Abstract
In mammals, birds, snakes and many lizards and fish, sex is determined genetically (either male XY heterogamy or female ZW heterogamy), whereas in alligators, and in many reptiles and turtles, the temperature at which eggs are incubated determines sex. Evidently, different sex-determining systems (and sex chromosome pairs) have evolved independently in different vertebrate lineages. Homology shared by Xs and Ys (and Zs and Ws) within species demonstrates that differentiated sex chromosomes were once homologous, and that the sex-specific non-recombining Y (or W) was progressively degraded. Consequently, genes are left in single copy in the heterogametic sex, which results in an imbalance of the dosage of genes on the sex chromosomes between the sexes, and also relative to the autosomes. Dosage compensation has evolved in diverse species to compensate for these dose differences, with the stringency of compensation apparently differing greatly between lineages, perhaps reflecting the concentration of genes on the original autosome pair that required dosage compensation. We discuss the organization and evolution of amniote sex chromosomes, and hypothesize that dosage insensitivity might predispose an autosome to evolving function as a sex chromosome.
Keywords: Y degradation, X inactivation, comparative genomics, marsupials, monotremes
Introduction
Sex chromosomes are critical in many vertebrate species for fating an embryo to develop as a male or female. All therian (eutherian and marsupial) mammals have an XY male/XX female sex chromosome system (or some variant of it), in which maleness is normally determined by the dominant Y-borne SRY gene that triggers a cascade of events leading to testis development. In contrast, birds, snakes and many lizards have a female heterogametic sex chromosome system with ZZ males and ZW females.
Because the heterogametic sex has only one X (or Z), genes borne by this chromosome are at only half the dose compared with the homogametic sex (XX or ZZ). It has always been expected that dosage compensation (DC) of X/Z gene expression between the sexes (Lyon, 1961), and with the autosomes, was critical for life (Nguyen and Disteche, 2006). Indeed, the imbalance in one sex of X and autosomal genes is fatal to human embryos and Drosophila larvae (Deng and Meller, 2006; Deng et al., 2009). Here we review characteristics of sex chromosomes and DC in amniote vertebrates, and discuss the possibility that an autosome pair might be predisposed to evolving into sex chromosomes because it contains genes that are dosage insensitive.
Y degradation and DC
The evolution of sex chromosomes has attracted much interest over the century since Muller (1914) proposed that the X and Y evolved from an homologous pair of autosomes. He suggested that after one member of the pair acquired a testis-determining factor, other sex-specific alleles accumulated near the new sex-determining locus, and there was selection for suppression of recombination to keep the sex-specific cassette of genes together. In the absence of recombination, the Y degrades because of genetic drift (Muller's ratchet) and inefficient selection (Charlesworth, 1991). Drift is exacerbated by the smaller population size of Ys (present in the population at a frequency only ¼ that of the autosomes), and selection is inefficient because it acts on the Y as a whole, rather than at individual loci. Deleterious mutations could be dragged to fixation on an otherwise ‘good' Y (genetic hitchhiking), and favourable alleles could be lost from the population if they arose on a ‘bad' Y. The Y is also subject to higher mutation rates because of additional cell divisions in the testis. Thus, gene loss and degradation of the Y (or W in ZW systems) progressed (Charlesworth, 1991).
This now widely accepted theory that sex chromosomes evolved from autosomes is supported by comparative genomics, which reveals that the sex chromosomes in one vertebrate lineage are orthologous to autosomes in another. For example, the chicken Z chromosome is largely homologous to human chromosomes 5, 9 and 18 (Nanda et al., 2002), and the ancient therian X chromosome maps to the short arm of chicken chromosome 4 (Kuroiwa et al., 2002; Kohn et al., 2004; Waters et al., 2005) and the added region to part of chicken chromosome 1 (Nanda et al., 1999). The snake ZW is homologous to chicken chromosome 2 and the bird ZW is homologous to snake chromosome 2 (Matsubara et al., 2006). In fact, early comparisons of ZW systems in different snake families led Ohno (1967) to his famous hypothesis that they represent varying degrees of progressive W-chromosome degradation. Homologies shared by the mammal X and Y imply a similar evolutionary pathway from a pair of autosomes, and were proposed to have originated by an analogous process (Charlesworth, 1991).
As genes were lost from the proto-Y (or proto-W), corresponding genes on the proto-X (or proto-Z) remained as single copy in the heterogametic sex, whereas the homogametic sex retained two copies. Monosomy (the presence of only one copy of a chromosome), resulting in haploinsufficiency, is deleterious in most organisms (Birchler, 2010), and genetic interaction studies have shown that minor differences in expression levels of a few connected genes can lead to the collapse of genetic networks (Gupta et al., 2006). Indeed, the only viable monosomic condition in humans is Turner syndrome (45 XO female). Thus, it was thought that balancing X-chromosome gene dose between the sexes, and with the autosomes, must be critical for survival.
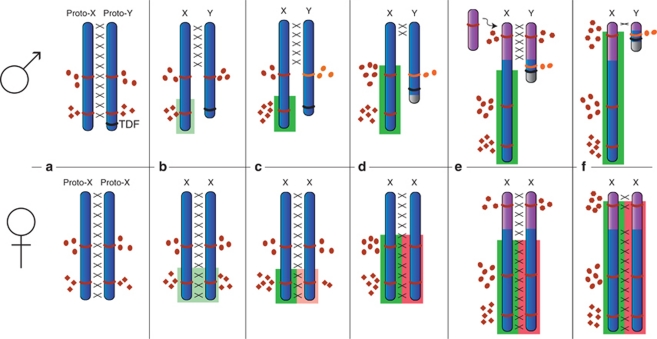
It was hypothesized that mammal sex chromosome DC evolved by the upregulation of transcription from the single X chromosome in XY males to match expression from the autosomes (i.e., X:AA=1), and then reduced expression from one of the two hyper-expressed Xs in females to also match the autosomes, which is achieved by X-chromosome inactivation (XCI) (Figure 1). Nguyen and Disteche (2006) provided microarray data that showed increased global expression from the active X chromosome in humans to match the autosomes (Nguyen and Disteche, 2006). However, Xiong et al. (2010) recently used RNA-seq (transcriptome sequencing) data to compare the X:AA ratio in human and mouse, reporting a ratio of ∼0.5 that would indicate a lack of global X upregulation and challenge Ohno (1967) hypothesis that genes on the single active X are overexpressed to re-establish equal gene dose with the autosomes. The absence of upregulation of X genes in males would obviate the need for transcriptional silencing of one X in females and admit an alternative hypothesis that, as genes on the Y lost function, the protein products of the X homologues in males evolved to function at half the gene dose. These hyper-functioning X proteins would then result in selective pressure for their inactivation on one X in females, resulting in a male:female expression ratio of 1:1 from the X.
