Letter to the Editor:
Although front-line chemotherapy regimens containing cytarabine induce complete remission in 65–80% of patients with newly-diagnosed AML, persistence of disease in a substantial proportion of patients leads to disease recurrence. A recent report showed positive correlation between expression of FKBP5, a 51KD protein with peptidyl-propyl-isomerase activity and cellular sensitivity to cytarabine (1). Genetic variants in FKBP5 have been studied extensively for association with depression, and mood disorders Although role of FKBP5 in steroid signaling through HSP90 has been well established (2), recent reports have shown an additional function as a negative regulator of the AKT signaling (3–5). FKBP5 promotes dephosphorylation of AKT-Ser473 by acting as a scaffolding protein for AKT and PH-domain-leucine-rich-repeat-protein-phosphatase (PHLPP). This decreased AKT phosphorylation can contribute to increased cytotoxicity by enhancing apoptosis in response to chemotherapeutic agents such as cytarabine (3–5). Thus, inter-patient variation in FKBP5 expression/activity due to genetic polymorphisms could influence response to chemotherapy, hence in this study we determined the clinical significance of FKBP5 SNPs in pediatric AML patients treated with cytarabine containing chemotherapy (6).
Patient population included 232 subjects (<22 years) treated on St. Jude AML02 clinical trial (clinicaltrials.gov: NCT00136084). Details of the study design and clinical outcome are described elsewhere (6). At diagnosis, patients were provisionally classified as having low-risk AML if their leukaemic cells had t(8;21)/AML1-ETO, inv(16)/CBFβ-MHY11, or t(9;11)/MLL-AF9. High-risk cases included those with −7,FLT3-ITD, t(6;9), megakaryoblastic leukaemia, treatment-related AML, or AML arising from myelodysplastic syndrome. All other patients were provisionally classified as having standard-risk AML. Patients were randomized to receive first Induction therapy containing high or low dose cytarabine plus daunorubicin and etoposide as described previously (6). Subsequent therapy was adapted according to presenting features and MRD as assessed by flow cytometry. The in vitro cytarabine sensitivity of leukemic cells was determined in 76 patients by MTT cytotoxicity assays. Briefly, bone marrow was obtained at diagnosis, and mononuclear cells were isolated using Ficoll-Hypaque density-gradient centrifugation and resuspended in modified RPMI-1640 medium as previously described (7). If necessary, samples were further enriched to achieve more than 80 percent blasts by the use of magnetic cell sorting (Miltenyi Biotec). The cells were treated with varying concentrations of cytarabine (range 0.002 to 2.5 ng/µL) to determine the LC50 value.
We genotyped 25 SNPs within FKBP5 (selected from the literature and HAPMAP database) in DNA from 187 AML samples (that were available). Outcome measures such as EFS, OS, relapse and treatment related death were not significantly different between genotyped vs. non-genotyped group (P>0.1 for all). Four of 25 FKBP5 SNPs were not present in the AML patient cohort. Linkage analysis using Haploview-software identified two linkage disequilibrium (LD) blocks: LD block-1 with 5 SNPs (rs9296158, rs9368878, rs1360780, rs737054 and rs4713902) and LD block-2 (6 SNPs (rs6912833, rs9394309, rs9380525, rs9470080, rs7763535 and rs4713916). The remaining 10 SNPs were not in LD with any other SNP. All the SNPs except rs1043895 were in Hardy Weinberg equilibrium.
The association of the number of minor alleles with in vitro ara-C LC50 and day 22 MRD levels was measured by rank-order correlation and p values were determined by 10,000 permutations. The analysis of day 22 MRD was stratified by treatment arm. EFS and OS were defined as described previously. The Kaplan-Meier method was used to estimate EFS and OS. Cox proportional hazard regression models were used to explore the association of genotype with EFS and OS univariately and while adjusting for other variables previously selected by statistical criteria and clinical knowledge. Fisher’s exact test was used to compare genotypes across clinically defined groups and self-reported race groups. The method of Benjamini and Hochberg was used to compute FDR-adjusted p-values (reported with the letter q) for the full-cohort univariate analyses screening association of SNPs with EFS, OS, LC50, and MRD (8).
