Abstract
Rodentia is the most species-rich mammalian order and includes several important laboratory model species. The amount of new information on karyotypic and phylogenetic relations within and among rodent taxa is rapidly increasing, but a synthesis of these data is currently lacking. Here, we have integrated information drawn from conventional banding studies, recent comparative painting investigations and molecular phylogenetic reconstructions of different rodent taxa. This permitted a revision of several ancestral karyotypic reconstructions, and a more accurate depiction of rodent chromosomal evolution.
Keywords: rodentia, rodents, comparative cytogenetics, chromosomal evolution
Introduction
Rodents have a cosmopolitan distribution with range extensions often associated with human movement. After their divergence from a common ancestor with Lagomorpha ∼65 My ago, rodents have undergone an impressive radiation leading to the high number of species observed today (Huchon et al., 2002; Benton and Donoghue, 2007). Rodents currently represent the most abundant mammalian order—they comprise about 42% of all living mammals, and include 2277 defined species (Carleton and Musser, 2005). Some rodents are used extensively in biomedical research and this has stimulated interest in the study of this group.
Modern taxonomy recognizes 5 suborders (Anomaluromorpha, Castorimorpha, Hystricomorpha, Myomorpha and Sciuromorpha) and 33 families. Sciuromorpha (including sciurids, mountain beaver and dormice), Myomorpha (muroids, jerboas and jumping mice) and Hystricomorpha (gundis, porcupines and caviamorphs) have all received good support as monophyletic taxa based on morphological and molecular data analysis. The evidence for the recognition of Castorimorpha and Anomaluromorpha is less persuasive. The degree of karyotype scrutiny varies within suborders but support nonetheless exists for the division of Rodentia into five subordinal groups.
Karyotypic features of rodents
Karyotypes of rodents were initially investigated by conventional cytogenetics that provided information on chromosome number and morphology. Interesting observations included the extreme variability of diploid chromosome number (from 2n=10 to 2n=102) and the presence of B-chromosomes in some species (Supplement Data 1). Moreover, before the development of chromosome banding, Matthey (1972) studied numerous rodent groups and described various cases of chromosomal polymorphisms and unusual sex chromosome systems. He led much of the thinking on the occurrence of unequal rates of chromosome evolution in different rodent groups, the existence of ancestral karyotypes, and the direction of karyotypic evolution.
These ideas were subsequently supported to a large extent by comparative G-, Q-, R-banding studies that revealed chromosomes/chromosomal regions with similar banding pattern that led authors to assume homology by descent. These data also showed high levels of chromosomal conservation in certain taxa. For instance, the sciurids were proposed to have conserved karyotypes similar to the hypothesized ancestral karyotype of Rodentia (Petit et al., 1984; Viegas-Péquignot et al., 1986). The karyotypes of castorimorph and anomaluromorph rodents were also considered conserved (Ward et al., 1991). In contrast, there was evidence to suggest that the karyotypes of myomorphs are highly reorganized (Graphodatsky, 1989). Additionally, significant heterochromatic variation (Patton and Sherwood, 1982; Graphodatsky, 1989; Svartman et al., 2005 among others) was noted, as were fascinating sex determining systems (for example, Fredga, 1983) and the frequent presence B-chromosomes (Trifonov et al., 2002).
New insights into karyotypic evolution in different rodent clades
Cross-species chromosome painting is currently the method of choice for comparative cytogenetic studies in rodents. Labeled whole chromosome probes are used to highlight the regions of homology by fluorescent in situ hybridization. The first successful chromosome painting was reported by Scherthan et al. However, only a few human probes were localized on mouse chromosomes and it is likely that the highly rearranged nature of the mouse genome contributed to the limited success in chromosome painting between mouse and human (Ferguson-Smith et al., 1998). More detailed comparison between human and mouse were facilitated by the availability of whole genome sequences for both species (Guigo et al., 2003). The first genome-wide comparison between two rodent genomes (Mus musculus and Rattus norvegicus) by comparative chromosome painting was made in 1999 resulting in the almost simultaneous publication of papers by Grutzner et al. (1999), Guilly et al. (1999) and Stanyon et al. (1999). Since then, over 100 rodent genomes from all major taxa have been analyzed resulting in the availability of comparative chromosomal maps and a more detailed analysis of karyotypic evolution in several of the major taxonomic groups (Table 1, Supplement Data 2).
Table 1. Chromosome painting of Rodentia.
| Suborder | Family/ subfamily | Species | Set of painting probes | Number of autosomal conserved segments revealed. Comments | Reference |
|---|---|---|---|---|---|
| Sciuromorpha | Gliridae | Eliomys melanurus | Homo sapiens | 45 | Sannier et al., 2011 |
| Eliomys munbyanus | Homo sapiens | Paints HSA 2, 5, 6, 14, 15, 18 were hybridized | Sannier et al., 2011 | ||
| Eliomys quercinus | Homo sapiens | Paints HSA 1, 2, 4, 12, 15, 20, 22 were hybridized | Sannier et al., 2011 | ||
| Sciuridae | Menetes berdmorei | Homo sapiens | ⩾34 | Richard et al., 2003 | |
| Sciurus carolinensis | Homo sapiens | 38 | Stanyon et al., 2003; Li et al., 2004 | ||
| Petaurista albiventer | Homo sapiens | 36 | Li et al., 2004 | ||
| Tamias sibiricus | Homo sapiens | 35 | Li et al., 2004 | ||
| Callosciurus erythraerus | Sciurus carolinensis | 21 | Li et al., 2004 | ||
| Petaurista albiventer | Sciurus carolinensis | 21 | Li et al., 2004 | ||
| Tamias sibiricus | Sciurus carolinensis | 21 | Li et al., 2004 | ||
| Marmota himalayana | Sciurus carolinensis | 21 | Li et al., 2006a | ||
| Xerus cf. rythropus | Sciurus carolinensis | 19 | Li et al., 2006a | ||
| Marmota himalayana | Homo sapiens | 35 | Li et al., 2006a | ||
| Xerus cf. rythropus | Homo sapiens | 35 | Li et al., 2006a | ||
| Tamias sibiricus | Homo sapiens | 36 | Romanenko et al., 2010 | ||
| Marmota baibacina | Tamias sibiricus | 21 | Beklemisheva et al., 2011 | ||
| Marmota kastschenkovii | Tamias sibiricus | 21 | Beklemisheva et al., 2011 | ||
| Spermophilus erythrogenys | Tamias sibiricus | 23 | Beklemisheva et al., 2011 | ||
| Spermophilus major | Tamias sibiricus | 23 | Beklemisheva et al., 2011 | ||
| Spermophilus suslicus | Tamias sibiricus | 23 | Beklemisheva et al., 2011 | ||
| Spermophilus undulatus | Tamias sibiricus | 23 | Beklemisheva et al., 2011 | ||
| Sciurus vulgaris | Tamias sibiricus | 20 | Beklemisheva et al., 2011 | ||
| Tamias sibiricus | Homo sapiens | 36 | Beklemisheva et al., 2011 | ||
| Tamias sibiricus | Castor fiber | 40 | Beklemisheva et al., 2011 | ||
| Castorimorpha | Castor fiber | Homo sapiens | 43 | Graphodatsky et al., 2008 | |
| Castor fiber | Homo sapiens | 44 | Beklemisheva et al., 2011 | ||
| Castor fiber | Tamias sibiricus | 42 | Beklemisheva et al., 2011 | ||
| Anomaluromorpha | Pedetes capensis | Homo sapiens | 46 | Graphodatsky et al., 2008 | |
| Hystricomorpha | Caviidae | Cavia porcellus | Homo sapiens | ⩾71 | Our unpublished data |
| Cavia tschudii | Cavia porcellus | 30 | Our unpublished data | ||
| Cavia tschudii | Homo sapiens | ⩾71 | Our unpublished data | ||
| Bathyergidae | Cryptomys (Fukomys) mechowi | Heterocephalus glaber | 43 | Deuve et al., 2006 | |
| Heliophobius argenteocinereus | Heterocephalus glaber | 45 | Deuve et al., 2008 | ||
| Bathyergus janetta | Heterocephalus glaber | 43 | Deuve et al., 2008 | ||
| Bathyergus siullus | Heterocephalus glaber | 43 | Deuve et al., 2008 | ||
| Georychus capensis | Heterocephalus glaber | 43 | Deuve et al., 2008 | ||
| Fukomys damarensis | Heterocephalus glaber | 47 | Deuve et al., 2008 | ||
| Fukomys darlingi | Heterocephalus glaber | 45 | Deuve et al., 2008 | ||
| Octodontidae | Tympanoctomys barrerae | Octodon degus | Some paints gave satisfactorily results of hybridization only | Svartman et al., 2005 | |
| Thryonomyidae | Thryonomys swinderianus | Heterocephalus glaber | 33 | Deuve et al., 2008 | |
| Myomorpha | Dipodidae | Sicista betulina | Homo sapiens | 62 | Graphodatsky et al., 2008 |
| Muridae: Murinae | Mus musculus | Homo sapiens | Only chromosome-specific probes 16, 17 and X were used | Scherthan et al., 1994 | |
| Rattus norvegicus | Mus musculus | Only six chromosome-specific probes were used | Scalzi and Hozier, 1998 | ||
| Mus musculus | Rattus norvegicus | Only 10 chromosome-specific probes were used | Guilly et al., 1999 | ||
| Rattus norvegicus | Mus musculus | 37 | Guilly et al., 1999 | ||
| Rattus norvegicus | Mus musculus | 31 | Grutzner et al., 1999 | ||
| Rattus norvegicus | Mus musculus | 35 | Stanyon et al., 1999 | ||
| Mus musculus | Rattus norvegicus | 35 | Stanyon et al., 1999 | ||
| Mus musculus | Cricetulus griseus | 38 | Yang et al., 2000 | ||