Figure 1.
Evolution of the eutherian sex chromosome differentiation and dosage compensation (X-chromosome inactivation). (a) One member of a pair of autosomes (the proto-Y) acquires a testis-determining factor (TDF). Male beneficial genes accumulate near this new TDF and recombination (represented by crosses) with the proto-X is suppressed. Most genes (two represented here; brown) remain in the pseudoautosomal region (PAR) and are expressed equally from the X and Y (brown ovals and diamonds represent products from different genes). In females, the two X-chromosome pair and recombine normally. (b) Recombination is further suppressed between the X and Y. In the absence of recombination, the proto-Y degrades, resulting in the loss of function of Y genes that can tolerate some haploinsufficiency (here represented by the gene expressing diamonds). In males, there is selection for upregulation of this gene from the single X (light green box behind the X) giving a gene dose of four. In females, the two X chromosomes express at this upregulated level, resulting in a gene dose of eight. The gene represented by the ovals cannot tolerate any decreased dosage, so is retained on the Y as a functional copy. (c) More upregulation of X genes in males occurs to restore gene dose to its original level of 6 (green box behind chromosome). In females, this gene is also upregulated on the X, which results in selection to downregulate the other X (pink box behind the chromosome). The gene that cannot tolerate dosage change (ovals) remains functional from its Y allele, but its prolonged retention on the Y gives it a chance to begin to acquire a male-specific function (light brown). (d) Further restriction of recombination leads to further degeneration of the Y and insertion of heterochromatin (grey). The Y copy of the gene producing ovals has gained a male-specific role (orange), and loses its original function. Overexpression of X genes in females (green box behind chromosome) results in complete transcriptional inactivation of homologous regions of the other X (red box behind chromosome). (e) An autosomal segment is added to both sex chromosomes (added to one and recombined onto the other), extending the PAR and contributing additional genes (gene product represented by brown hexagons). (f) Genes on the added segment are then differentiated or lost on the Y and the X copy is upregulated. The established inactivation mechanism can rapidly spread into the added region to compensate for Y gene loss and upregulation of the X in males.
Comparing amniote sex chromosomes
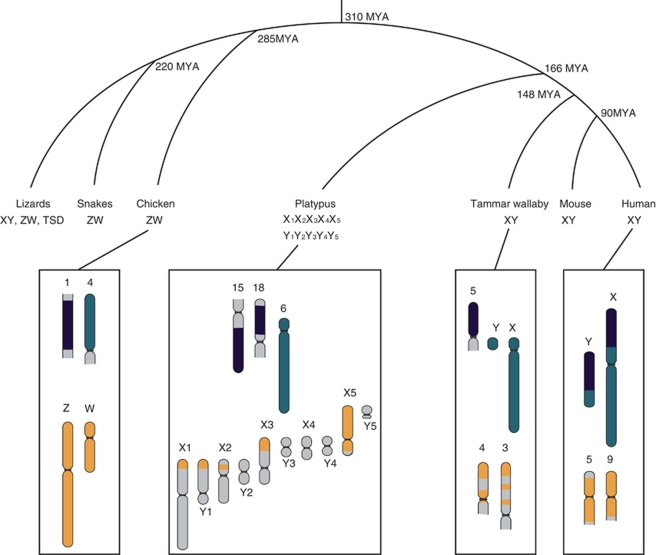
There are three main groups of extant mammals (eutherians, marsupials and monotremes). Eutherian (‘placental') mammals (Infraclass Eutheria) are divided into four superordinal clades: Euarchontoglires (or Supraprimates), Laurasiatheria, Xenarthra and Afrotheria (Murphy et al., 2001). Eutherian mammals diverged from marsupials (Infraclass Metatheria) 148 million years ago (MYA), and Subclass Theria (eutherians and marsupials) diverged from the monotreme mammals (Subclass Prototheria) 166 MYA (Bininda-Emonds et al., 2007). Taken together, birds (part of Archosauria) and lizards/snakes (part of Squamatidae) comprise Sauropsida. Sauropsida and mammals comprise amniotes, which radiated 310 MYA (Kumar and Hedges, 1998) (Figure 2).
Figure 2.
Vertebrate relationships with estimated divergence dates (branch lengths not to scale). The sex chromosome systems observed in each lineage are shown (TSD=temperature-dependent sex determination). Orthology of sex chromosomes and autosomes is represented by different colours for representative mammals and birds. The orange regions represent an original ZW system in amniotes that has been retained on sex chromosomes in birds, a gekko and monotremes, or independently selected for sex chromosome function in these lineages. The green region became X and Y chromosomes only recently in therian mammals (marsupials and eutherians). The black region remains autosomal, except for eutherian mammals, in which it was added to the X and Y. A full color version of this figure is available at the Heredity journal online.
Eutherian sex chromosomes
The human X chromosome bears 1669 genes (http://www.ncbi.nlm.nih.gov) and is ∼155 Mb, representing about 5% of the haploid genome (Ross et al., 2005). The eutherian X is well conserved in size, gene order and gene content between even the most distantly related eutherian species (Delgado et al., 2009). However, rearrangements have been detected between some eutherian sex chromosomes (reviewed in Wilson and Makova, 2009).
The X chromosome is not devoted to femaleness. The human X encodes a mixture of widely expressed housekeeping genes, as well as genes that have specialized functions such as visual pigments and blood clotting factors. However, there is an elevated frequency (compared with the autosomes) of genes with sex and reproduction-related functions (Saifi and Chandra, 1999). It was suggested that the accumulation of male advantage genes on the human X chromosome is the result of rapid selection in the hemizygous male (Rice, 1987). A new recessive allele will be expressed in males (since there is only one copy), and if it has an advantageous function in the sperm or gonad, it will be immediately selected for. This selection process will occur regardless of the function in females since there will be no phenotypic effect on the heterozygous females, and the proportion of homozygous females will be negligible (reviewed in Graves, 2006).