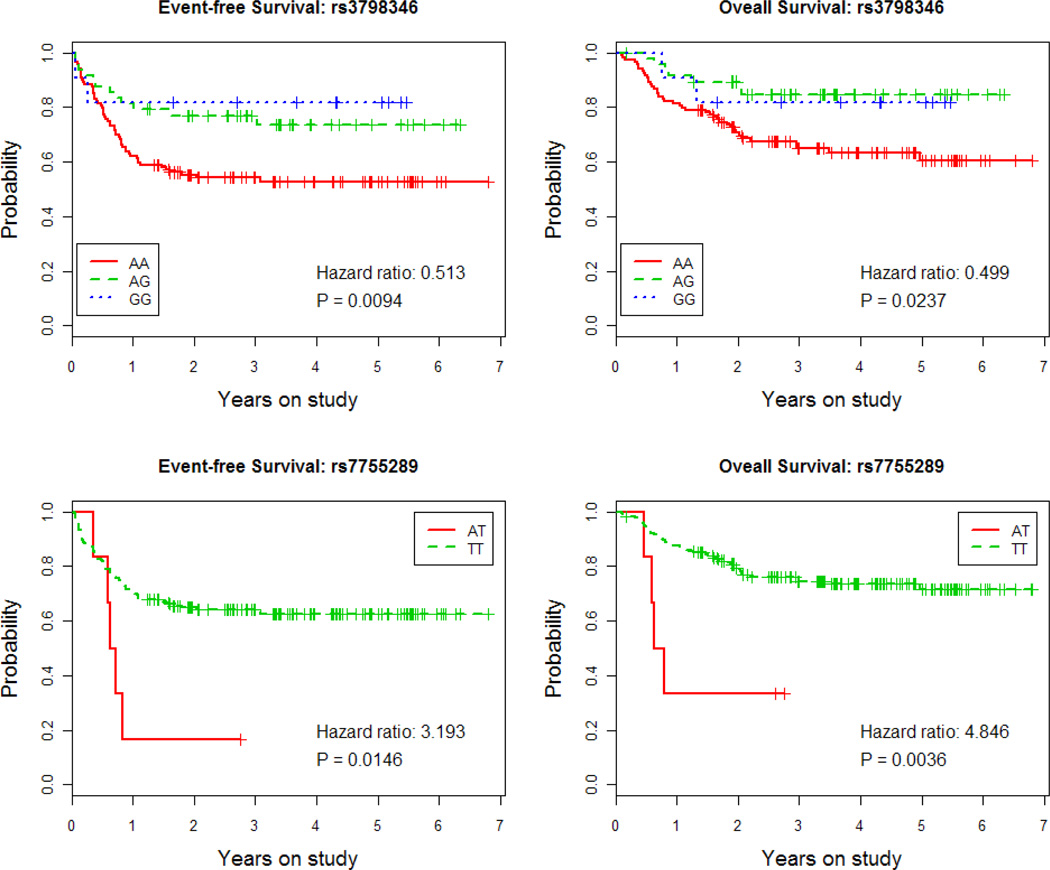
FKBP5 SNPs were screened for association with EFS (n=187), OS (n=187), response to induction I as characterized by day 22 MRD-levels, (n=176) and in vitro diagnostic blast ara-C LC50 (n=68) the significant associations are summarized in Table.1. The most interesting FKBP5 polymorphism was intronic SNP rs3798346 (A>G). The variant G allele was associated with an improvements in EFS (P = 0.0094, hazard ratio=0.513, 95% CI=0.31–0.85, Figure 1A) and OS (P = 0.037, hazard ratio=0.499, 95% CI=0.27–0.91, Figure 1B) in univariate analysis. Similar results were obtained in the multivariable analyses that accounted for all factors selected on the basis of statistical criteria and the literature in our previous report (6) (Table 2). The variant allele was significantly associated with reduction in the cumulative incidence of relapse (P = 0.0097, hazard ratio = 0.37, 95% CI = 0.18 – 0.79) but did not show a significant association with treatment-related mortality (P = 0.6, hazard ratio = 0.7, 95% CI = 0.18–2.77). The variant allele showed a non-significant association with in vitro ara-C cytotoxicity, day 22 MRD levels, WBC counts, provisional risk and FLT3 status (P>0.1 each). The genotype frequencies showed some evidence of departure from Hardy-Weinberg equilibrium (P = 0.06) because the variant G allele is significantly more prevalent among whites than non-whites with the allele frequency of 0.24 vs. 0.073, respectively (P = 0.0003). Still, the observed associations retained significance in analyses restricted only to whites. The G allele was associated with better EFS and OS in univariate analyses (P = 0.0173 and 0.035, respectively, data not shown) and in multivariable analyses (P = 0.023 and 0.005, respectively, data not shown).
