Abstract
Sin resolvase is a site-specific serine recombinase that is normally controlled by a complex regulatory mechanism. A single mutation, Q115R, allows the enzyme to bypass the entire regulatory apparatus, such that no accessory proteins or DNA sites are required. Here we present a 1.86Å crystal structure of the Sin Q115R catalytic domain, in a tetrameric arrangement stabilized by an interaction between Arg115 residues on neighboring subunits. The subunits have undergone significant conformational changes from the inactive dimeric state previously reported. The structure provides a new high-resolution view of a serine recombinase active site that is apparently fully assembled, suggesting roles for the conserved active site residues. The structure also suggests how the dimer-tetramer transition is coupled to assembly of the active site. The tetramer is captured in a different rotational substate than that seen in previous hyperactive serine recombinase structures, and unbroken crossover site DNA can be readily modeled into its active sites.
Keywords: serine recombinase, resolvase, site-specific recombination, DNA binding protein, crystal structure
Introduction
Site-specific recombinases are meticulous enzymes that bind specific nucleotide sequences and are able to break, rearrange and rejoin DNA segments in a highly controlled manner.(Grindley et al., 2006) These recombinases do not require ATP or other high-energy cofactors because the energy of the broken phosphodiester bonds is stored in covalent protein – DNA intermediates, and because the reactants and products are chemically isoenergetic. However, precise regulation is needed to prevent potentially deleterious outcomes. Some methods of regulation utilized by these recombinases involve accessory proteins and binding sites as well as cues from the topologies of their substrates.(Stark and Boocock, 1995) Two distinct families of site-specific recombinase exist, the serine recombinases and the tyrosine recombinases, which are so named after the active site nucleophile responsible for the initial DNA cleavage event. Sin, the focus of this study, is a serine recombinase that was first identified on the Staphylococcus aureus multiresistance plasmid pI9789, but has since been found on other mobile elements.(Derbise et al., 1995; Highlander et al., 2007) Its hypothesized role in vivo is to resolve plasmid dimers, which may arise during replication, into monomers.(Paulsen et al., 1994; Rowland and Dyke, 1989)
Serine recombinases generally bind their individual crossover sites as dimers, but the strand exchange reaction itself occurs within a tetramer (Figure 1). Double-strand breaks are introduced by the attack of each protomer’s active site serine on a scissile phosphate group, leading to 2-nucleotide 3′ overhangs. Two subunits are then hypothesized to rotate 180° relative to the other two, aligning the cleaved DNA ends with new partners.(Dhar et al., 2009a; Dhar et al., 2009b; Grindley et al., 2006; Li et al., 2005) The religation reaction is chemically the reverse of the cleavage reaction, with the 3′ hydroxyl groups attacking the phosphoserine linkages. Many lines of evidence imply that regulation is achieved by controlling the dimer – tetramer transition,(Grindley et al., 2006; Smith et al., 2010) and recent data for Sin indicate that even the initial DNA cleavage reaction requires tetramerization.(Mouw et al., 2010) However, more structural data have been needed to better understand the conformational changes that take place during the dimer-to-tetramer transition, and how they stimulate catalytic activity.