The X chromosome is also enriched in intelligence-related genes, mutations in which result in various X-linked mental retardation syndromes (Zechner et al., 2001). This bias was hypothesized to be the result of sexual selection, with smarter males being selected by females, either because of their superior ability to care for young or because intelligence, being an expensive trait (like peacocks' tails), is an indicator of superior genes (reviewed in Graves et al., 2002). Interestingly, many of the genes involved in mental retardation syndromes are also sex and reproduction-related genes, and thus have been dubbed ‘brains and balls' genes (for a full review on the brains and balls hypothesis we refer readers to Graves et al. (2002)).
In contrast to the X, the human Y chromosome is small (∼60 Mb), gene poor and largely heterochromatic. The male-specific region of the Y bears 156 transcription units, which includes 78 protein coding units. Many of these are amplified, so they collectively code for only 27 distinct proteins (Skaletsky et al., 2003). Of these 27 genes, 20 have partners on the X from which they evolved (Waters et al., 2007). All that remains of once extensive homology with the X are the X/Y shared genes, and two small terminal pseudoautosomal regions, the larger of which is critical for proper pairing and segregation at male meiosis. Unlike the X, there is considerable variation of Y structure and gene content between eutherian species, due to independent degradation in different lineages, which has resulted in different but overlapping subsets of genes on the Y (reviewed in Graves, 2006).
Marsupial sex chromosomes
The basic marsupial X chromosome is smaller than the eutherian X chromosome, representing only 3% of the haploid genome. The opossum (Monodelphis domestica) X is estimated to be 80 Mb and contains at least 442 protein-coding genes (Hubbard et al., 2009). The marsupial X shares extensive homology with the long arm and proximal short arm of the human X (Graves, 1995), which defines an ancient X conserved region (Figure 2). The marsupial X is smaller than the eutherian X because the remainder of the region homologous to human Xp is located on an autosome (Wilcox et al., 1996) (Figure 2). In all other vertebrates, orthologues of this autosomal block are also separate from orthologues of the X conserved region, so it represents a single fusion to the eutherian X after the marsupial/eutherian split (148 MYA), but before the eutherian radiation (105 MYA). This region of the eutherian X chromosome was called the X added region (Graves, 1995). The newly added region was also added to the eutherian Y, and was subjected to the same forces that caused degradation of the original proto-Y, as such the eutherian Y also consists of conserved and added regions (YCR and YAR) (Graves, 1995).
The Y chromosome of a model marsupial (tammar wallaby, Macropus eugenii) bears at least 10 genes (PD Waters, personal communication; reviewed in Waters et al., 2007), each with a partner on the X. All of these genes were retained from the original proto-Y, and orthologues were also detected on the Tasmanian devil Y (Sarcophilus harrisii, a distantly related Australian marsupial), defining a core set of 10 ancient genes on the Australian marsupial Y (PD Waters, personal communication). Thus, the marsupial YCR retains more ancient Y genes than the human YCR, which includes only four genes (SRY, RBMY, KDM5D and RSP4Y). Indeed, most human male-specific region of the Y genes are relics from the recent autosomal addition (YAR) (Waters et al., 2001). It is conceivable that the acquisition of the YAR permitted loss of all but the four YCR genes in human, assuming that YAR contained genes with functional redundancies that permitted pervasive YCR gene loss.
Intriguingly, the eutherian X homologues of all 10 Australian marsupial X–Y shared genes are implicated in human brain/central nervous system function, and all but four are implicated in human and/or mouse male reproduction. This implies that there was preferential retention, on the marsupial Y, of genes with ‘brains and balls' function, although these functions were likely to have been specialized or altered. This contrasts sharply with the 15 genes retained on the added region of the human Y chromosome, of which only one has an X homologue associated with X-linked mental retardation.
Mammalian sex chromosomes: prototheria
Monotreme mammals are represented by the platypus and four species of echidna. The platypus genome is smaller than that of therian mammals, comprising six large and many small chromosomes, which are reminiscent of reptilian macro- and micro-chromosomes (Warren et al., 2008). Their phylogenetic position (Figure 2) makes monotremes an out-group for all other mammals; hence, studying the platypus sex chromosome system could provide insight into sex determination and DC in therian mammals (Rens et al., 2004).
There are 10 unpaired chromosomes in the male platypus, which form a translocation chain at meiosis held together by chiasmata between homologous regions. In females, five of these chromosomes are absent and the remaining five are present in duplicate. Thus, the sex chromosomes of males were described as X1Y1 X2Y2 X3Y3 X4Y4 X5Y5, and in females as X1X1 X2X2 X3X3 X4X4 X5X5 (Grützner et al., 2004; Rens et al., 2004). The echidna has an equally complex sex chromosome system, and it was shown that the sex chromosomes of the platypus and echidna share homology but are not identical; the echidna has five X and four Y chromosomes, sharing seven with the platypus. In the echidna, an autosome replaces one of the platypus X chromosomes, and the X5 homologue in echidna has a central position in the chain instead of being at the end, as in platypus (Dohm et al., 2007; Rens et al., 2007).
The completion of the platypus genome sequence and the availability of bacterial artificial chromosome clones with identifiable gene content (Warren et al., 2008) permitted more detailed studies of the platypus sex chromosomes. Surprisingly, it was found that none of the platypus X chromosomes share homology with the therian X chromosome (Veyrunes et al., 2008). Instead, the conserved region of the therian X chromosome is homologous to platypus chromosome 6 (Veyrunes et al., 2008), whereas the added region of the eutherian sex chromosomes is orthologous to parts of chromosomes 15q and 18p (Veyrunes et al., 2008) (Figure 2). The observation that therian X chromosome is represented by an autosome in monotremes unambiguously dates the origin of the therian sex chromsomes to after the divergence of monotermes from therians (166 MYA), but before the therian radiation (148 MYA).
Surprisingly, it was observed that the platypus X5 is largely homologous to the chicken Z, with chicken Z orthologues also detected on X3q, X2p and X1p/Y1q (Rens et al., 2007; Veyrunes et al., 2008) (Figure 2). DMRT1, the putative sex-determining gene in birds (Smith et al., 2009), is located on platypus chromosome X5 (Grützner et al., 2004), but it is doubtful that it could be a sex-determining gene in platypus, because it is present in two copies in female and single copy in males, which is opposite to its dosage in chicken (two copies in males). Accordingly, the platypus sex-determining gene remains unknown (Grafodatskaya et al., 2007).
Sex chromosomes of birds, snakes and lizards
Although the XY pair of therian mammals and the ZW pair of birds are superficially similar, comparative gene mapping showed that they are non-homologous (Kohn et al., 2004; Matsubara et al., 2006). The chicken Z chromosome shares homology with human chromosomes 5, 9 and 18, and the human X shares homology with chicken chromosomes 4 and 1 (Nanda et al., 1999, 2002).