Table 1.
Association of FKBP5 SNPs with EFS, OS, day 22 MRD and in vitro ara-C cytotoxicity to ara-C in pediatric AML patients.
| FKBP5 SNPs |
Allele frequency in AML cohort |
P value | SNP Location |
|---|---|---|---|
| Event Free Survival (N=187) | |||
| rs7755289 | 0.019 | 0.0146 | Intronic |
| rs3798346 | 0.197 | 0.0094 | Intronic |
| Overall Survival (N=187) | |||
| rs7755289 | 0.019 | 0.0036 | intronic |
| rs3798346 | 0.197 | 0.0237 | intronic |
| Day 22 MRD (N=176) | |||
| rs7755289 | 0.014 | 0.018 | intronic |
| rs16878591 | 0.032 | 0.011 | intronic |
| rs34866878 | 0.052 | 0.045 | Non synonymous |
| ara-LC50 (N=69) | |||
| (LD block-2) rs7763535 | 0.24 | 0.03 | Promoter and intronic SNPs |
Figure 1.
Association of FKBP5 SNPs with clinical response in AML patients treated with ara-C containing chemotherapeutic regimens. Kaplan Meier survival curves for association of rs3739346 and rs7755289 SNPs with EFS and OS in AML patients.
Table.2.
Association of FKBP5 SNPs rs3798346 and rs7755289 with EFS and OS in univariate and multivariable proportional hazards regression analyses.
| Event-Free Survival | Overall Survival | ||||
|---|---|---|---|---|---|
| Analysis | Parameter | Hazard Ratio (95% CI) |
P value | Hazard Ratio (95% CI) |
P value |
| Univariate | rs3798346 | 0.513 (0.31–0.849) | 0.0094 (q= 0.13) |
0.499 (0.273–0.911) | 0.0237 (q= 0.16) |
| rs7755289 | 3.193 (1.258–8.104) | 0.0146 (q=0.14) |
4.846 (1.677–14.006) | 0.0036 (q = 0.13) |
|
| Multivariable 1 | rs3798346 | 0.5 (0.27–0.94) | 0.031 | 0.41 (0.2–0.87) | 0.021 |
| High-Dose Arm | 1.27 (0.73–2.22) | 0.399 | 1.71 (0.88–3.3) | 0.111 | |
| Day 22 MRD >=1% | 3.01 (1.66–5.46) | 0 | 3.51 (1.73–7.13) | 0.001 | |
| Core-binding factor | 0.21 (0.07–0.61) | 0.004 | 0.2 (0.04–0.89) | 0.034 | |
| Age at diagnosis >=10 | 1.48 (0.83–2.63) | 0.187 | 1.83 (0.93–3.6) | 0.078 | |
| 11q23 other than t(9;11) | 1.78 (0.82–3.86) | 0.144 | 3.3 (1.37–7.95) | 0.008 | |
| M7 without t(1;22) | 2.87 (1.07–7.69) | 0.037 | 6.85 (2.32–20.28) | 0.001 | |
| FLT3-ITD | 1.5 (0.74–3.02) | 0.259 | 1.76 (0.77–3.98) | 0.179 | |
| Multivariable 2 | rs7755289 | 1.9 (0.63–5.75) | 0.256 | 2.76 (0.8–9.47) | 0.107 |
| High-Dose Arm | 1.29 (0.75–2.21) | 0.363 | 1.51 (0.79–2.9) | 0.216 | |
| Day 22 MRD >=1% | 3.04 (1.65–5.59) | 0 | 2.52 (1.25–5.08) | 0.01 | |
| Core-binding factor | 0.23 (0.08–0.68) | 0.008 | 0.21 (0.05–0.93) | 0.039 | |
| Age at diagnosis >=10 | 1.16 (0.65–2.07) | 0.608 | 1.7 (0.85–3.41) | 0.133 | |
| 11q23 other than t(9;11) | 1.49 (0.68–3.28) | 0.319 | 2.19 (0.87–5.5) | 0.095 | |
| M7 without t(1;22) | 2.57 (0.89–7.4) | 0.081 | 6.63 (2.06–21.3) | 0.001 | |
| FLT3-ITD | 1.72 (0.85–3.49) | 0.131 | 2.27 (0.98–5.27) | 0.056 | |
We also observed that the variant A allele of FKBP5 SNP rs7755289 (T>A;intron 8) was significantly associated with worse EFS (P = 0.014, hazard ratio=3.193, 95% CI=1.258–8.104, Figure 1C) and OS (P = 0.0036, hazard ratio=4.846, 95% CI=1.68–14, Figure 1D). Additionally, the A allele was associated with increased day 22 MRD (P = 0.017), increased cumulative incidence of relapse (p = 0.045, hazard ratio = 3.4, 95% CI = 1.03 – 11.22) and an increased cumulative incidence of treatment-related mortality (P = 0.012, hazard ratio = 5.57, 95% CI = 1.44 – 21.47). However as this SNP occurred with the allele frequency of only ~0.2, the low sample size restricted us from performing further analysis. Although the above mentioned SNPs were the most interesting SNPs we also observed association of SNP rs16878591 (P=0.011) with day 22 MRD levels and SNPs within LD block 2 with in vitro ara-C LC50 values (P=0.03)(Table 1).