| Rattus norvegicus | Mus musculus | 48 | Helou et al., 2001 | ||
| Rattus norvegicus | Mus musculus | 64 (combined Zoo-FISH, FISH and RH data) | Nilsson et al., 2001 | ||
| Rattus rattus rattus | Mus musculus | 36 | Cavagna et al., 2002 | ||
| Rattus rattus rattus | Rattus norvegicus | 20 | Cavagna et al., 2002 | ||
| Rattus rattus frugivorous | Mus musculus | 37 | Cavagna et al., 2002 | ||
| Rattus rattus frugivorous | Rattus norvegicus | 20 | Cavagna et al., 2002 | ||
| Mus platythrix | Mus musculus | 26 | Matsubara et al., 2003 | ||
| Rhabdomys pumilio | Mus musculus | 39 | Rambau and Robinson, 2003 | ||
| Apodemus sylvaticus | Mus musculus | 37 | Stanyon et al., 2004 | ||
| Mus musculus | Apodemus sylvaticus | There was no data about the number of autosomal conserved segments revealed | Stanyon et al., 2004 | ||
| Apodemus agrarius | Mus musculus | 36 | Matsubara et al., 2004 | ||
| Apodemus argenteus | Mus musculus | 36 | Matsubara et al., 2004 | ||
| Apodemus gurkha | Mus musculus | 36 | Matsubara et al., 2004 | ||
| Apodemus peninsulae | Mus musculus | 36 | Matsubara et al., 2004 | ||
| Apodemus semotus | Mus musculus | 36 | Matsubara et al., 2004 | ||
| Apodemus speciosus | Mus musculus | 37 | Matsubara et al., 2004 | ||
| Apodemus sylvaticus | Mus musculus | 37 | Matsubara et al., 2004 | ||
| Mus musculus | Mesocricetus auratus | 43 | Romanenko et al., 2006 | ||
| Nannomys minutoides | Mus musculus | 26 | Veyrunes et al., 2006 | ||
| Mus musculus | Nannomys minutoides | 25 | Veyrunes et al., 2006 | ||
| Coelomys pahari | Nannomys minutoides | 29 | Veyrunes et al., 2006 | ||
| Coelomys pahari | Mus musculus | 34 | Veyrunes et al., 2006 | ||
| Nannomys mattheyi | Mus musculus | 26 | Veyrunes et al., 2006 | ||
| Tokudaia tokunoshimensis | Mus musculus | 32 | Nakamura et al., 2007 | ||
| Tokudaia osimensis | Mus musculus | 33 | Nakamura et al., 2007 | ||
| Micromys minutus | Mus musculus | 49 | Nakamura et al., 2007 | ||
| Millardia meltada | Mus musculus | 37 | Nakamura et al., 2007 | ||
| Mus musculus | Peromyscus maniculatus | 38 | Mlynarski et al., 2008 | ||
| Apodemus peninsulae | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Rattus norvegicus | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Rattus norvegicus | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Maxomys surifer | Rattus norvegicus | 25 | Badenhorst et al., 2011 | ||
| Rattus exulans | Rattus norvegicus | 20 | Badenhorst et al., 2011 | ||
| Rattus exulans | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Rattus tanezumi | Rattus norvegicus | 20 | Badenhorst et al., 2011 | ||
| Rattus tanezumi | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Rattus losea | Rattus norvegicus | 20 | Badenhorst et al., 2011 | ||
| Rattus losea | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Bandicota savilei | Rattus norvegicus | 21 | Badenhorst et al., 2011 | ||
| Bandicota savilei | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Berylmys berdmorei | Rattus norvegicus | 20 | Badenhorst et al., 2011 | ||
| Berylmys berdmorei | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Berylmys bowersi | Rattus norvegicus | 20 | Badenhorst et al., 2011 | ||
| Berylmys bowersi | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Leopoldamys edwardsi | Rattus norvegicus | 20 | Badenhorst et al., 2011 | ||
| Leopoldamys edwardsi | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Niviventer fuvescens | Rattus norvegicus | 22 | Badenhorst et al., 2011 | ||
| Niviventer fuvescens | Maxomys surifer | 25 | Badenhorst et al., 2011 | ||
| Muridae: Deomyinae | Acomys dimidiatus | Mus musculus | 39 | Nakamura et al., 2007 | |
| Muridae: Otomyinae | Otomys irroratus | Mus musculus | 42 | Engelbrecht et al., 2006 | |
| Calomyscidae: Calomyscinae | Calomyscus sp. | Mesocricetus auratus | 36 | Romanenko et al., 2007a | |
| Cricetidae: Cricetinae | Allocricetulus eversmanni | Mesocricetus auratus | 26 | Romanenko et al., 2007a | |
| Cricetulus griseus | Mus musculus | 47 | Yang et al., 2000 | ||
| Cricetulus griseus | Mesocricetus auratus | 25 | Romanenko et al., 2006 | ||
| Mesocricetus auratus | Cricetulus griseus | 23 | Romanenko et al., 2006 | ||
| Mesocricetus auratus | Mus musculus | 43 | Romanenko et al., 2006 | ||
| Cricetulus barabensis | Mesocricetus auratus | 25 | Romanenko et al., 2007a | ||
| Cricetulus longicaudatus | Mesocricetus auratus | 25 | Romanenko et al., 2007a | ||
| Cricetulus migratorius | Mesocricetus auratus | 25 | Romanenko et al., 2007a | ||
| Cticetus cricetus | Mesocricetus auratus | 25 | Romanenko et al., 2007a | ||
| Mesocricetus brandtii | Mesocricetus auratus | 23 | Romanenko et al., 2007a | ||
| Mesocricetus raddei | Mesocricetus auratus | 21 | Romanenko et al., 2007a | ||
| Phodopus campbelli | Mesocricetus auratus | 34 | Romanenko et al., 2007a | ||
| Phodopus roborowskii | Mesocricetus auratus | 35 | Romanenko et al., 2007a | ||
| Phodopus sungorus | Mesocricetus auratus | 34 | Romanenko et al., 2007a | ||
| Tscherskia triton | Mesocricetus auratus | 30 | Romanenko et al., 2007a | ||
| Mesocricetus auratus | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Cricetulus griseus | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Tscherskia triton | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Cricetidae: Neotominae | Peromyscus maniculatus | Mus musculus | Only chromosome-specific probes 3, 7 and 9 were used | Dawson et al., 1999 | |
| Peromyscus maniculatus | Mus musculus | 39 | Mlynarski et al., 2008 | ||
| Peromyscus eremicus | Mesocricetus auratus | 31 | Romanenko et al., 2007a | ||
| Cricetidae: Arvicolinae | Eothenomys militus | Eothenomys proditor | 27 | Li et al., 2006b | |
| Microtus clarkei | Eothenomys proditor | 27 | Li et al., 2006b | ||
| Microtus oeconomus | Microtus agrestis | 29 | Sitnikova et al., 2007 | ||
| Microtus oeconomus | Mesocricetus auratus | 40 | Sitnikova et al., 2007 | ||
| Ellobius lutestens | Microtus agrestis | 34 | Romanenko et al., 2007b | ||
| Ellobius lutestens | Mesocricetus auratus | 44 | Romanenko et al., 2007b | ||
| Ellobius talpinus | Microtus agrestis | 35 | Romanenko et al., 2007b | ||
| Ellobius talpinus | Mesocricetus auratus | 43 | Romanenko et al., 2007b | ||
| Ellobius tancrei | Microtus agrestis | 35 | Unpublished data | ||
| Microtus oeconomus | Microtus agrestis | 27 | Sitnikova et al., 2007 | ||
| Microtus oeconomus | Mus musculus | 47 | Sitnikova et al., 2007 | ||
| Dicrostonyx torquatus | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Ellobius talpinus | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Microtus oeconomus | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Microtus rossiaemeridionalis | Mus musculus | Microdissected chromosome-specific probes 3, 6, 18, 19 were used | Trifonov et al., 2010 | ||
| Microtus arvalis ‘arvalis' | Microtus agrestis | 28 | Lemskaya et al., 2010 | ||
| Microtus daghestanicus | Microtus agrestis | 28 | Lemskaya et al., 2010 | ||
| Microtus dogramacii | Microtus agrestis | 28 | Lemskaya et al., 2010 | ||
| Microtus gregalis | Microtus agrestis | 29 | Lemskaya et al., 2010 | ||
| Microtus guentheri guentheri | Microtus agrestis | 28 | Lemskaya et al., 2010 | ||
| Microtus maximowiczii | Microtus agrestis | 30 | Lemskaya et al., 2010 | ||
| Microtus rossiaemeridionalis | Microtus agrestis | 29 | Lemskaya et al., 2010 | ||
| Microtus socialis | Microtus agrestis | 32 | Lemskaya et al., 2010 | ||
| Cricetidae: Sigmodontinae | Akodon cursor | Mus musculus | ⩾31 | Hass et al., 2008 | |
| Akodon cursor | Akodon paranaensis | 31 | Ventura et al., 2009 | ||
| Akodon cursor | Akodon sp. n. | 10 | Ventura et al., 2009 | ||
| Akodon montensis | Mus musculus | ⩾26 | Hass et al., 2008 | ||
| Akodon montensis | Akodon paranaensis | 21 | Ventura et al., 2009 | ||
| Akodon montensis | Akodon cursor | 11 | Ventura et al., 2009 | ||
| Akodon montensis | Akodon sp. n. | 11 | Ventura et al., 2009 | ||
| Akodon paranaensis | Mus musculus | ⩾28 | Hass et al., 2008 | ||
| Akodon serrensis | Mus musculus | ⩾24 | Hass et al., 2008 | ||
| Akodon sp. n. | Akodon paranaensis | 24 | Ventura et al., 2009 | ||
| Akodon sp. n. | Akodon cursor | 16 | Ventura et al., 2009 | ||
| Necromys lasiurus | Mus musculus | 27 | Hass et al., 2011 | ||
| Oligoryzomys avescens | Mus musculus | ⩾26 | Hass et al., 2008 | ||
| Sigmodon arizonae | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Sigmodon fulviventer | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Sigmodon hirsutus | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Sigmodon leucotis | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Sigmodon mascotensis | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Sigmodon ochrognathus | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Sigmodon peruanus | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Sigmodon toltecus | Sigmodon hispidus | 29 | Swier et al., 2009 | ||
| Thaptomys nigrita | Mus musculus | 30 | Hass et al., 2011 |
Sciuromorpha
The suborder Sciuromorpha is well supported as a monophyletic taxon by both morphological and molecular data (Murphy et al., 2001; Waddell et al., 2001; Churakov et al., 2010) and is subdivided into three families—Aplodontiidae, Sciuridae and Gliridae.