The avian Z chromosome is similar in size, morphology and G-band pattern across most families of birds, and chromosome painting reveals molecular homology (Shetty et al., 1999). In contrast, the W chromosome differs between the distantly related ratites (large flightless birds, for example, emu) and carinates (for example, chicken). Carinate Z and W chromosomes are highly differentiated, with a small and heterochromatic W chromosome, whereas the ratite W chromosome is almost indistinguishable from the Z chromosome and lacks heterochromatin, indicating an early stage of sex chromosome differentiation (Graves and Shetty, 2001).
Like the birds, snakes have a ZW system in which the Z chromosome is similar in size across species, but the W ranges from homomorphy to highly differentiated, an observation that led Ohno (1967) to his hypothesis that they represent different stages of W degradation. The snake ZW pair is superficially similar to the bird ZW, but recent gene mapping demonstrated that the bird and snake Zs were non-homologous (Matsubara et al., 2006), perhaps differing by a single reciprocal translocation and sharing repetitive sequences (O'Meally et al., 2010).
Some lizards have ZZ/ZW female heterogametic systems (which differ between species), whereas others have XX/XY male heterogametic systems (also different between species). Many reptiles, alligators and turtles lack sex chromosomes, and determine sex by the temperature at which the eggs are incubated (temperature-dependent sex determination). Some reptiles blur the line completely, with genetic sex determination that can be reversed at temperature extremes. For example, the Australian central bearded dragon has differentiated Z and W chromosomes (ZZ male, ZW female). However, ZZ undifferentiated eggs follow a female developmental pathway at high incubation temperatures; thus, temperature over-rides genetic male differentiation (Quinn et al., 2007).
The homology of the chicken ZW pair to autosomes in snakes and turtles leaves open the question of which state was ancestral to the reptile–bird clade. Of considerable interest is a gecko species (Gekko hokouensis) with a ZW system that shares homology with the bird ZW system (Kawai et al., 2009). Thus, there are at least three sex chromosome systems, in distantly related vertebrate groups (monotremes, birds and geckos), which share extensive homology. This raises the possibility that there was a sex chromosome system in the amniote ancestor that was largely equivalent to the orthologous regions shared by extant monotreme, bird and gecko sex chromosomes. This ancient ZW system was supplanted by new systems in therian mammals, snakes and many other reptiles (reviewed in Waters and Marshall Graves, 2009). However, the G. hokouensis sex chromosome system is most likely to have been evolved recently (Pokorna et al., 2011), supporting an alternative hypothesis that the original autosome pair from which the monotreme, bird and gecko sex chromosomes derived might have been chosen independently several times because they are intrinsically suitable to function as sex chromosomes (Graves and Peichel, 2010).
Regulating sex chromosome gene dosage
The many independently evolved vertebrate sex chromosome systems each raise the problem of dosage difference of X- or Z-borne genes between the sexes. The universal necessity for DC seems apparent from the independently evolved mechanisms in mammals through to fruit flies and worms.
Eutherian XCI
XCI represents transcriptional repression of one X in female somatic cells (Graves and Gartler, 1986) enforced by chromatin changes. Early chromatin changes include the loss of histone modifications associated with transcription, and gain of specific repressive modifications (reviewed in Heard, 2005). Exactly how these marks are recruited is not fully understood, but it is controlled by a master locus called the X inactivation centre (Okamoto and Heard, 2009). Within the X inactivation centre there are several genes that code for long non-coding RNAs (Erwin and Lee, 2009). The most important is XIST (X-inactive-specific transcript), which codes for a large untranslated RNA. In mouse, XIST is transcribed exclusively from the future inactive X (Xi), and spreads in cis from the X inactivation centre to coat the whole X, triggering silencing (reviewed in Okamoto and Heard, 2009). However, in humans XIST upregulation at the blastomere stage precedes XCI, implying that the choice of which X becomes inactivated occurs downstream of XIST upregulation (Okamoto et al., 2011).
Further stabilization of the inactive state occurs with additional layers of chromatin modifications (Chaumeil et al., 2002), including association of the Xi with macroH2A and DNA methylation at CpG islands (reviewed in Okamoto and Heard, 2009). These modifications form a multilayered silencing complex that maintains transcriptional repression of the Xi (Chow and Heard, 2009), and could play a role in the stabilization and somatic heritability of the inactive state (Kohlmaier et al., 2004). The epigenetic signature established on facultative heterochromatin of the Xi is different to that of constitutive heterochromatin (Beck et al., 2011).
Marsupial XCI
As for the human and mouse X, the marsupial Xi replicates late in the S phase (Graves, 1967). However, marsupial XCI appears to be incomplete, tissue specific (reviewed in Cooper et al., 1993) and less stable (Kaslow and Migeon, 1987), and Barr body formation is inconsistent between tissues and species (McKay et al., 1987). A striking difference between marsupial and eutherian XCI is that there is no evidence for an X inactivation centre in marsupials, and no XIST gene. It appears that XIST evolved in the eutherian ancestor, possibly from an ancient protein-coding gene (reviewed in Romito and Rougeulle, 2011).
Until recently, little was known about epigenetic modifications associated with the Xi in marsupial mammals. Differential acetylation had been observed on the X chromosomes in female tammar wallaby (Wakefield et al., 1997), as was enrichment of an active histone methylation mark (presumably on the active X) (Wakefield et al., 1997; Koina et al., 2009), and no differential DNA methylation differences were detected (Piper et al., 1993; Loebel and Johnston, 1996). However, interesting observations have come from several recent studies of representative Australian and American marsupials, using immunofluorescence to examine histone marks that are associated with XCI in human and mouse. The inactive marsupial X lost epigenetic modifications associated with transcription during interphase (Chaumeil et al., 2011) and metaphase (Koina et al., 2009; Rens et al., 2010), and the nuclear territory that harbours Xi was devoid of RNA Pol II (Chaumeil et al., 2011). These observations established that the marsupial Xi is situated in a transcriptionally inert nuclear compartment, as it is in eutherian mammals.