In previous reports FKBP5 expression has been shown to positively influence response to cytarabine and gemcitabine. More recently FKBP5 has been identified as scaffolding protein that facilitates PHLPP-mediated dephosphorylation of AKT-Ser473, thus indicating that higher expression of FKBP5 might contribute to enhanced chemosensitivity. (3–5)siRNA-mediated FKBP5 knock down increases resistance to cytarabine and other agents as etoposide, paclitaxel and doxetaxel(1, 3–5). Thus, FKBP5 SNPs may also be associated with response to other agents used in combination with cytarabine in AML patients. In conclusion, our preliminary results suggest that the FKBP5 polymorphisms mentioned above may also be relevant for AML treatment response. These results should be confirmed with functional studies and independent clinical studies. Identification of pharmacogenetic markers of response such as FKBP5 SNP such as rs3798346 might help in further understanding inter-patient variation in response to chemotherapy.
Acknowledgements
We acknowledge support from NIH R01CA132946 (LAMBA) and Cancer Center Support (CORE) P30 CA021765 grants from the National Institutes of Health, and by the American Lebanese Syrian Associated Charities (ALSAC). Help from Biomedical genomics center (BMGC), University of Minnesota in performing genotyping is highly appreciated.
Footnotes
Conflict of interest: Authors have no conflict of interest to declare.
References
- 1.Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68(17):7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinwal UK, Koren J, 3rd, Borysov SI, Schmid AB, Abisambra JF, Blair LJ, et al. The Hsp90 cochaperone, FKBP51, increases Tau stability and polymerizes microtubules. J Neurosci. 2010;30(2):591–599. doi: 10.1523/JNEUROSCI.4815-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, Lou Z, Wang L. The role of FKBP5 in cancer aetiology and chemoresistance. Br J Cancer. 2011;104(1):19–23. doi: 10.1038/sj.bjc.6606014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16(3):259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei H, Lou Z, Wang L. Emerging role of FKBP51 in AKT kinase/protein kinase B signaling. Cell Cycle. 2010;9(1):6–7. doi: 10.4161/cc.9.1.10290. [DOI] [PubMed] [Google Scholar]
- 6.Rubnitz JE, Inaba H, Dahl G, Ribeiro RC, Bowman WP, Taub J, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11(6):543–552. doi: 10.1016/S1470-2045(10)70090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamba JK, Crews K, Pounds SB, Cao X, Gandhi V, Plunkett W, et al. Identification of predictive markers of cytarabine response in acute myeloid leukemia by integrative analysis of gene-expression profiles with multiple phenotypes. Pharmacogenomics. doi: 10.2217/pgs.10.191. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini YaH Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;Series B:57. [Google Scholar]