Abstract
It is generally accepted that cancers result from the aggregation of somatic mutations. The emergence of next-generation sequencing (NGS) technologies during the past half-decade has enabled studies of cancer genomes with high sensitivity and resolution through whole-genome and whole-exome sequencing approaches, among others. This saltatory advance introduces the possibility of assembling multiple cancer genomes for analysis in a cost-effective manner. Analytical approaches are now applied to the detection of a number of somatic genome alterations, including nucleotide substitutions, insertions/deletions, copy number variations, and chromosomal rearrangements. This review provides a thorough introduction to the cancer genomics pipeline as well as a case study of these methods put into practice.
Keywords: cancer, genomics, sequencing, computation
Introduction
Over the course of the 10 years that have passed since the publication of the first human genome sequence, the landscape of cancer research has changed with remarkable speed. The completion of the human genome project marked the beginning of a new era of scientific research ― one in which the genetic determinants of human disease could be elucidated for a range of conditions based on the appearance of unique genomic alterations in groups of patients.
The fundamental hypothesis driving disease genomics is that there is a constellation of mutations that appear in affected persons, but not in unaffected persons. Based on the constantly expanding library of common genomic polymorphisms, the genomics community now has a large “normal” population against which mutations identified in diseased populations can be compared. Single nucleotide polymorphisms (SNPs) are those mutations seen in at least 1 percent of the population, while single nucleotide variants (SNVs) include all mutations, common and rare. In cancer genomics, there is one more level of comparison, since each affected person has two types of tissue: tumor tissue and normal tissue. By identifying those mutations that appear in tumor tissue but not in normal tissue, the pool of total identified somatic variants is further refined. The compendium of mutations found in strictly disease tissues can be evaluated for mechanistic impact.
Current understanding of cancer biology allows for the classification of cancer mutations into two categories: “drivers” and “passengers” [1]. Driver mutations are those that grant cells a survival advantage, while the passenger mutations are those that have been acquired at some point during clonal evolution but do not provide a substantial survival advantage. A major challenge of cancer research is to differentiate these two types of mutations. While driver mutations are best confirmed in experimental models, cancer genomics can aid in the identification of putative candidates.
As shown below, while NGS of cancers affords a powerful pipeline for the discovery of disease causing genomic variants, there are numerous difficulties that increase the complexity of research efforts. This review endeavors to present a highly practical overview of the discovery process in cancer genomics.
Reading the Genome
The assembly of a reference genome by the Human Genome Project was accomplished using capillary-based dideoxy-terminator sequencing methods termed “Sanger” sequencing [2]. In “shotgun de novo Sanger sequencing,” genomic DNA is fragmented, cloned into a plasmid vector, and then used to transform E. coli ― effectively using bacteria to amplify the DNA fragments. In the Sanger sequencing reaction, stochastically incorporated fluorescently labeled dideoxynucleotides (ddNTPs) terminate the DNA extension reaction, and the sequence is determined via electrophoretic separation of end-labeled ssDNA in a capillary-based gel [3]. In this method, 96 or 384 capillaries provide one read each per sequencing run [4].
Using overlaps in sequenced random fragments, much longer sequences can be assembled. Imagine, for example, that a sequence that reads WXY where W, X, and Y represent long stretches of DNA. If another sequence read UVW, it would be reasonable to assemble the union of these two sequences to read UVWXY. Assembly of the sequence UVWXY depends on W being long enough so that it would be very unlikely to appear randomly. Therefore, shotgun de novo Sanger sequencing requires the same sequence to appear in multiple DNA fragments. Through this laborious process, the NIH funded sequencing effort assembled a 90 percent complete working draft of the human genome more than a decade ago, which has since been carried closer to completion [2].
Next-generation, or second-generation, sequencing (NGS) encompasses a number of different methodologies that have emerged since 2005 [4,5,6,8,9]. In numerous NGS methods, fragmented genomic DNA ligated to universal adaptors are amplified into PCR colonies or “polonies.” Each polony contains many copies of the same fragment, and all of the polonies can be sequenced in parallel using arrays allowing millions of reads per array [4]. Other NGS methods do not use polonies, but instead read single DNA sequences [8]. While older NGS technologies read sequence from one end of a given segment, newer methods allow for paired-end reads. Once a sequence is read with NGS, it is aligned to the most current reference human genome (currently in its 19th iteration as hg19). This mapping provides the basis for all further analysis [9,10].