Comparative chromosome painting subsequently allowed a more precise comparison of sciuromorph genomes and currently 17 species (of 307) have been examined by this technique (Table 1), mostly belonging to Sciuridae. Three species of Gliridae have also been studied. These investigations relied predominantly on human (Homo sapiens, HSA) paints, although two sciuromorph-specific sets of painting probes were developed from the flow-sorted chromosomes of Sciurus carolinensis and Tamias sibiricus and used in comparative painting experiments (Li et al., 2004; Beklemisheva et al., 2011).
Studies of ground squirrels confirmed the general tendency for sciurid genome conservation and these data permitted a revision of the putative sciurid karyotype (Richard et al., 2003). The HSA 1/8 and HSA 2/17 associations previously found in sciuromorphs (and considered to represent the sciurid ancestral condition) are absent in the Eurasian ground squirrels—sousliks and woodchucks—while HSA 10/13 and HSA 8/4/8/12/22 are disrupted in four Spermophilus species. Some ground squirrels (Xerus, Menetes) and the flying squirrel (Petaurista) have highly conserved karyotypes that are probably very similar to the ancestral squirrel karyotype and do not differ significantly from that of Rodentia (Richard et al., 2003; Stanyon et al., 2003; Li et al., 2004, 2006a; Beklemisheva et al., 2011). In contrast to the general conservation of syntenic groups, most sciurid genomes are characterized by variation in the size and distribution of heterochromatin; additionally, multiple centromeric shifts have been reported in some species (Beklemisheva et al., 2011). Importantly, however, because of the slow rate of karyotype change and some convergence of characters, cytogenetic evidence failed to resolve close associations within Sciuridae.
The availability of comparative banding and painting data led to suggestions of a putative ancestral karyotype for Sciuridae (Richard et al., 2003; Li et al., 2004; Beklemisheva et al., 2011). The consensus is that this comprised 38 elements corresponding to the following human chromosomes and/or segmental associations: HSA 9/11, 1/10p, 3/21, 16q/19q, 7/16p, 20/15/14, 1pq/8q, 10q/13, 2q/17, 7/22qprox/12qdist, 8p/4q/8p/12pq/22qdiss, 3/19p (Supplement Data 3) (Li et al., 2004, 2006a; Beklemisheva et al., 2011). The karyotypes of Gliridae are also relatively conserved. Comparison of three Eliomys species made by fluorescence in situ hybridization (FISH) using human probes and RBG-banding (R-bands by BrdU using Giemsa) revealed the retention of several eutherian ancestral syntenies in their genomes (Sannier et al., 2011). As karyotypes of other glirids have not yet been studied, and because the different Eliomys karyotypes are conserved, one might consider the E. melanurus syntenies to reflect the chromosomal signatures for glirids in general. Importantly, however, representatives of Aplodontiidae, as well as other glirid genera, have not been included in comparative FISH experiments, and it is possible that these data could shed additional light on the composition of an ancestral karyotype for Sciuromorpha.
Anomaluromorpha
The monophyly of anomaluromorphs was recently confirmed by DNA sequences (Montgelard et al., 2008; Blanga-Kanfi et al., 2009). Nine extant species are currently recognized in the suborder. The taxon is poorly studied at a chromosomal level (the diploid chromosome number is known for only one species). There are no descriptions or comparisons of banded chromosomes between representatives of this group. The karyotype of only one species—Pedetes capensis—has been investigated using human painting probes (Graphodatsky et al., 2008). Surprisingly, some characteristic human chromosomal associations were not detected in P. capensis, that is, the HSA 7/16, and 16/19 as well as core glires HSA 1/10, 9/11 (Supplement Data 3). As the basal position of Sciuromorpha is well supported by different phylogenies (Murphy et al., 2001; Huchon et al., 2002; Adkins et al., 2003; Debry, 2003; Blanga-Kanfi et al., 2009; Churakov et al., 2010), fissions of the ancestral chromosomal signatures (or break points in close proximity to these regions) suggest considerable reorganization of the P. capensis genome, and possibly in the genomes of all anomalurids.
Castorimorpha
The Castorimorpha has traditionally been included in the sciuromorpha-like rodent group because of similar morphological traits (see Carleton and Musser (2005) and references therein). The latest molecular data suggest, however, that these species should be considered a separate taxon independent from Sciuromorpha (Blanga-Kanfi et al., 2009; Horn et al., 2011). According to current taxonomy, Castorimorpha includes beavers, pocket and kangaroo mice, pocket gophers—some 102 species in all.
Karyotypes of only two species have been described using banding techniques—Castor fiber and C. canadensis (Genest et al., 1979; Atlas of mammalian chromosomes, 2006). Castor fiber is the only species investigated by comparative chromosome painting (Beklemisheva et al., 2011). Characteristic placental associations such as HSA 7/16, 14/15, 16/19 (as well as some regarded as core to Glires, that is, HSA 1/10, 9/11) were not detected in the beaver genome supporting their placement (and that of the whole suborder) into a distinct clade (Supplement Data 3) (Graphodatsky et al., 2008).
Hystricomorpha
The representatives of this suborder are poorly studied by comparative cytogenetics. Although some species were described using banding techniques (see Atlas of Mammalian Chromosomes, 2006) there are only three recent publications involving hystricomorph species in chromosome painting studies.
Seven Bathyergidae and one species of Thryonomyidae were compared using naked mole rat probes (Deuve et al., 2006, 2008). The investigators' defined autosome–gonosome translocations, fusions and fissions as the major trends of karyotypic evolution in both families. Unfortunately, the lack of a link to human chromosomes does not permit conclusions on possible human associations specific to Bathyergidae and Thryonomyidae. The complexity of chromosomal evolution within Cavia and allies was initially reported by Viegas-Péquignot et al. (1986). Reciprocal chromosome painting between human and Guinea pig (Cavia porcellus) subsequently localized human paining probes to hystricomorph chromosomes (unpublished data). Only three adjacent chromosomal syntenies, signatures common to most placentals, are retained in the C. porcellus genome: HSA 3/21 and HSA 12/22 (twice). HSA 4/8/4 was also identified, but reciprocal painting showed that it was formed by different segments of human chromosomes 4 and 8 to those in other rodents. The fact that the placental signatures HSA 4/8p, 7/16, 14/15, and glirid signatures HSA 1/10, 3/19, 7/16, 8p/4/8p, 9/11, 14/15, 16/19, are absent in Cavia, indicates that its genome has undergone significant reorganization through fusions and fissions (Supplement Data 3) confirming that the Hystricomorpha represent yet another rodent suborder with unusually high rates of genome evolution. Interestingly, comparative chromosome painting demonstrated that the karyotypes of two Guinea pigs—C. porcellus and C. tschudii—are identical (unpublished data) suggesting that additional painting data are needed to establish, which associations are characteristic for suborder.
Myomorpha
Nearly one-third of all rodent species belong to the suborder Myomorpha making this taxon particularly appealing for evolutionary studies. The majority of studied species belong to two large families within Muroidea—the Cricetidae and Muridae. Only few representatives of other families have been included in comparative cytogenetic investigations. Nonetheless, comparative cytogenetic data show that high karyotypic reshuffling is characteristic for Myomorpha (for example, Stanyon et al., 1999) but that the elevated rate of chromosomal change was not accompanied by a rapid evolution of morphological features. Generally, the group has been reasonably well investigated by conventional cytogenetics, while 71 species have been investigated by comparative chromosome painting. With one exception (Sicista betulina), all studied species belong to the superfamily Muroidea.