The pattern of inactive marks on the marsupial Xi was strikingly different from that associated with the eutherian mammal Xi. Some modifications were completely absent from the marsupial Xi (Koina et al., 2009; Chaumeil et al., 2011), whereas others were restricted to specific windows of the cell cycle (Chaumeil et al., 2011). Of considerable surprise was the accumulation of marks on Xi that are associated with pericentromeric heterochromatin (Mahadevaiah et al., 2009; Rens et al., 2010; Chaumeil et al., 2011; Zakharova et al., 2011). Thus, it seems that the marsupial Xi bears an epigenetic signature similar to that of pericentromeric heterochromatin, and is unlike the XIST-associated facultative heterochromatin in eutherians. Chaumeil et al. (2011) hypothesized that marsupial XCI represents an ancient DC system that was exapted from neighbouring constitutive heterochromatin, to which layers of molecular complexities were subsequently added in eutherian mammals. These epigenetic differences might account for the incomplete and locus-specific inactivation observed on the marsupial X. It is observed that transcription from loci on the Xi is partial, never 100% monoallelic (inactivated) or 100% biallelic (escapee). Rather, there is a locus-specific probability that a gene on the Xi will be transcribed (Al Nadaf et al., 2010).
DC in monotremes, birds and snake/lizard
Perhaps, then, complete DC between sexes, such as therian mammal XCI (where global X expression ratios are 1:1 between the sexes), is a recent evolutionary sophistication, and incomplete DC is more widespread in amniotes, and therefore ancient. Consequently, it is of great interest to study DC in birds and monotremes.
In platypus, asynchronous replication of loci on X-specific regions of X1, X3 and X5 (compared with autosomal loci) was observed, although cytologically different condensation was observed only for homologues on X3 (Ho et al., 2009). The percentage of cells with asynchronous replication was lower than that observed for the mouse X, suggesting partial DC. Consistent with this was RNA-in situ hybridization, which indicated that some X-specific genes were compensated by regulating the probability that they are either mono- or bi-allelically transcribed (Deakin et al., 2008). This is similar to the more recent observations in marsupials (Al Nadaf et al., 2010), although the sex chromosomes are not homologous.
It is currently unknown which epigenetic changes are associated with the partial X inactivation system in platypus. Rens et al. (2010) observed no differences in staining on metaphase chromosomes for active modifications (H4K8ac, H4K16ac, H3K9ac and H2AK5ac) on X1 and X5; however, region-specific hyperacetylation for all chromosomes corresponded to R-banded regions of euchromatin. In addition, all sex chromosomes and autosomes were equally stained for repressive modifications (H3K27me3, H3K9me3, H4K20me3 and HP1α). Thus, the data collected so far indicate that epigenetic modifications associated with therian XCI do not appear to be involved in monotreme DC.
Early allozyme work (Baverstock et al., 1982) revealed no DC for three genes on the chicken Z. However, microarray expression analysis in chicken (Ellegren et al., 2007; Itoh et al., 2007; Melamed and Arnold, 2007) and zebra finch (Itoh et al., 2007), and RNA-seq analysis in crow (Wolf and Byrk, 2011) estimate male-to-female (M:F) expression ratios of Z genes to be between 1.2 and 1.6. Melamed and Arnold (2007) provided evidence for regional differences in Z chromosome DC, with significantly higher M:F ratios on Zq than Zp. When a running average curve was constructed, a broad peak of high ratios was observed towards distal Zq (non-compensated genes), and a ‘valley' of low ratios was observed 25–35 Mb from the telomere on Zp (compensated genes) that correlated with a hypermethylated region of the Z in males. However, these regional patterns were not observed in zebra finch (Itoh et al., 2010). Using fold-change and amplitude analysis to assess neighbourhoods of DC on the chicken Z, Mank and Ellegren (2009) argued that the dip in M:F ratios 28 Mb from Zpter was not a region of DC, but rather a region that contained two genes with a strong female bias. There was no dip in the amplitude (which would have approached zero if DC were equalizing Z dosage), and removing the two female-biased genes from the fold-change analysis resulted in a fold-change that was statistically identical to that of the remainder of the Z chromosome. They concluded that there are no regions of DC on the chicken Z, but rather Z genes are regulated on an individual gene-by-gene basis independently in different tissues and at different developmental time points (Mank and Ellegren, 2009).
Almost nothing is known about DC, or its distribution, in reptiles. Early studies of replication timing in different snake families revealed no asynchronous replication of the Z chromosomes in males (Bianchi et al., 1969). Allozyme studies of FH in snakes indicated that this locus was on the Z, and the similar staining intensity of FH products in extracts from males and females (King and Lawson, 1996) suggested some form of DC. However, some of the most basic questions are still to be answered. Do reptiles have an incomplete bird-like compensation system? Is there stochastic inactivation of one allele in some nuclei and not others, as is the case for platypus and tammar wallaby? Of particular interest would be DC in G. hokouensis, which shares sex chromosome homology birds and monotremes.
Does dosage insensitivity predispose an autosome to evolve sex chromosome function?
A survey of sex chromosomes in tetrapods has revealed instances of sex chromosome homology between distantly related groups, and posed the question of whether this homology represents identity by descent or the independent evolution of sex chromosomes from the same autosome (Graves and Peichel, 2010). At least in some fish lineages, the conclusion seems inescapable that certain autosomes repeatedly evolved as sex chromosomes in different lineages. Are these autosomes particularly good at being sex chromosomes? What properties do they share?
The apparently less stringent DC systems in platypus and chicken suggest that DC is not as critical to life as previously thought, and that dosage differences on a global scale are better tolerated on the bird Z chromosome and platypus X chromosomes than on the therian X chromosome. Indeed, the question of whether we have given undue importance to DC has been raised (Mank et al., 2011).
In humans XXX and XXY aneuploidy is tolerated, but probably only because in both cases just one X chromosome remains active (Ohno, 1969). The additional dosage in XXX females of the 150 escapee genes must not, therefore, be phenotypically damaging. In contrast, XO (Turner's) individuals have a more severe phenotype that must result from haploinsufficiency of the few genes shared with the Y. In fact, >95% of XO foetuses rapidly abort, and there is suggestion that all surviving XO individuals begin as mosaics. This, along with the relatively tight X-chromosome inactivation system observed in eutherian mammals, suggests that there are many genes on the therian X chromosome that require strict DC.
In chickens, triploids that are either ZZZ (phenotypically normal males) or ZZW (phenotypically female birds until sexual maturity, when they adopt a masculinized phenotype) can be obtained (Thorne and Sheldon, 1993). In contrast, diploid ZO or ZZW chickens are never observed, suggesting that these genotypes are embryonic lethal (Graves, 2003), and that correct dose of the Z is important. However, this dosage sensitivity may be due to regionally dosage compensated genes in the male hypomethylated region of the Z, and dosage imbalance might be less important for the majority of genes on other regions of the chicken Z, obviating the need for global DC.