The general advantages of second-generation sequencing over Sanger sequencing are three-fold. First, since the preparations are done in vitro, bottlenecks like transformation of E. coli are avoided. Second, there is increased parallelism in second-generation methods because they are based on arrays rather than capillaries, which significantly reduces sequencing time. Third, since the polonies are all bound to the same array, they can be treated with single reagent volumes rather than multiple independent volumes, thereby dramatically cutting costs [4].
Polonies are generated from single molecules, rather than working with a population of molecules as in Sanger sequencing. Thus, in NGS, there is a digital readout of tumor mosaicism that is not captured in Sanger sequencing. This can be advantageous as the mosaicism is anticipated in tumors, but at the same time, low-frequency mosaicism is difficult to differentiate from stochastic or systematic errors. Collective NGS benefits are offset by increased error rates as compared to Sanger sequencing, as well as shorter read lengths [4]. Though each NGS read has a relatively high per-base error rate compared to Sanger sequencing, a comparable consensus genotype can be determined by reading a given base many times, i.e., deep-sequencing.
Both whole-genome and whole-exome sequencing can be carried out using Sanger or NGS. Whole-exome sequencing is a targeted strategy to capture the 1 to 2 percent of the human genome that is protein coding and contains the vast majority of disease-causing mutations. While mutations identified in non-coding regions may in fact be drivers of tumor progression, research efforts focus primarily on mutations in exons or at exon-intron boundaries because they are more easily interpreted.
For most users, the NGS sequencing process entails isolating DNA from patient samples and sending them to a core facility for library preparation and sequencing. DNA library preparation can be carried out by the submitting lab in order to reduce costs and increase control over samples. Depending on the local sequencing pipeline, the facility will return summary data from the sequencing runs along with raw reads without aligning the reads. From there, publicly available tools like bowtie, BWA, Maq, and SOAP2 are used to generate sequence alignments, and, subsequently, variants are called with programs such as SAMtools and GATK [6,7,11-14].
'Calling' Cancer Mutation
Though a researcher presented with a sequencing run summary and a list of potential variants may feel prepared to begin asking the biological questions that motivated the study, there are numerous considerations that require immediate attention. Indeed, a cursory examination of the list will likely reveal a large number of potential variants in tumors and also in blood.
Before examining the mutation data, it is pertinent to ask whether the sequencing itself was sufficiently redundant (or “deep”) to allow confident mutation identification. There are two simple metrics to evaluate depth of coverage: mean coverage and percent of bases covered at least N times. As a general guideline in exome capture, a mean coverage greater than 100 times and percent of bases covered at least 20 times greater than 90 percent are desirable for the tumor sample due to normal tissue contamination and tumor mosaicism. The purity of blood samples allows for lower required redundancy.
For patient data where both tumor and blood pass first inspection, the next task is to filter automated sequencing calls. A single tumor/blood pair can yield more than 20,000 hits, but there are numerous criteria used to derive a working subset. For every called variant, the quality score ― the –log10 probability that a variant call occurred by error as based on the individual base qualities ― provides the first threshold [15,16]. Different quality score thresholds of greater than 60 to greater than 100 may be used; the experience gained from initial sequencing efforts helps set the scoring threshold in future experiments. Non-synonymous, frameshift, splicing and insertion/deletion mutations are typically prioritized over synonymous changes because they are more easily interpretable.
Given a genomic coordinate where there is a putative difference between tumor and blood, a p-value from the Fisher exact test can be used to evaluate whether the sequencing read distributions differ significantly. The Fisher exact test, in this case, is used to identify non-random associations between the number of each type of read (variant or no-variant) and the sample (normal or tumor). The threshold for the Fisher exact test can be set using the inverse of the problem of interest. Specifically, one does not expect to see variants in the blood/normal sample that are not in the tumor. Thus, the p-value threshold can be set at a level that rejects the blood variant calls [16].
Based on the expectation that disease-causing mutations appear in a small percentage of the population, it is often advantageous to further limit analysis to novel mutations. dbSNP and the 1000 Genomes Project (and soon the NIH Exome Project) provide a large catalog of common variation across populations [17,18]. The strength of mutation’s tumor driving potential is expected to be inversely correlated with its frequency in these catalogs of common variants.