It is important to note that the ‘catastrophic' reorganization of myomorph genomes (characterized by a significant change of the whole genome, including the formation of a new linkage groups characteristic only for the given taxon, see Graphodatsky, 1989) made their study by the direct hybridization to human painting probes problematic. Consequently, a variety of myomorph probes was developed in order to compare karyotypes within the group (Table 1). However, in spite of these resources, in most instances laboratory mouse paining probes were used to make comparisons among the Myomorpha because these are commercially available. This led to their use as a common reference for the various species. In comparison to the most other muroids, however, mouse chromosomes are highly rearranged and this has detracted to some extent from their use in comparative studies in preference to those derived from species with conserved genomes (largely because of the difficulties in the interpretation of hybridization results).
Cricetidae
The family comprises 681 species grouped in 6 subfamilies. Representatives of four subfamilies have been included in comparative chromosome painting experiments (Table 1) but a reconstruction of the karyotypic relationships within Cricetidae has not been attempted.
Cricetinae
The subfamily contains 19 species from 7 genera. The first data obtained using comparative chromosome painting with golden hamster (Mesocricetus auratus, MAU) painting probes (Romanenko et al., 2006, 2007b) revealed that the karyotypes of some closely related species differed greatly. Chromosome painting data are currently available for 14 species of 6 hamster genera including M. auratus and Cricetulus griseus (Yang et al., 2000). Comparative painting and banding within the group permitted the analysis of chromosomal evolution and karyotype relationships within the subfamily resulting in findings that are in broad agreement with molecular data (Neumann et al., 2006). It was determined that Mesocricetus, Tscherskia, Phodopus and Cricetus represent a monophyletic clade (Neumann et al., 2006; Romanenko et al., 2007b). Moreover, different chromosomal rearrangements are characteristic for different lineages. For example, the derivation of Phodopus karyotypes necessitates the complex fission and fusion of ancestral chromosomes. Robertsonian fusions, and appearance of additional heterochromatin blocks, characterized the karyotype evolution of Mesocricetus, while inversions are important in shaping the chromosomes of Allocricetulus, Cricetulus and Cricetus.
In the case of the greater long-tailed hamster, Tscherskia, comparative chromosome painting data provided important characters for the separate status of the genus. Tscherskia triton was long considered part of Cricetus because of morphological similarities. Conventional banding analysis has shown several partial chromosomal homologies between T. triton and other Cricetus species (Radjabli, 1975). However, painting showed extensive intra- and extra-chromosomal rearrangement in T. triton strongly supporting a separate position for the genus within Palaearctic hamsters (Romanenko et al., 2007b). This conclusion was subsequently confirmed by molecular data (Lebedev et al., personal communication).
On the basis of the defined signatures, we propose an Ancestral Cricetinae Karyotype with 2n=48–54. This variation in diploid number is the result of uncertainty concerning the number of segments of M. musculus (MMU) 14 and 15, that is, their presence in the karyotype as one or two fragments (Supplement Data 4, 5). Another problem is the association MMU 11/5/14. It is currently not possible to ascertain whether there was only one chromosome combining segments MMU 5, 11 and 14, or two chromosomes homologous to MMU 5/14 and MMU5/11. However, as Arvicolinae species (see below) have two segments homologous to MMU 5 in their karyotypes, the presence of MMU 11/5/14 seems more likely. Consequently, an Ancestral Cricetinae Karyotype with 2n=48 containing MMU 1/17, 2, 3, 4, 4, 5/16, 6, 6/17, 7, 7/19, 8, 8/2/13, 9, 10, 10/17, 11/5/14, 11/17/16, 12, 12/17, 13/15, 15/1/17, 17/1/10/17, 18, X and Y is proposed.
Arvicolinae
The subfamily includes voles and lemmings. Diploid numbers range from 2n=17 in Ellobius lutescens and Microtus oregoni to 2n=64 in M. longicaudus. G-banded chromosomes of 50 arvicoline species were summarized in the Atlas of Mammalian Chromosomes (2006). The subfamily includes species with several striking cytogenetic features: the presence of B-chromosomes in some, unusual systems of sex chromosomes in others (Dicrostonyx, Ellobius and Microtus) and giant sex chromosomes in Microtus agrestis (Supplement Data 1).
Most representatives of Microtus have been included in comparative painting investigations. The limited variation in external morphology has been a significant challenge in Microtus classification and this has made cytogenetic data important for solving problems of vole taxonomy. A comparison of eight Microtus species using M. agrestis painting probes allowed reconstruction of a putative ancestral karyotype and insights to karyotype evolution within the taxon (Lemskaya et al., 2010). Surprisingly, cross-species chromosome painting in Microtus revealed no rearrangements that clearly support the branching pattern depicted in the molecular tree (see Lemskaya et al., 2010). Karyotypes of grey voles are generally characterized by the conservation of large ancestral syntenies suggesting that Robertsonian translocations predominate in the karyotype evolution of these species (Li et al., 2006b; Lemskaya et al., 2010).
Reorganization of several ancestral chromosomes occurred during formation of modern Ellobius karyotypes. The genus comprises five species (Carleton and Musser, 2005) whose diploid numbers vary from 17 (E. lutestens) to 54 (E. talpinus). The species E. lutestens, E. talpinus, and E. tancrei were compared using chromosome painting (Romanenko et al., 2007a; unpublished data) and its clear that E. lutestens has undergone a ‘catastrophic' reshuffling of its chromosomes during its evolution. In spite of the high number of fusions, fissions and inversions detected, it was nonetheless possible to identify conserved elements that could be considered ancestral for Ellobius. In the case of E. tancrei and E. alaicus, a ‘Robertsonian fan' was described (Lyapunova et al., 1980). Chromosome painting showed that E. tancrei (2n=30–54) has a complex karyotypic structure formed by racial hybridization, and that chromosomal diversity was accompanied by independent and repeated Robertsonian rearrangements (single and multiple), and possibly by whole-arm reciprocal translocations (Bakloushinskya et al., 2010).
On the basis of the signatures revealed in different arvicolines, the ancestral karyotype of the Arvicolinae (2n=56) appears to be identical to that proposed for Ellobius (Romanenko et al., 2007a). It comprises: MMU 1/14/1, 1/17, 1/17/7/5/10/17, 2, 2, 2/13, 3, 3, 4, 4, 5/11, 5/16, 6, 6/12/17, 7, 7/19, 8, 8, 9, 10, 11/17/16, 12, 13/15, 14, 15, 17/1/10/17, 18, X and Y. The ancestral Microtus karyotype (AMiK, Lemskaya et al., 2010) can therefore be derived by one fusion, that of MMU 6 and MMU 6/12/17, which resulted in the formation of MMU 6/17/12/6.
Sigmodontinae
A high degree of karyotype conservation was revealed for eight species of Sigmodon (Swier et al., 2009). In contrast, the karyotypes of Akodon, Necromys and Thaptomys are highly rearranged. For example, Robertsonian and tandem fusion rearrangements, pericentric inversions and/or centromere repositionings, paracentric inversions, translocations and insertions were observed in Akodon species (Hass et al., 2008; Ventura et al., 2009). Cross-species FISH using murine probes suggest that MMU 8/13 may be a signature for the Sigmodontinae (Hass et al., 2011; Supplement Data 3). Syntenies such as MMU 3/18 and 6/12 are combined in Akodon and Necromys. However, as most painting data for the group are incomplete we cannot draw definitive conclusion on the composition of a putative Sigmodontinae ancestral karyotype.
Neotominae
Although previously included in Sigmodontinae, 16 genera (many of New World rats and mice) are grouped in the Neotomyinae within the New World Cricetidae. Of these, conventional banding analysis showed a high degree of karyotypic conservation within Peromyscus: all species have 2n=48. The number of chromosomal arms ranges from 52 to 92 because of variation attributable to heterochromatin additions and pericentric inversions (Robbins and Baker, 1981; Rogers et al., 1984). On the basis of the painting data, it seem reasonable to consider the P. eremicus karyotype as being close to the putative ancestral state for Muroidea (Romanenko et al., 2007b). However, there is some disagreement on the murine signatures found in P. eremicus and P. maniculatus (Romanenko et al., 2007b; Mlynarski et al., 2008) and broader taxon sampling is necessary for reconstructing the ancestral karyotype of the subfamily.
Calomyscidae
Mouse-like hamsters of the genus Calomyscus represent a striking example of speciation underscored by cytogenetic characters—morphologically similar species of mouse-like hamsters have different diploid and fundamental numbers, and specific sets of translocations (Graphodatsky et al., 2000). Current cytogentic data confirm conclusions based on molecular studies that show Calomyscus to be the most basal clade within Muroidea (Jansa and Weksler, 2004; Romanenko et al., 2007b).
Muridae
Muridae comprises ∼730 species and is larger than any other mammalian family. The examination of murid chromosomes using conventional cytogenetics allowed the detection of some notable features in their karyotypes including (i) extensive variation in diploid numbers—from 2n=14 (Taterillus tranieri) to 2n=74 (Gerbillus latastei), (ii) considerable interspecific differences in the amount and distribution of heterochromatin (Graphodatsky, 1989), (iii) the presence of supernumerary chromosomes in many species (Trifonov et al., 2002) and (iv) sex chromosomes systems that differ from the conventional XX/XY.