The absence of a requirement for a tight global DC system (in response to the loss of gene function on the sex-specific chromosome) might be what predisposes a pair of autosomes to being intrinsically good candidates for sex chromosomes, such that the same pair became sex chromosomes in birds, in G. hokouensis and monotremes. Interestingly, chromosome 9, which is orthologous to large parts of the chicken Z, is highly structurally polymorphic in humans (Humphray et al., 2004). Trisomies of human 9p (Wilson et al., 1985), and monosomies of 5q and 18q (which all share homology with the chicken Z), survive to birth (Nusbaum et al., 2005; Boultwood et al., 2010). Moreover, the snake Z chromosome is known to share homology with human 3p21–p24, 7p14–p15, 10p11–p15 and 17q21–q24 (Matsubara et al., 2006). Monosomy of at least parts of these regions also survive to birth, albeit with phenotypes of varying severity (Wieczorek et al., 1997; Schwarzbraun et al., 2006; Barber, 2008; Puusepp et al., 2009; Lindstrand et al., 2010). This suggests that dosage imbalance from the autosome pair that became the chicken Z, and the autosome pair that became the snake Z, is somewhat tolerable in amniotes.
Perhaps, then, the tight dosage control of the therian X chromosome requires a special explanation. What genes on the therian X might be more sensitive to dosage imbalances? The bias towards retention of X-linked mental retardation/sex and reproduction-related (‘brains and balls') genes on the ancient YCR may provide an explanation.
When the proto-Y first acquired a testis-determining factor, there would have been no system of DC (see Figure 1). As the Y degenerated, the X inactivation system developed and spread as genes that had lost their Y partner were progressively recruited. There appears to be a lag between loss of the Y allele and inactivation of the X allele, as evidenced by recently unpartnered human X genes like UBE1X and STS, which remain exempt from inactivation (Carrel et al., 1999; Graves, 2006). We hypothesize that ‘brains and balls' genes are less able to tolerate dosage imbalances than genes with functions in other tissues. These genes would, therefore, be retained on the Y for longer periods of time (to maintain dosage) than were other classes of genes, prolonging opportunity for recruitment into a male-specific function (Figure 1). Ultimate loss of brain function from the Y homologue would not have occurred until full DC of the X allele. These constraints of dosage imbalance were probably not as strict for dosage-sensitive ‘brains and balls' genes on the added region of the eutherian sex chromosomes because they could be quickly recruited into an established XCI system, permitting their rapid loss from the Y; this explains why there is no retention bias of genes that originally functioned in X-linked mental retardation/sex and reproduction on the YAR.
Therefore, the evolution of a pair of autosomes into sex chromosomes, and their subsequent differentiation, could depend partially on whether or not dosage imbalance of the genes it contains can be tolerated, and the consequent requirement (or lack thereof) for the evolution of a global DC system. In the absence of an established DC mechanism, sensitivity of a gene to haploinsufficiency would result in retention of its homologue on the sex-specific chromosome for an extended period of time, during which it could be exapted into a sex-specific function.
This hypothesis makes predictions about the function of sex-linked genes in birds and reptiles. If the autosome pair that became the therian X required strict DC, we would expect monosomy of the orthologous region in chicken (chromosome 4p) to be lethal. And if global DC is not required for the autosome pair that became the chicken/gecko/platypus sex chromosomes, there should be no biased retention of dosage-sensitive genes on the W (in chicken and gecko) or Y (in monotremes) as a result of maintaining dosage between the sexes. Finally, should the autosome pair that became the snake Z also be relatively dosage insensitive, we would expect a partial DC system in snake similar to that observed for chicken.
Data Archiving
There was no data to deposit.
Acknowledgments
This work is supported by an Australian Research Fellowship to PDW (DP0987091) and an Australian Research Council discovery project grant to PDW and JAMG (DP1094868).
The authors declare no conflict of interest.
References
- Al Nadaf S, Waters PD, Koina E, Deakin JE, Jordan KS, Graves JAM. Activity map of the tammar X chromosome shows that marsupial S inactivation is incomplete and escape is stochastic. Genome Biol. 2010;11:R122. doi: 10.1186/gb-2010-11-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JC. Terminal 3p deletions: phenotypic variability, chromosomal non-penetrance, or gene modification. Am J Med Genet A. 2008;146A:1899–1901. doi: 10.1002/ajmg.a.32387. [DOI] [PubMed] [Google Scholar]
- Baverstock PR, Adams M, Polkinghorne RW, Gelder M. A sex-linked enzyme in birds—Z-chromosome conservation but no dosage compensation. Nature. 1982;296:763–766. doi: 10.1038/296763a0. [DOI] [PubMed] [Google Scholar]
- Beck DB, Bonasio R, Kaneko S, Li G, Margueron R, Oda H, et al. Chromatin in the nuclear landscape. Cold Spring Harb Symp Quant Biol. 2011;75:11–22. doi: 10.1101/sqb.2010.75.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi NO, Becak W, De Bianchi MS, Becak ML, Rabello MN. Chromosome replication in four species of snakes. Chromosoma. 1969;26:188–200. doi: 10.1007/BF00326454. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Birchler JA. Reflections on studies of gene expression in aneuploids. Biochem J. 2010;426:119–123. doi: 10.1042/BJ20091617. [DOI] [PubMed] [Google Scholar]
- Boultwood J, Pellagatti A, McKenzie AN, Wainscoat JS. Advances in the 5q− syndrome. Blood. 2010;116:5803–5811. doi: 10.1182/blood-2010-04-273771. [DOI] [PubMed] [Google Scholar]
- Carrel L, Cottle AA, Goglin KC, Willard HF. A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Guggiari M, Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Waters PD, Koina E, Gilbert C, Robinson TJ, Graves JAM. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals. PLoS One. 2011;6:e19040. doi: 10.1371/journal.pone.0019040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Cooper D, Johnston PG, Graves JAM. X-inactivation in marsupials and monotremes. Semin Cell Dev Biol. 1993;4:117–128. [Google Scholar]
- Deakin JE, Hore TA, Koina E, Marshall Graves JAM. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 2008;4:e1000140. doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado CL, Waters PD, Gilbert C, Robinson TJ, Graves JAM. Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosome Res. 2009;17:917–926. doi: 10.1007/s10577-009-9079-1. [DOI] [PubMed] [Google Scholar]
- Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem Sci. 2006;31:526–532. doi: 10.1016/j.tibs.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Deng X, Nguyen DK, Hansen RS, Van Dyke DL, Gartler SM, Disteche CM. Dosage regulation of the active X chromosome in human triploid cells. PLoS Genet. 2009;5:e1000751. doi: 10.1371/journal.pgen.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Tsend-Ayush E, Reinhardt R, Grutzner F, Himmelbauer H. Disruption and pseudoautosomal localization of the major histocompatibility complex in monotremes. Genome Biol. 2007;8:R175. doi: 10.1186/gb-2007-8-8-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Hultin-Rosenberg L, Brunstrom B, Dencker L, Kultima K, Scholz B. Faced with inequality: chickens do not have a general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Lee JT. New twists in X-chromosome inactivation. Curr Opin Cell Biol. 2009;20:349–355. doi: 10.1016/j.ceb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafodatskaya D, Rens W, Wallis MC, Trifonov V, O'Brien PC, Clarke O, et al. Search for the sex-determining switch in monotremes: mapping WT1, SF1, LHX1, LHX2, FGF9, WNT4, RSPO1 and GATA4 in platypus. Chromosome Res. 2007;15:777–785. doi: 10.1007/s10577-007-1161-y. [DOI] [PubMed] [Google Scholar]
- Graves JAM. DNA synthesis in chromosomes of cultured leucocytes from two marsupial species. Exp Cell Res. 1967;46:37–57. doi: 10.1016/0014-4827(67)90407-7. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex and death in birds: a model of dosage compensation that predicts lethality of sex chromosome aneuploids. Cytogenet Genome Res. 2003;101:278–282. doi: 10.1159/000074349. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Gartler SM. Mammalian X chromosome inactivation: testing the hypothesis of transcriptional control. Somat Cell Mol Genet. 1986;12:275–280. doi: 10.1007/BF01570786. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Gecz J, Hameister H. Evolution of the human X—a smart and sexy chromosome that controls speciation and development. Cytogenet Genome Res. 2002;99:141–145. doi: 10.1159/000071585. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Peichel CL. Are homologies in vertebrate sex determination due to shared ancestry or to limited options. Genome Biol. 2010;11:205. doi: 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM, Shetty S. Sex from W to Z: Evolution of vertebrate sex chromosomes and sex determining genes. J Exp Zool. 2001;290:449–462. doi: 10.1002/jez.1088. [DOI] [PubMed] [Google Scholar]
- Grützner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O'Brien PC, Jones RC, et al. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature. 2004;432:913–917. doi: 10.1038/nature03021. [DOI] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, et al. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Ho KK, Deakin JE, Wright ML, Graves JAM, Grutzner F. Replication asynchrony and differential condensation of X chromosomes in female platypus (Ornithorhynchus anatinus) Reprod Fertil Dev. 2009;21:952–963. doi: 10.1071/RD09099. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphray SJ, Oliver K, Hunt AR, Plumb RW, Loveland JE, Howe KL, et al. DNA sequence and analysis of human chromosome 9. Nature. 2004;429:369–374. doi: 10.1038/nature02465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, et al. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Replogle K, Kim YH, Wade J, Clayton DF, Arnold AP. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 2010;20:512–518. doi: 10.1101/gr.102343.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow DC, Migeon BR. DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupials: evidence for multistep maintenance of mammalian X dosage compensation. Proc Natl Acad Sci USA. 1987;84:6210–6214. doi: 10.1073/pnas.84.17.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A, Ishijima J, Nishida C, Kosaka A, Ota H, Kohno S, et al. The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma. 2009;118:43–51. doi: 10.1007/s00412-008-0176-2. [DOI] [PubMed] [Google Scholar]
- King RB, Lawson R. Sex-linked inheritance of fumarate hydratase alleles in natricine snakes. J Hered. 1996;87:81–83. [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn M, Kehrer-Sawatzki H, Vogel W, Graves JAM, Hameister H. Wide genome comparisons reveal the origins of the human X chromosome. Trends Genet. 2004;20:598–603. doi: 10.1016/j.tig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Koina E, Chaumeil J, Greaves IK, Tremethick DJ, Graves JAM. Specific patterns of histone marks accompany X chromosome inactivation in a maruspial. Chromosome Res. 2009;17:115–126. doi: 10.1007/s10577-009-9020-7. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Kuroiwa A, Uchikawa M, Kamachi Y, Kondoh H, Nishida-Umehara C, Masabanda J, et al. Chromosome assignment of eight SOX family genes in chicken. Cytogenet Genome Res. 2002;98:189–193. doi: 10.1159/000069803. [DOI] [PubMed] [Google Scholar]
- Lindstrand A, Malmgren H, Verri A, Benetti E, Eriksson M, Nordgren A, et al. Molecular clinical characterization of patients with overlapping 10p deletions. Am J Med Genet A. 2010;152A:1233–1243. doi: 10.1002/ajmg.a.33366. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Johnston PG. Methylation analysis of a marsupial X-linked CpG island by bisulfite genomic sequencing. Genome Res. 1996;6:114–123. doi: 10.1101/gr.6.2.114. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol. 2009;19:1478–1484. doi: 10.1016/j.cub.2009.07.041. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity. 2009;102:312–320. doi: 10.1038/hdy.2008.116. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65:2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, et al. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci USA. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay LM, Wrigley JM, Graves JAM. Evolution of mammalian X-chromosome inactivation: sex chromatin in monotremes and marsupials. Aust J Biol Sci. 1987;40:397–404. doi: 10.1071/bi9870397. [DOI] [PubMed] [Google Scholar]
- Melamed E, Arnold AP. Regional differences in dosage compensation on the chicken Z chromosome. Genome Biol. 2007;8:R202. doi: 10.1186/gb-2007-8-9-r202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. A gene for the fourth chromosome of Drosophila. J Exp Zool. 1914;17:325–336. [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Nanda I, Haaf T, Schartl M, Schmid M, Burt DW. Comparative mapping of Z-orthologous genes in vertebrates: implications for the evolution of avian sex chromosomes. Cytogenet Genome Res. 2002;99:178–184. doi: 10.1159/000071591. [DOI] [PubMed] [Google Scholar]