Picking the Battles
At this point, synonymous mutations, low quality variants, and variants found in both the tumor and the normal samples have been excluded, effectively eliminating large swaths of the variant pool. There may, however, be a non-negligible number of misreads from the NGS process requiring manual curation.
Using the sequence alignment files described earlier, it is possible to visualize the read alignments around a variant’s genomic location in order to eliminate false positive calls. This task may be accomplished with publicly available software like the Integrated Genome Browser or custom designed programs [19].
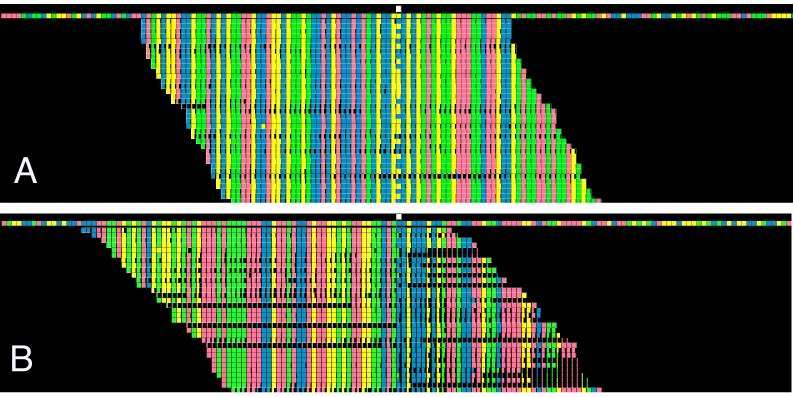
In Figure 1, the reference genome appears in the colored row at the top of the figure. The white box above the reference genome marks the location of the genomic locus of interest. Aligned reads appear in the rows below the reference genome. The colors of the boxes in each position represent the base recognized by NGS. In this example, red boxes represent adenine, blue represent cytosine, green represent guanine, and yellow represent thymine. The degree to which the box is filled in with color is proportional to the quality score of that base on a given read, meaning a nearly black box indicates a very low quality read for that specific base.
Figure 1.
Sample visualization of read alignments. In panels A and B, columns of interest are denoted by white boxes at the top of the respective diagrams. The first colored horizontal line represents the reference genome in each case. Red boxes represent adenine, blue boxes represent cytosine, green boxes represent guanine, and yellow boxes represent thymine. In (A), variant call is judged to be a correct read. In (B), variant call is judged to be a misread.
Figure 1A shows many high-quality reads of a tumor sample that contains a variant. To interpret this figure, focus on the locus of interest in the reference genome as highlighted by the white box at the top of the image. The reference genome shows a yellow box at this locus, indicating that the expected base is thymine. In numerous reads below the reference genome, the same column has filled-in blue boxes representing cytosine. It can be concluded from these data that there is a thymine to cytosine variation in the genome at this locus. The tumor variant does not necessarily need to be represented in the majority of reads, as the tumor samples are inherently mosaic and frequently contaminated with normal tissue.
Conversely, Figure 1B, which shows a blue box at the locus of interest, has a number of reads with partially filled-in green boxes representing guanine. The incomplete filling of the green boxes indicates that these are low quality reads. In addition, there are many instances where the boxes adjacent to the called variants are mostly black. This pattern is associated with low base quality (incorrect base calls) and incorrect alignment. Together, these data suggest that the variant shown in Figure 1B is a false positive.
Visual analysis of read alignment further trims the list of potentially significant mutations but simultaneously highlights the relatively high per-read error rate of NGS. For this reason, mutations called by NGS need to be confirmed via targeted Sanger sequencing or another validation method. Targeted Sanger sequencing requires forward and reverse polymerase chain reaction (PCR) amplification of the region of interest. Typically, greater than 100 base separation between the locus of interest and the end of the primer is recommended, with a maximum read length of approximately 1,000 bases and an ideal length of approximately 600 to 700 nucleotides [4]. Those familiar with working with human samples will be aware of some of the complexities of using PCR with human DNA. Specifically, repeat regions and common variations found in the human genome can cause the PCR amplification to fail. There are many publicly available tools designed to help avoid these pitfalls, including SNPmasker [20] and Primer3 [21]. After a successful PCR confirmed by DNA gel electrophoresis of both blood/normal and tumor DNA samples, the products are sent to a core facility for sequencing. Since it may not be clear a priori which mutations will ultimately be of interest, it is reasonable to attempt to confirm as many calls with targeted Sanger sequencing as reasonably possible.