Representatives of the Muridae were the first rodents studied by chromosome painting (Scherthan et al., 1994) and today some 29 species from three subfamilies have been investigated (Table 1) using different sets of probes. Generally, murid genomes have been extensively reorganized during evolution. However, some species with conserved genomes have been identified. For example, a single chromosomal rearrangement distinguishes Apodemus (Matsubara et al., 2004; Stanyon et al., 2004) and relatively high genome conservation was established for species within Rattus and Tokudaia, representatives of the tribe Rattini (Guilly et al., 1999; Stanyon et al., 1999; Cavagna et al., 2002; Nakamura et al., 2007; Badenhorst et al., 2011).
The most likely Ancestral Murinae Karyotype had 2n=46 and contained following associations of mouse chromosomes: MMU 1, 2, 2/13, 3, 4, 5/6, 5/11, 7/19, 8, 8, 9, 10/17, 10/17, 11/16, 12/17, 13/15, 14, 14, 15, 16, 17/1/17, 18, X and Y. However there could be three segments of MMU 5 and MMU 10, and MMU 4 and MMU 9 may have been present in two fragments and not in one (Supplement Data 5), thereby collectively increasing the ancestral 2n to 54.
Overview of karyotype evolution in rodents
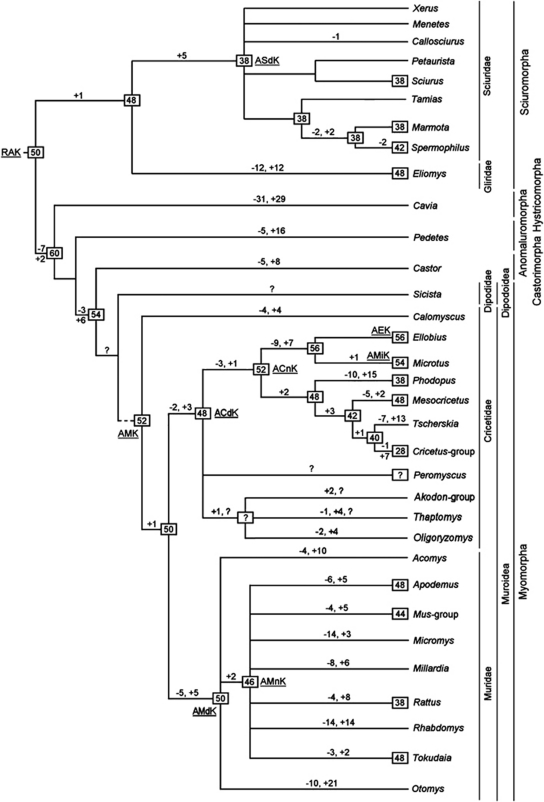
In order to reconstruct the putative ancestral karyotypes at some of the major nodes of the Rodentia tree, we combined all available painting data and attempted to identify shared syntenic associations between lineages. These shared ancestral (and hence symplesiomorphic) elements were considered to be a part of the ancestral karyotype under consideration. In those instances where syntenic arrangements were different between closely related taxa (for example, chromosomal segments 1 and 2 were fused in species A, but disrupted in its sister species B) outgroup comparisons were used (Dobigny et al. 2004) to determine the ancestral state. In this example, the fused presence of chromosomal segments 1 and 2 in a distantly related species C, suggests that this is the ancestral condition shared with species A.
An Ancestral Karyotype of Rodentia (RAK) was proposed (Graphodatsky et al., 2008) that suggested the associations HSA 8/12 and 15/20 may define rodents. Originally, HSA 1/10p and 9/11 were considered ancestral for Glires (the cohort combining Rodentia and Lagomorpha). However, the most recent glirid painting data showed the absence of HSA 9/11 and 3/19 in Eliomys. In the light of these findings, we concur with Sannier et al. (2011) that the occurrence HSA 9/11 cannot be unequivocally explained. It could be a result of convergence in some lagomorphs, sciurids and myomorphs, or it may represent an ancestral feature that was lost in certain branches. The association HSA 3/19 was found in all Carnivora and one eulipotyphlan species, as well as in all studied species of Sciuridae and Anomaluromorpha. Consequently questions about its presence in the RAK remain open. Syntenies such as HSA 3/21, 4/8p, 7/16, 12/22, 14/15, 16/19 are shared with other eutherians (Ferguson-Smith and Trifonov, 2007). We consequently placed the RAK at the base of the tree and tracked its reorganization during rodent evolution.
Molecular phylogenies are not universally consistent, but they all consider Sciuromorpha as a basal clade within Rodentia (Huchon et al., 2002; Adkins et al., 2003; Debry, 2003; Blanga-Kanfi et al., 2009; Churakov et al., 2010). The fusion HSA 8/12 (resulting in the HSA 8/4/8/12/22 association) could be the single rearrangement distinguishing the Ancestral Sciuromorpha Karyotype from RAK. All sciurids share following associations: HSA 1/8, 2/17, 7/22, 10/13, 15/20 (Richard et al., 2003; Li et al., 2004, 2006a; Graphodatsky et al., 2008; Beklemisheva et al., 2011). Thus, only five fusions are needed to explain the derivation of the Sciuridae Ancestral Karyotype from that of the putative ancestral Sciuromorpha karyotype. Many more rearrangements (12 fissions and 12 fusions) are needed to form a putative ancestral Rodentia karyotype from that of the ancestral Gliridae (Sannier et al., 2011).
Suborders Anomaluromorpha, Hystricomorpha, Myomorpha and Castorimorpha form a single clade based on the presence of HSA 1/7, the disruption of the HSA 7/16 synteny, and the fissions of HSA 1, 4, 5, 6, 11, and 15 in karyotypes of all studied representatives. There is also a possibility that the HSA 10/16 association may be ancestral for the clade. The HSA 8 and HSA 19 association was found in different rodent species (that is, Castor fiber, P. capensis), but this involved different non-homologous fragments of HSA 8 so it cannot be considered ancestral for the group. The absence of HSA 1/10 was previously proposed as a signature for a clade comprising Anomaluromorpha+Myomorpha+Castorimorpha but this association was subsequently detected in karyotypes of M. musculus and R. norvegicus (Ensembl Mouse web site (http://www.ensembl.org); Nilsson et al., 2001). HSA 1/10 was not present in the Cavia karyotype. Finally, we conclude that the putative ancestral karyotype of Anomaluromorpha, Hystricomorpha, Myomorpha and Castorimorpha had a 2n=60 (or 2n=62 if the HSA 1/10 association in myomorphs and sciuromorphs is convergent) that consisted of HSA 1, 1/7, 1/10, 2, 2, 3, 3/19, 3/21, 4, 4, 4/8, 5, 5, 6, 6, 7, 8, 9/11, 10/16, 11, 12/22 (twice), 13, 14/15, 15, 16/19, 17, 18, 20, X and Y.
The following rearrangements offer a striking confirmation of the close evolutionary relationship of Myomorpha and Castorimorpha: HSA 5/17 (it is absent in Mus and Rattus, but present in Sicista), HSA 11/15 and fission of HSA 14/15 (Graphodatsky et al., 2008). However, the use of different methods of analysis (DNA sequences for myomorph genomes and chromosome painting for castorimorphs) could give inconsistent results because of resolution differences of the analyses. For example, HSA 5/17 was not detected in the mouse and rat karyotypes by FISH and a disruption of ancestral eutherian synteny HSA 14/15 occurred in hystricomorphs (Cavia). Moreover, all representatives of Hystricomorpha, Myomorpha and Castorimorpha studied have three fragments of HSA 12 in their karyotypes. These features corroborate our suggestion that Hystricomorpha is intermediate between Anomaluromorpha and Myomorpha+Castorimorpha. Although such an arrangement contradicts the latest data based on painting with human probes, sequencing of nuclear genes and the distribution of short interspersed elements all place Hystricomorpha in a basal position to the Anomaluromorpha+Myomorpha+Castorimorpha clade (Ferguson-Smith and Trifonov, 2007; Blanga-Kanfi et al., 2009; Churakov et al., 2010; Horn et al., 2011). We therefore consider the fission of HSA 14/15 and the disruption of HSA 12 onto three fragments as convergent events that took place independently in hystricomorphs, myomorphs and castorimorphs.
Despite some gaps in Cavia and human whole genome homology maps, we were able to demonstrate a ‘catastrophic' reorganization of the hystricomorph karyotype—as many as 29 fusions and 31 fissions were detected when compared with the RAK. The evolution of the C. fiber karyotype was accompanied by a smaller number of rearrangements—these included disruptions to HSA 1/7, 1/10, 3/19, 9/11, 16/19 and the presence of 8 fusions.
Within Myomorpha the karyotype of only one species (Sicista betulina) was studied using human painting probes (Graphodatsky et al., 2008). This species falls within Dipodoidea, which represents a basal branch in Myomorpha (Jansa and Weksler, 2004; Steppan et al., 2004). Two other species (M. musculus and R. norvegicus) were compared with human using non-painting approaches (that is, based on full genome sequencing data) but the resolution of the methods differs greatly thus precluding the construction of an ancestral karyotype for Myomorpha.
As a result of the extent of genomic reshuffling in muroid rodents, conserved syntenies are referenced to mouse rather than to human chromosomes. Reciprocal chromosome painting between mouse/golden hamster and golden hamster/field vole (Romanenko et al., 2006; Lemskaya et al., 2010) provided an opportunity to include cricetid and arvicolin comparisons in the myomorph investigation. Here, we examined the karyotype evolution of Muroidea based solely on mouse chromosome associations.