- Nanda I, Shan Z, Schartl M, Burt DW, Koehler M, Nothwang H, et al. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat Genet. 1999;21:258–259. doi: 10.1038/6769. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Nusbaum C, Zody MC, Borowsky ML, Kamal M, Kodira CD, Taylor TD, et al. DNA sequence and analysis of human chromosome 18. Nature. 2005;437:551–555. doi: 10.1038/nature03983. [DOI] [PubMed] [Google Scholar]
- O'Meally D, Patel HR, Stiglec R, Sarre SD, Georges A, Graves JAM, et al. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 2010;18:787–800. doi: 10.1007/s10577-010-9152-9. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex Chromosomes and Sex-Linked Genes. Springer-Verlag: New York, NY; 1967. [Google Scholar]
- Ohno S. Evolution of sex chromosomes in mammals. Annu Rev Genet. 1969;3:495–524. [Google Scholar]
- Okamoto I, Heard E. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res. 2009;17:659–669. doi: 10.1007/s10577-009-9057-7. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Piper AA, Bennett AM, Noyce L, Swanton MK, Cooper DW. Isolation of a clone partially encoding hill kangaroo X-linked hypoxanthine phosphoribosyltransferase: sex differences in methylation in the body of the gene. Somat Cell Mol Genet. 1993;19:141–159. doi: 10.1007/BF01233530. [DOI] [PubMed] [Google Scholar]
- Pokorna M, Giovannotti M, Kratochvil L, Kasai F, Trifonov VA, O'Brien PC, et al. Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma. 2011;120:455–468. doi: 10.1007/s00412-011-0322-0. [DOI] [PubMed] [Google Scholar]
- Puusepp H, Zilina O, Teek R, Mannik K, Parkel S, Kruustuk K, et al. 5.9 Mb microdeletion in chromosome band 17q22–q23.2 associated with tracheo-esophageal fistula and conductive hearing loss. Eur J Med Genet. 2009;52:71–74. doi: 10.1016/j.ejmg.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JAM. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316:411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- Rens W, Grutzner F, O'Brien PC, Fairclough H, Graves JAM, Ferguson-Smith MA. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc Natl Acad Sci USA. 2004;101:16257–16261. doi: 10.1073/pnas.0405702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W, O'Brien PC, Grutzner F, Clarke O, Graphodatskaya D, Tsend-Ayush E, et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 2007;8:R243. doi: 10.1186/gb-2007-8-11-r243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci USA. 2010;107:17657–17662. doi: 10.1073/pnas.0910322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romito A, Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie. 2011;93:1935–1942. doi: 10.1016/j.biochi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifi GM, Chandra HS. An apparent excess of sex- and reproduction-related genes on the human X chromosome. Proc Biol Sci. 1999;266:203–209. doi: 10.1098/rspb.1999.0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbraun T, Windpassinger C, Ofner L, Vincent JB, Cheung J, Scherer SW, et al. Genomic analysis of five chromosome 7p deletion patients with Greig cephalopolysyndactyly syndrome (GCPS) Eur J Med Genet. 2006;49:338–345. doi: 10.1016/j.ejmg.2005.10.133. [DOI] [PubMed] [Google Scholar]
- Shetty S, Griffin DK, Graves JAM. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 1999;7:289–295. doi: 10.1023/a:1009278914829. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Thorne MH, Sheldon BL.1993Triploid intersex and chimeric chickens: useful models for studies of avian sex determinationIn: Reed KC, Graves JAM (eds).Sex Chromosomes and Sex Determining Genes Harwood Academic Publishers: Switzerland; 201–208. [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield MJ, Keohane AM, Turner BM, Graves JAM. Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc Natl Acad Sci USA. 1997;94:9665–9668. doi: 10.1073/pnas.94.18.9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JAM, Birney E, Ponting CP, Grutzner F, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PD, Delbridge ML, Deakin JE, El-Mogharbel N, Kirby PJ, Carvalho-Silva DR, et al. Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus anatinus): implications for mammalian sex chromosome evolution. Chromosome Res. 2005;13:401–410. doi: 10.1007/s10577-005-0978-5. [DOI] [PubMed] [Google Scholar]
- Waters PD, Duffy B, Frost CJ, Delbridge ML, Graves JAM. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet Cell Genet. 2001;92:74–79. doi: 10.1159/000056872. [DOI] [PubMed] [Google Scholar]
- Waters PD, Marshall Graves JAM. Monotreme sex chromosomes—implications for the evolution of amniote sex chromosomes. Reprod Fertil Dev. 2009;21:943–951. doi: 10.1071/RD09250. [DOI] [PubMed] [Google Scholar]
- Waters PD, Wallis MC, Marshall Graves JAM. Mammalian sex—origin and evolution of the Y chromosome and SRY. Semin Cell Dev Biol. 2007;18:389–400. doi: 10.1016/j.semcdb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Bolt J, Schwechheimer K, Gillessen-Kaesbach G. A patient with interstitial deletion of the short arm of chromosome 3 (pter-->p21.2::p12-->qter) and a CHARGE-like phenotype. Am J Med Genet. 1997;69:413–417. doi: 10.1002/(sici)1096-8628(19970414)69:4<413::aid-ajmg15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Wilcox SA, Watson JM, Spencer JA, Graves JAM. Comparative mapping identifies the fusion point of an ancient mammalian X-autosomal rearrangement. Genomics. 1996;35:66–70. doi: 10.1006/geno.1996.0323. [DOI] [PubMed] [Google Scholar]
- Wilson GN, Raj A, Baker D. The phenotypic and cytogenetic spectrum of partial trisomy 9. Am J Med Genet. 1985;20:277–282. doi: 10.1002/ajmg.1320200211. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Makova KD. Genomic analyses of sex chromosome evolution. Annu Rev Genomics Hum Genet. 2009;10:333–354. doi: 10.1146/annurev-genom-082908-150105. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Byrk J. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics. 2011;12:91. doi: 10.1186/1471-2164-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Chen X, Chen Z, Wang X, Shi S, Zhang J, et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- Zakharova IS, Shevchenko AI, Shilov G, Nesterova TB, VandeBerg JL, Zakian SM. Histone H3 trimethylation at lysine 9 marks the inactive metaphase X chromosome in the marsupial Monodelphis domestica. Chromosoma. 2011;120:177–183. doi: 10.1007/s00412-010-0300-y. [DOI] [PubMed] [Google Scholar]
- Zechner U, Wilda M, Kehrer-Sawatzki H, Vogel W, Fundele R, Hameister H. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution. Trends Genet. 2001;17:697–701. doi: 10.1016/s0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]