Mutations to Mechanism
The great intellectual challenge in cancer genomics lies in relating confirmed mutations to protein function. In a best-case scenario, the disease cohort will have multiple patients with mutations in the same gene. This scenario may be considered low-hanging fruit for follow-up analysis. It is worth considering, however, the probability that n mutations in a single gene will appear in a cohort of X samples at random. Existing datasets or statistical estimates can be used to obtain an estimate of how often a gene is mutated in the general population. Logically, two mutations in a single gene in a cohort of eight patients is more striking than two mutations in a cohort of 20 patients, and the Fisher exact test, among other statistical tools, will yield a more precise estimate of significance of a finding. Another method to assess significance is to simulate numerous draws of X patients from an existing dataset of non-diseased samples and count the number of times n mutations in this single gene occurs, creating a probability distribution by Monte Carlo simulation [22].
In silico, there are a number of methods by which the import of a given confirmed mutation is estimated. It is important to understand that none of these methods are in and of themselves sufficient evidence, but all can contribute to the development of a hypothesis. PolyPhen2 uses sequence and structural features along with a classification algorithm to present the probability a given mutation will be deleterious [23]. Sorting Intolerant from Tolerant (SIFT) uses sequence homology to predict effect of amino acid substitution on protein function [24]. Conservation, a useful metric for evaluating the importance of a residue, provides a historical record of amino acid variability. The functional significance of a residue is hypothesized to be proportional to its degree of conservation [25]. Conservation at a given locus does not require software-based evaluation. The UCSC genome browser, among other tools, shows conservation across species and can be used to visualize wider regions [26]. In addition to conservation, the function of multiple protein domains has been elucidated. UniProtKB/Swiss-Prot is an excellent resource for identifying protein domains [27].
It is often of interest to study proteins in a network or pathway context. The Kyoto Encyclopedia of Genes and Genomes (KEGG) provides a searchable collection of manually collated pathways [28], while Gene Ontology (GO) annotation is a resource for defining gene product properties [29]. GO annotation includes cellular context, molecular function, and the essential biological processes of a protein. Other pieces of publicly available software use literature scans and available datasets to determine protein-protein interactions. Two such examples are STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) and GeneMANIA [30,31]. While very useful, caution is required when using these resources as multiple datatypes and datasets have been integrated to create the shown interaction networks. Ultimately, evidence of interaction needs to be confirmed within the cell or tissue type of interest.
Power of Cancer Genomics
As is now readily apparent, hypothesis generation in cancer genomics involves a moderately difficult experimental process coupled with a great deal of informatics work. Skills in a scripting language such as Perl or Python prove invaluable in processing the text-based data in an efficient manner and, while not obvious, there are also non-trivial computational considerations. Chief among these is the very large storage requirement for genomics data. Even with these factors, cancer genomics has enabled new avenues of promising research and will undoubtedly continue to do so in the future.
A recent analysis of multiple myeloma, a B-lymphoid malignancy, provides an effective case study for the concepts presented in this review [32]. In this study, NGS was used to sequence the whole-genome of 23 patients and the whole-exomes of 16 patients (with one patient overlap). Previous studies of multiple myeloma have identified activation of the MYC, FDFR3, KRAS, and NRAS genes as well as of the NF-κB pathway, and it was hypothesized that sequencing would reveal biologically relevant patterns otherwise unobserved.
After assignment of a statistical threshold based on background mutation rates and a false discovery rate of ≤ 0.10, 10 genes including KRAS (10 patients) and NRAS (9 patients) showed significant rates of non-silent mutations. Six of these genes were novel associations in cancer. As discussed previously, there are numerous methods by which the import of mutations can be assessed, including computational techniques, regional conservation, functional domains, and, perhaps most importantly, frequency of mutation in the study cohort. The authors observed four mutations in the DIS3 gene, all of which appear in a highly conserved region that, based on crystal structures, face a catalytic pocket. From an investigator’s perspective, these observations are highly suggestive of functional significance. In addition, five patients showed mutations in the uncharacterized FAM46 gene.