The analysis of chromosomal signatures in different muroid karyotypes suggests a 2n=52 for the Ancestral Muroidea Karyotype (AMK) (Figure 1). The AMK differs from the one proposed earlier based on a wide range of hamster species comparison but that included few murids (Romanenko et al., 2007b). It is also possible that MMU 5/14 and MMU 11, which were syntenic in the AMK were disrupted in the Calomyscus branch. In this case, the AMK was identical to the common ancestral karyotype of Cricetidae and Muridae. The 2n=52 karyotype of Calomyscus sp. differs from the proposed AMK by four fusions and four fissions.
Figure 1.
Putative ancestral karyotypes: (a) AMK–ancestral Muroidea karyotype, (b) ACdK—ancestral Cricetidae karyotype, (c) AMdK—ancestral Muridae karyotype. Different colors correspond to separate mouse chromosomes. Dashed gray frame and arrows mark elements whose state in the AKs was not ambiguously determined. fis, fission; fus, fusion. See comments in the text.
The evolution of the Ancestral Cricetidae Karyotype (Figure 1) was accompanied by a small number of fissions (MMU 17 twice) and fusions (MMU 17/1+MMU 10/17, MMU 1+17 and MMU 6+17) of ancestral chromosomes. If MMU4 was present as two segments, then Ancestral Cricetidae Karyotype would have been 2n=50.
In view of the disagreements in painting results obtained for two Neotomyinae species (Mlynarski et al., 2008; Romanenko et al., 2007b), it was not possible to unequivocally define the type and number of rearrangements for Peromyscus. The partial hybridization of M. musculus probes to four Akodon species does not provide sufficient data to reconstruct the ancestral karyotype for Sigmodontinae, and to define the number and types of rearrangements for the different branches.
The ancestral karyotype common to the Arvicolinae and Cricetinae probably had 2n=52 or 50 (it depends on the number of segments homologous to MMU14) and contained the following associations: MMU 1/17, 1/17, 2, 3, 4, 4, 5/16, 6, 6/17, 7, 7/19, 8, 8/2/13, 9, 10, 10/17, 11/5/14, 11/17/16, 12, 12/17, 13/15, 14, 15, 17/1/10/17, 18, X and Y. The subsequent formation of the Ancestral Cricetinae Karyotype was accompanied by two fusions (Romanenko et al., 2007b). As mentioned above, the putative ancestral karyotype of Arvicolinae may be identical to that of Ellobius and can be derived from the Ancestral Cricetidae Karyotype by nine fissions and seven fusions (Romanenko et al., 2007a).
The Ancestral Muridae Karyotype (Figure 1) differs from that of the common ancestor of Cricetidae and Muridae by at least five fissions and five fusions. The chromosome number of Ancestral Muridae Karyotype ranges from 2n=50 to 2n=56 because of variable interpretations of the number of segments homologous to MMU 4, 5 and 10. Although the sequence-based Muridae phylogeny is controversial (Conroy and Cook, 1999; Martin et al., 2000; Michaux et al., 2001; Jansa and Weksler, 2004; Steppan et al., 2004; Blanga-Kanfi et al., 2009), we propose that two fusions (synteny MMU 5/6 and fusion of two segments homologous to different parts of MMU 9) occurred during the formation of the Ancestral Murinae Karyotype (with 2n=46–52). The various generic associations suggested in Murinae by sequence-based phylogenies could not be verified using painting data. The types and numbers of rearrangements that are thought to lead to the ancestral karyotypes of each genus are shown in Figure 2. These data prompted us to revise the diploid chromosome number previously proposed for the Mus-group (2n=46 to 2n=44 and which combines subgenera Coelomys, Nannomys, Mus and Pyromys). It seems more plausible that the ancestor of the subgenera had three segments homologous to MMU 5 as reported by Veyrunes et al. (2006). Considering that most Murinae have the MMU 13/15 association in their karyotypes, we suggested that MMU 13/15 was present in the ancestral karyotype of the Mus-group, and not the MMU 13/15/13 configuration suggested by Veyrunes et al. (2006).
Figure 2.
Putative scheme of chromosome evolution in Rodentia to the genus level. RAK—ancestral Rodentia karyotype; ACdK—ancestral Cricetidae karyotype; ACnK—ancestral Cricetinae karyotype; AEK—ancestral Ellobius karyotype; AMdK—ancestral Muridae karyotype; AMK—ancestral Muroidea karyotype; AMiK—ancestral Microtus karyotype; AMnK—ancestral Murinae karyotype; ASdK—Sciuridae ancestral karyotype. Presumable ancestral diploid number characters for node are shown in black frames. Minus sign indicates chromosome fissions, plus sign indicates chromosome fusions, and question mark indicates unresolved positions. See comments in the text.
A high number of species and elevated rates of chromosomal change make rodent karyotypes particularly informative for building and improving existing phylogenies. However, it is clear that the incorporation of new species in future molecular cytogenetic studies and the application of new molecular markers would result in better understanding of rodent evolution.
Rates of karyotype evolution
Although the numbers of autosomal segments scored in comparative chromosome painting experiments include hemiplasic (Avise and Robinson, 2008) and homoplasic segments, they nonetheless remain good indicators of the level of genome conservation. The numbers of human autosomal conserved segments detected in sciurid genomes vary from 35 to 36 (Table 1). Generally, the genomes of sciurids are highly conserved and most closely reflect the putative ancestral genome of all rodents. Investigations of glirids, castorimorphs and anomaluromorphs detected slightly higher numbers of human autosomal segments in their karyotypes. Hystricomorph painting indicated a high level of Cavia genomic reshuffling (>71 conserved segments). The only representative myomorph species (Sicista betulina; Graphodatsky et al., 2008) had 62 autosomal conserved segments when analyzed by FISH using human probes.
Generally, the rates of karyotype evolution differ in various branches of rodent phylogenetic tree. Even within a single family it is not unusual to find genera with low rates of reorganization (for example, Apodemus species) and those whose genomes are extensively rearranged (Mus species). Rough estimates show that rates may vary as much as 10 times across different branches of the muroid tree (Veyrunes et al., 2006).
Conclusions
In recent years, modern cytogenetics has contributed significantly to studies of evolutionary relationships among mammals. Chromosome painting has resulted in novel discoveries and has extended previous conclusions drawn from conventional comparative banding data. Nowhere is this more apparent than in rodents where high species diversity and extensive genome reshuffling has produced unparallel opportunities for studying chromosomal evolution in mammals.
It is clear, however, that future studies should focus on problematic and uninvestigated branches in Rodentia, particularly in Myomorpha. These should include pivotal lineages such as Laonastes and gundis, basal murids and jerboas and combine refinements in methodology that would permit the detection of smaller rearrangements (such as multicolor banding and mapping using bacterial artificial chromosomes). Importantly, the absence of reciprocal painting in most studies makes it currently difficult to unambiguously define chromosomal characters because of the questionable homology of supposedly syntenic fragments. Finally, although prospective studies will undoubtedly benefit greatly from the whole genome analysis of different rodents (see the Genome 10K Project proposed by the Genome 10K Community of Scientists, 2009), their selection will, in part, be directed by detailed karyotypic descriptions resulting from molecular cytogenetic investigations such as those outlined in this paper.
