomme F. Phylogeography and postglacial expansion of Mus musculus domesticus inferred from mitochondrial DNA coalescent from Iran to Europe. Mol Ecol. 2008;17:627–641. doi: 10.1111/j.1365-294X.2007.03601.x. [DOI] [PubMed] [Google Scholar]
- Ramalhinho MG, Braz C, Catalan J, Mathias ML, Britton-Davidian J. AgNOR variability among Robertsonian races of the house mouse from the island of Madeira: implications for patterns of Rb fusion formation and genetic differentiation. Biol J Linn Soc. 2005;84:585–591. [Google Scholar]
- Raskina O, Barber JC, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res. 2008;120:351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- Robinson TJ, Ruiz-Herrera A, Avise J. Hemiplasy and homoplasy in the karyotypic phylogenies of mammals. Proc Natl Acad Sci. 2008;105:14477–14481. doi: 10.1073/pnas.0807433105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe LB, Janaswami PM, Barter ME, Birkenmeier EH. Genetic mapping of 18S ribosomal RNA-related loci to mouse chromosomes 5, 6, 9, 12, 17, 18, 19, and X. Mamm Genome. 1996;7:886–889. doi: 10.1007/s003359900262. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Okazaki T, Kominami R. Variation among mouse ribosomal RNA genes within and between chromosomes. Mol Biol Evol. 1987;4:594–601. doi: 10.1093/oxfordjournals.molbev.a040469. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kurihara Y, Moriwaki K. Variation in the distribution of silver-staining Nucleolar Organizer Regions on the chromosomes of the wild mouse, Mus musculus. Mol Biol Evol. 1990;7:271–282. doi: 10.1093/oxfordjournals.molbev.a040598. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Miyashita N, Moriwaki K, Kominami R, Muramatsu M, Kanehisa T, et al. Evolutionary implications of heterogeneity of the nontranscribed spacer region of ribosomal DNA repeating units in various subspecies of Mus musculus. Mol Biol Evol. 1986;3:126–137. doi: 10.1093/oxfordjournals.molbev.a040384. [DOI] [PubMed] [Google Scholar]
- Veltsos P, Keller I, Nichols RA. Geographical localised bursts of ribosomal DNA mobility in the grasshopper Podisma pedestris. Heredity. 2009;103:54–61. doi: 10.1038/hdy.2009.32. [DOI] [PubMed] [Google Scholar]
- Wang C-JR, Harper L, Cande Z. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell. 2006;18:529–544. doi: 10.1105/tpc.105.037838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winking H, Nielsen K, Gropp A. Variable positions of NORs in Mus musculus. Cytogenet Cell Genet. 1980;26:158–164. doi: 10.1159/000131437. [DOI] [PubMed] [Google Scholar]
- Yonekawa H, Sato JJ, Suzuki H, Moriwaki K.2012Origin and genetic status of Mus musculus molossinus: a typical example for reticulate evolution in the genus MusIn: Macholan M, Baird S, Munclinger P, Piálek J (eds).Evolution of the House Mouse Cambridge University Press: Cambridge, UK [Google Scholar]
- Zavala Guillén AK, Hirai Y, Tanoue T, Hirai H. Transcriptional repression mechanisms of nucleolus organizer regions (NORs) in humans and chimpanzees. Chrom Res. 2004;12:225–237. doi: 10.1023/b:chro.0000021911.43225.eb. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.