Gene Set Enrichment Analysis (GSEA) is a simple, but powerful tool that identifies coordinated changes in specified groups of genes [33]. The authors used GSEA to show a correlation between FAM46 expression and the set of ribosomal proteins. Given prior knowledge that DIS3 is involved in the regulation of RNA levels and the correlation between FAM46 and regulators of translation, the authors searched their pool of mutations that did not pass significance testing and found five other genes related to protein translation and stability. At final count, 16 of the 48 patients had mutations affecting translation and homeostasis. GSEA was also used to link multiple singly occurring mutations to the NF-κB pathway, as well as to histone modifying enzymes.
Conclusions
Research in cancer genomics is entering a period of great promise, but also of great expectation. Ongoing efforts to catalog mutations found in cancer are now being coupled with a hunt for new therapeutic targets. While this search may reveal numerous, low frequency driver mutations, new in silico tools enable the consolidation of variants into specific pathways. An increasing focus on these pathways will require the application of more nuanced algorithms better able to capture the network and evolutionary dynamics of tumor cells.
At the same time, ongoing sequencing efforts will continue to generate massive quantities of data. A major challenge in cancer genomics is the standardization, storage, and public availability of these data. While large consortia helped forge the field of cancer genomics, saltatory technological developments have opened the door to sequencing for smaller research groups. With this development, more teams are now pursuing parallel research goals, stressing the need for continued collaboration and communication. Similarly, as methods for data analysis increase in sophistication and complexity, there must be a focus on accessibility enabling unhindered information flow between computational and biological scientists.
As shown in the case of multiple myeloma, NGS enables a powerful discovery pipeline. Access to this pipeline, however, is governed by an understanding of the core methods and limitations in cancer sequencing. As sequencing costs continue to decline, there will be an expanded effort to sequence matched tumor-normal DNA, but this growth must be accompanied by increased fluency in the terminology, techniques, and challenges of cancer genomics. Soon, data management and computation will replace access to sequencing as the major bottleneck in the discovery pipeline.
Acknowledgments
I thank Marcus Bosenberg, Murim Choi, and Colin Tominey for productive conversations about the current trajectory of cancer genomics and comments on the manuscript.
Glossary
- NGS
next-generation sequencing
- SNP
single nucleotide polymorphism
- SNV
single nucleotide variant
- PCR
polymerase chain reaction
- SIFT
Sorting Intolerant from Tolerant
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene Ontology
- GSEA
Gene Set Enhancement Analysis
References
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Nat Acad Sci USA. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- DePristo M, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G. et al. Real-Time DNA Sequencing from Single Polymerase Moleculers. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Ann Rev Genomics Hum Genet. 2009;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Guo J, Yu L, Turro NJ, Ju J. An integrated system for DNA sequencing by synthesis using novel nucleotide analogues. Acc Chem Res. 2010;43:551–563. doi: 10.1021/ar900255c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam T, Yiu S, Kristiansen K. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-Calling of Automated Sequencer Traces Using Phred-I Accuracy Assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Yue P, Björklund P, Zhao B, Nelson-Williams C. et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331(6018):768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Altshuler DL, Abecasis GR, Bentley DR, Chakravarti A, Clark AG. et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helpt GA, Blanchard SG, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;1025(20):2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreson R, Puurand T, Remm M. SNPmasker: automatic masking of SNPs and repeats across eukaryotic genomes. Nucleic Acid Res. 2006;34:651–655. doi: 10.1093/nar/gkl125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editors. Totowa, NJ: Humana Press; 2000. Primer3 on the WWW for General Users and for Biologist Programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Galante P, Parmigiani RB, Zhao Q, Caballero OL, de Souza JE, Navarro FCP. et al. Distinct patterns of somatic alterations in a lymphoblastoid and a tumor genome derived from the same individual. Nucleic Acid Res. 2011;39(14):6056–6068. doi: 10.1093/nar/gkr221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P. et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4(8):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Valdar WSJ. Scoring residue conservation. Structure, Function, and Genetics. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT. et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31(1):51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S. et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC. et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette M. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]