Acknowledgments
This study was funded in part by programs MCB and SB RAS Programs and research grants of Russian Fund for Basic Research.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Adkins RM, Walton AH, Honeycutt RL. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol. 2003;26:409–420. doi: 10.1016/s1055-7903(02)00304-4. [DOI] [PubMed] [Google Scholar]
- Atlas of mammalian chromosomes 2006. Stephen J. O'Brien, Joan C. Menninger, William G. Nash (eds). A John Wiley & Sons: Hoboken, NJ, USA [Google Scholar]
- Avise JC, Robinson TJ. Hemiplasy: a new term in the lexicon of phylogenetics. Syst Biol. 2008;57:503–507. doi: 10.1080/10635150802164587. [DOI] [PubMed] [Google Scholar]
- Badenhorst D, Dobigny G, Adega F, Chaves R, O'Brien PCM, Ferguson-Smith MA, et al. Chromosome evolution in Rattini (Muridae, Rodentia) Chromosome Res. 2011;19:709–727. doi: 10.1007/s10577-011-9227-2. [DOI] [PubMed] [Google Scholar]
- Bakloushinskya IYu, Romanenko SA, Graphodatsky AS, Matveevsky SN, Lyapunova EA, Kolomiets OL.2010The role of chromosome rearrangements in the evolution of mole voles of the genus Ellobius (Rodentia, Mammalia) Russ J Genet 461143–1145.(In Russian). [PubMed] [Google Scholar]
- Beklemisheva VR, Romanenko SA, Biltueva LS, Trifonov VA, Vorobieva NV, Serdukova NA, et al. Reconstruction of karyotype evolution in core Glires. I. The genome homology revealed by comparative chromosome painting. Chomosome Res. 2011;19:549–565. doi: 10.1007/s10577-011-9210-y. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry RW, Huchon D. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol Biol. 2009;9:71. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton MD, Musser GG.2005Order RodentiaIn: Wilson DE, Reeder DM (eds).Mammal Species of the World: A Taxonomic and Geographic Reference Johns Hopkins University Press: Baltimore; 745–1601. [Google Scholar]
- Cavagna P, Stone G, Stanyon R. Black rat (Rattus rattus) genomic variability characterized by chromosome painting. Mamm Genome. 2002;13:157–163. doi: 10.1007/BF02684021. [DOI] [PubMed] [Google Scholar]
- Churakov G, Sadasivuni MK, Rosenbloom KR, Huchon D, Brosius J, Schmitz J. Rodent evolution: back to the root. Mol Biol Evol. 2010;27:1315–1326. doi: 10.1093/molbev/msq019. [DOI] [PubMed] [Google Scholar]
- Conroy CJ, Cook JA. MtDNA evidence for repeated pulses of speciation within arvicoline and murid rodents. J Mamm Evol. 1999;6:221–245. [Google Scholar]
- Dawson WD, Young SR, Wang Z, Lui LW, Greenbaum IF, Davis LM, et al. Mus and Peromyscus chromosome homology established by FISH with three mouse paint probes. Mamm Genome. 1999;10:730–733. doi: 10.1007/s003359901080. [DOI] [PubMed] [Google Scholar]
- DeBry RW. Identifying conflicting signal in a multigene analysis reveals a highly resolved tree: the phylogeny of Rodentia (Mammalia) Syst Biol. 2003;52:604–617. doi: 10.1080/10635150390235403. [DOI] [PubMed] [Google Scholar]
- Deuve JL, Bennett NC, Britton-Davidian J, Robinson TJ. Chromosomal phylogeny and evolution of the African mole-rats (Bathyergidae) Chromosome Res. 2008;16:57–74. doi: 10.1007/s10577-007-1200-8. [DOI] [PubMed] [Google Scholar]
- Deuve JL, Bennett NC, O'Brien PC, Ferguson-Smith MA, Faulkes CG, Britton-Davidian J, et al. Complex evolution of X and Y autosomal translocations in the giant mole-rat, Cryptomys mechowi (Bathyergidae) Chromosome Res. 2006;14:681–691. doi: 10.1007/s10577-006-1080-3. [DOI] [PubMed] [Google Scholar]
- Dobigny G, Ducroz JF, Robinson TJ, Volobouev V. Cytogenetics and cladistics. Syst Biol. 2004;53:470–484. doi: 10.1080/10635150490445698. [DOI] [PubMed] [Google Scholar]
- Engelbrecht A, Doigny G, Robinson TJ. Further insights into the ancestral murine karyotype: the contribution of the Otomys-Mus comparison using chromosome painting. Cytogenet Genome Res. 2006;112:126–130. doi: 10.1159/000087524. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Trifonov V. Mammalian karyotype evolution. Nat Rev Genet. 2007;8:950–962. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Yang F, O'Brien PC. Comparative mapping using chromosome sorting and painting. ILAR J. 1998;39:68–76. doi: 10.1093/ilar.39.2-3.68. [DOI] [PubMed] [Google Scholar]
- Fredga K. Aberrant sex chromosome mechanisms in mammals. Evolutionary aspects. Differentiation. 1983;23:23–30. doi: 10.1007/978-3-642-69150-8_4. [DOI] [PubMed] [Google Scholar]
- Genest FB, Morisset P, Patenaude RP. The chromosomes of the Canadian Beaver Castor canadensis. Can J Genet Cytol. 1979;21:37–42. doi: 10.1139/g79-006. [DOI] [PubMed] [Google Scholar]
- Genome 10K Community of Scientists A proposal to obtain whole genome sequence for 10,000 vertebrate species. The Journal of Heredity. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graphodatsky AS.1989Conserved and variable elements of mammalian chromosomesIn: Halnan CRE (eds).Cytogenetics of Animals CAB International Press: UK; 95–123. [Google Scholar]
- Graphodatsky AS, Sablina OV, Meyer MN, Malikov VG, Isakova EA, Trifonov VA, et al. Comparative cytogenetics of hamsters of the genus Calomyscus. Cytogenet Cell Genet. 2000;88:296–304. doi: 10.1159/000015513. [DOI] [PubMed] [Google Scholar]
- Graphodatsky AS, Yang F, Dobigny G, Romanenko SA, Biltueva LS, Perelman PL, et al. Tracking genome organization in rodents by Zoo-FISH. Chromosome Res. 2008;16:261–274. doi: 10.1007/s10577-007-1191-5. [DOI] [PubMed] [Google Scholar]
- Grutzner F, Himmelbauer H, Paulsen M, Ropers HH, Haaf T. Comparative mapping of mouse and rat chromosomes by fluorescence in situ hybridization. Genomics. 1999;55:306–313. doi: 10.1006/geno.1998.5658. [DOI] [PubMed] [Google Scholar]
- Guigo R, Dermitzakis ET, Agarwal P, Ponting CP, Parra G, Reymond A, et al. Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc Natl Acad Sci USA. 2003;100:1140–1145. doi: 10.1073/pnas.0337561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilly MN, Fouchet P, de Chamisso P, Schmitz A, Dutrillaux B. Comparative karyotype of rat and mouse using bidirectional chromosome painting. Chromosome Res. 1999;7:213–221. doi: 10.1023/a:1009251416856. [DOI] [PubMed] [Google Scholar]
- Hass I, Muller S, Artoni RF, Sbalqueiro IJ. Comparative chromosome maps of neotropical rodents Necromys lasiurus and Thaptomys nigrita (Cricetidae) established by ZOO-FISH. Cytogenet Genome Res. 2011;135:42–50. doi: 10.1159/000330259. [DOI] [PubMed] [Google Scholar]
- Hass I, Sbalqueiro IJ, Muller S. Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetidae) from southern Brazil established by Zoo-FISH using Mus musculus (Muridae) painting probes. Chromosome Res. 2008;16:75–88. doi: 10.1007/s10577-007-1211-5. [DOI] [PubMed] [Google Scholar]
- Helou K, Walentinsson A, Levan C, Stahl F. Between rat and mouse ZOO–FISH reveals 49 chromosomal segments that have been conserved in evolution. Mammalian Genome. 2001;12:765–771. doi: 10.1007/pl00021067. [DOI] [PubMed] [Google Scholar]
- Horn S, Durka W, Wolf R, Ermala A, Stubbe A, Stubbe M, et al. Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species. PLoS One. 2011;6:e14622. doi: 10.1371/journal.pone.0014622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchon D, Madsen O, Sibbald MJ, Ament K, Stanhope MJ, Catzeflis F, et al. Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- Jansa SA, Weksler M. Phylogeny of muroid rodents: relationships within and among major lineages as determined by IRBP gene sequences. Mol Phylogenet Evol. 2004;31:256–276. doi: 10.1016/j.ympev.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Lemskaya NA, Romanenko SA, Golenishchev FN, Rubtsova NV, Sablina OV, Serdukova NA, et al. Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). III. Karyotype relationships of ten Microtus species. Chromosome Res. 2010;18:459–471. doi: 10.1007/s10577-010-9124-0. [DOI] [PubMed] [Google Scholar]
- Li T, O'Brien PC, Biltueva L, Fu B, Wang J, Nie W, et al. Evolution of genome organizations of squirrels (Sciuridae) revealed by cross-species chromosome painting. Chromosome Res. 2004;12:317–335. doi: 10.1023/B:CHRO.0000034131.73620.48. [DOI] [PubMed] [Google Scholar]
- Li T, Wang J, Su W, Nie W, Yang F. Karyotypic evolution of the family Sciuridae: inferences from the genome organizations of ground squirrels. Cytogenet Genome Res. 2006a;112:270–276. doi: 10.1159/000089881. [DOI] [PubMed] [Google Scholar]
- Li T, Wang J, Su W, Yang F. Chromosomal mechanisms underlying the karyotype evolution of the oriental voles (Muridae, Eothenomys) Cytogenet Genome Res. 2006b;114:50–55. doi: 10.1159/000091928. [DOI] [PubMed] [Google Scholar]
- Lyapunova EA, Vorontsov NN, Korobitsina KV, Ivanitskaya EYu, Borisov YuM, Yakimenko LV, et al. A Robertsonian fan in Ellobius talpinus. Genetica (The Hague) 1980;52:239–247. [Google Scholar]
- Martin Y, Gerlach G, Schlotterer C, Meyer A. Molecular phylogeny of European muroid rodents based on complete cytochrome b sequences. Mol Phylogenet Evol. 2000;16:37–47. doi: 10.1006/mpev.1999.0760. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Nishida-Umehara C, Kuriowa A, Tsuchiya K, Matsuda Y. Identification of chromosome rearrangements between the laboratory mouse (Mus musculus) and the Indian spiny mouse (Mus platythrix) by comparative FISH analysis. Chromosome Res. 2003;11:57–64. doi: 10.1023/a:1022010116287. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Nishida-Umehara C, Tsuchiya K, Nukaya D, Matsuda Y. Karyotypic evolution of Apodemus (Muridae, Rodentia) inferred from comparative FISH analyses. Chromosome Res. 2004;12:383–395. doi: 10.1023/B:CHRO.0000034103.05528.83. [DOI] [PubMed] [Google Scholar]
- Matthey R. Chromosomes and evalution. Triangle. 1972;11:107–112. [PubMed] [Google Scholar]
- Michaux J, Reyes A, Catzeflis F. Evolutionary history of the most speciose mammals: molecular phylogeny of muroid rodents. Mol Biol Evol. 2001;18:2017–2031. doi: 10.1093/oxfordjournals.molbev.a003743. [DOI] [PubMed] [Google Scholar]
- Mlynarski EE, Obergfell CJ, Rens W, O'Brien PC, Ramsdell CM, Dewey MJ, et al. Peromyscus maniculatus-Mus musculus chromosome homology map derived from reciprocal cross species chromosome painting. Cytogenet Genome Res. 2008;121:288–292. doi: 10.1159/000138900. [DOI] [PubMed] [Google Scholar]
- Montgelard C, Forty E, Arnal V, Matthee CA. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol Biol. 2008;8:321. doi: 10.1186/1471-2148-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Kuroiwa A, Nishida-Umehara C, Matsubara K, Yamada F, Matsuda Y. Comparative chromosome painting map between two Ryukyu spiny rat species, Tokudaia osimensis and Tokudaia tokunoshimensis (Muridae, Rodentia) Chromosome Res. 2007;15:799–806. doi: 10.1007/s10577-007-1163-9. [DOI] [PubMed] [Google Scholar]
- Neumann K, Michaux J, Lebedev V, Yigit N, Colak E, Ivanova N, et al. Molecular phylogeny of the Cricetinae subfamily based on the mitochondrial cytochrome b and 12S rRNA genes and the nuclear vWF gene. Mol Phylogenet Evol. 2006;39:135–148. doi: 10.1016/j.ympev.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Helou K, Walentinsson A, Szpirer C, Nerman O, Stahl F. Rat-mouse and rat-human comparative maps based on gene homology and high-resolution zoo-FISH. Genomics. 2001;74:287–298. doi: 10.1006/geno.2001.6550. [DOI] [PubMed] [Google Scholar]
- Patton JL, Sherwood SW. Genome evolution in pocket gophers (genus Thomomys). I. Heterochromatin variation and speciation potential. Chromosoma. 1982;85:149–162. doi: 10.1007/BF00294962. [DOI] [PubMed] [Google Scholar]
- Petit D, Couturier J, Viegas-Péquignot E, Lombard M, Dutrillaux B. Great degree of homeology between the ancestral karyotype of squirrels (Rodents) and that of Primates and Carnivora. Ann Genet. 1984;27:201–212. [PubMed] [Google Scholar]
- Radjabli SI.1975The karyotypic differentiation of Palaearctic hamsters (Rodentia, Cricetidae) Reports AS USSR 225697–700.(In Russian). [PubMed] [Google Scholar]
- Rambau RV, Robinson TJ. Chromosome painting in the African four-striped mouse Rhabdomys pumilio: detection of possible murid specific contiguous segment combination. Chromosome Res. 2003;11:91–98. doi: 10.1023/a:1022887629707. [DOI] [PubMed] [Google Scholar]
- Richard F, Messaoudi C, Bonnet-Garnier A, Lombard M, Dutrillaux B. Highly conserved chromosomes in an Asian squirrel (Menetes berdmorei, Rodentia: Sciuridae) as demonstrated by ZOO-FISH with human probes. Chromosome Res. 2003;11:597–603. doi: 10.1023/a:1024905018685. [DOI] [PubMed] [Google Scholar]
- Robbins LW, Baker RJ. An assessment of the nature of chromosomal rearrangements in 18 species of Peromyscus (Rodentia: Cricetidae) Cytogenet Cell Genet. 1981;31:194–202. doi: 10.1159/000131649. [DOI] [PubMed] [Google Scholar]
- Rogers DS, Greenbaum IF, Gunn SJ, Engstrom MD. Cytosystematic value of chromosomal inversion data in the genus Peromyscus (Rodentia, Cricetidae) J Mammal. 1984;65:457–465. [Google Scholar]
- Romanenko SA, Lemskaya NA, Beklemisheva VR, Perel'man PL, Serdyukova NA, Graphodatsky AS. Comparative cytogenetics of rodents. Russian J Genet. 2010;46:1138–1142. [PubMed] [Google Scholar]
- Romanenko SA, Perelman PL, Serdukova NA, Trifonov VA, Biltueva LS, Wang J, et al. Reciprocal chromosome painting between three laboratory rodent species. Mamm Genome. 2006;17:1183–1192. doi: 10.1007/s00335-006-0081-z. [DOI] [PubMed] [Google Scholar]
- Romanenko SA, Sitnikova NA, Serdukova NA, Perelman PL, Rubtsova NV, Bakloushinskaya IY, et al. Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). II. The genome homology of two mole voles (genus Ellobius), the field vole and golden hamster revealed by comparative chromosome painting. Chromosome Res. 2007a;15:891–897. doi: 10.1007/s10577-007-1171-9. [DOI] [PubMed] [Google Scholar]
- Romanenko SA, Volobouev VT, Perelman PL, Lebedev VS, Serdukova NA, Trifonov VA, et al. Karyotype evolution and phylogenetic relationships of hamsters (Cricetidae, Muroidea, Rodentia) inferred from chromosomal painting and banding comparison. Chromosome Res. 2007b;15:283–297. doi: 10.1007/s10577-007-1124-3. [DOI] [PubMed] [Google Scholar]
- Sannier J, Gerbault-Seureau M, Dutrillaux B, Richard FA. Conserved although very different karyotypes in Gliridae and Sciuridae and their contribution to chromosomal signatures in Glires. Cytogenet Genome Res. 2011;134:51–63. doi: 10.1159/000324691. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Cremer T, Arnason U, Weier HU, Lima-de-Faria A, Fronicke L. Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat Genet. 1994;6:342–347. doi: 10.1038/ng0494-342. [DOI] [PubMed] [Google Scholar]
- Scalzi JM, Hozier JC. Comparative genome mapping: mouse and rat homologies revealed by fluorescence in situ hybridization. Genomics. 1998;47:44–51. doi: 10.1006/geno.1997.5090. [DOI] [PubMed] [Google Scholar]
- Sitnikova NA, Romanenko SA, O'Brien PC, Perelman PL, Fu B, Rubtsova NV, et al. Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). I. The genome homology of tundra vole, field vole, mouse and golden hamster revealed by comparative chromosome painting. Chromosome Res. 2007;15:447–456. doi: 10.1007/s10577-007-1137-y. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Stone G, Garcia M, Froenicke L. Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics. 2003;82:245–249. doi: 10.1016/s0888-7543(03)00109-5. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Yang F, Cavagna P, O'Brien PC, Bagga M, Ferguson-Smith MA, et al. Reciprocal chromosome painting shows that genomic rearrangement between rat and mouse proceeds ten times faster than between humans and cats. Cytogenet Cell Genet. 1999;84:150–155. doi: 10.1159/000015244. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Yang F, Morescalchi AM, Galleni L. Chromosome painting in the long-tailed field mouse provides insights into the ancestral murid karyotype. Cytogenet Genome Res. 2004;105:406–411. doi: 10.1159/000078213. [DOI] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Svartman M, Stone G, Stanyon R. Molecular cytogenetics discards polyploidy in mammals. Genomics. 2005;85:425–430. doi: 10.1016/j.ygeno.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Swier VJ, Bradley RD, Rens W, Elder FF, Baker RJ. Patterns of chromosomal evolution in Sigmodon, evidence from whole chromosome paints. Cytogenet Genome Res. 2009;125:54–66. doi: 10.1159/000218747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov VA, Kosyakova N, Romanenko SA, Stanyon R, Graphodatsky AS, Liehr T. New insights into the karyotypic evolution in muroid rodents revealed by multicolor banding applying murine probes. Chromosome Res. 2010;18:265–275. doi: 10.1007/s10577-010-9110-6. [DOI] [PubMed] [Google Scholar]
- Trifonov VA, Perelman PL, Kawada SI, Iwasa MA, Oda SI, Graphodatsky AS. Complex structure of B-chromosomes in two mammalian species: Apodemus peninsulae (Rodentia) and Nyctereutes procyonoides (Carnivora) Chromosome Res. 2002;10:109–116. doi: 10.1023/a:1014940800901. [DOI] [PubMed] [Google Scholar]
- Ventura K, O'Brien PC, Yonenaga-Yassuda Y, Ferguson-Smith MA. Chromosome homologies of the highly rearranged karyotypes of four Akodon species (Rodentia, Cricetidae) resolved by reciprocal chromosome painting: the evolution of the lowest diploid number in rodents. Chromosome Res. 2009;17:1063–1078. doi: 10.1007/s10577-009-9083-5. [DOI] [PubMed] [Google Scholar]
- Veyrunes F, Dobigny G, Yang F, O'Brien PC, Catalan J, Robinson TJ, et al. Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse. Proc Biol Sci. 2006;273:2925–2934. doi: 10.1098/rspb.2006.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas-Péquignot E, Petit D, Benazzou T, Prod'homme M, Lombard M, Hoffschir F, et al. Phylogénie chromosomique chez les Sciuridae, Gerbillidae et Muridae, et étude d'espèces appartenant à d'autres familles de Rongeurs. Mammalia. 1986;50:164–202. [Google Scholar]
- Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- Ward OG, Graphodatsky AS, Wurster-Hill DH, Eremina VR, Park JP, Yu Q. Cytogenetics of beavers: a case of speciation by monobrachial central fusions. Genome. 1991;34:324–328. [Google Scholar]
- Yang F, O'Brien PCM, Ferguson-Smith MA. Comparative chromosome map of the laboratory mouse and Chinese hamster defined by reciprocal chromosome painting. Chromosome Res. 2000;8:219–227. doi: 10.1023/a:1009200912436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.