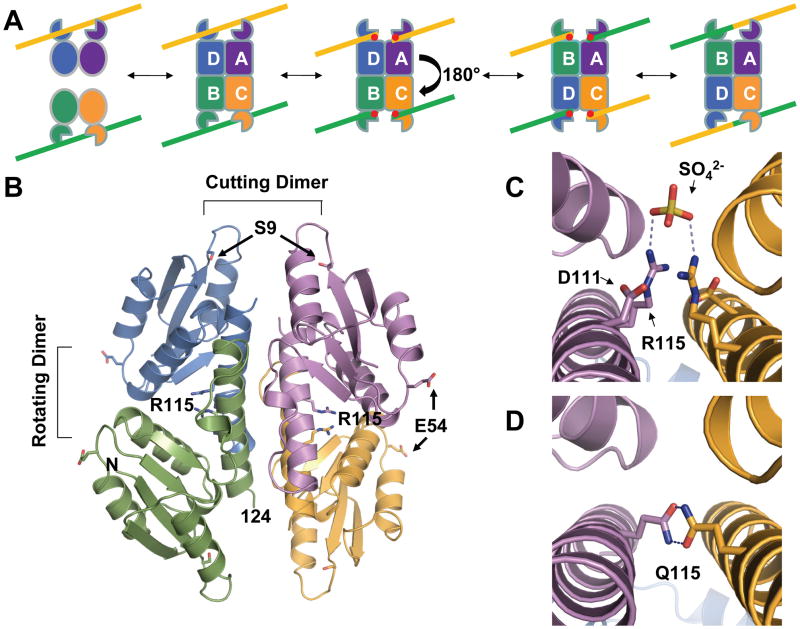
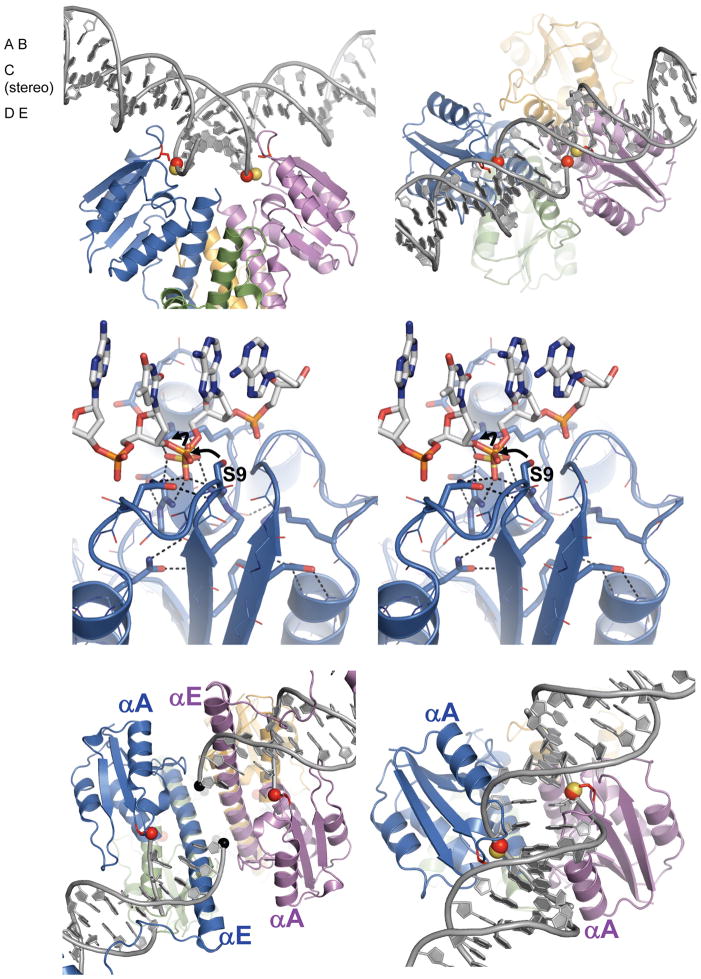
Figure 1. Structure of a Sin tetramer.
A. Cartoon showing the reaction pathway. Two inactive site I – bound dimers come together and form a catalytically active tetrameric species. DNA cleavage resulting in phosphoserine intermediates (red dots), is followed by a 180° rotation of one rotating dimer relative to the other, then religation, yielding the recombinant products. Formation of the tetrameric species can be triggered by accessory proteins bound to regulatory sites on the DNA, very high local concentration, or mutations in the protein. Here, an activating mutation, Q115R, was used. DNA cleavage, strand exchange, and rejoining occur within the tetramer.
B. Structure of an activated tetramer of the Sin Q115R catalytic domain. One tetramer of the three in the asymmetric unit is shown. Individual subunits are shown in purple, green, orange, and blue; Arg115 and E54 as matching sticks, and the Ser9 nucleophiles as red sticks. The N-terminus and residue 124, after which helix E becomes disordered, are labeled on the green subunit. (See also figure S1).
C. Closeup of the interactions between subunits of one rotating dimer (colored as in part B). Asp111 and Arg115 from each subunit and the sulfate ions bound by the arginines, are shown as sticks, as well as the sulfate ion bound by the two arginines. Putative hydrogen bonding interactions with the sulfate are shown as dashed lines. D. Model of inter-subunit interactions in the wild-type protein, which was made by substituting Arg115 with glutamine residues.
What triggers formation of the catalytically active tetramer? For all well-studied serine recombinases, formation of an appropriate protein-DNA synaptic complex is critical. The details of this complex vary with the system. For Sin, each full recombination site, termed res, consists of a crossover site and two regulatory sites. One dimer of Sin binds the crossover site, site I, another dimer of Sin binds the regulatory site, site II, and a non-specific DNA bending protein, HU, binds a second regulatory site.(Rowland et al., 2002) The two res sites are synapsed to create an elaborate synaptosome that traps three negative nodes and aligns the two crossover sites in parallel fashion.(Rowland et al., 2002) For WT Sin, assembly of the full synaptosome is required to trigger conversion of the two site I - bound dimers into a catalytically active tetramer. However, activated mutants of Sin (and other serine recombinases) have been identified that are active in the absence of the regulatory apparatus, and can recombine two separate linear site I’s.(Arnold et al., 1999; Burke et al., 2004; Haffter and Bickle, 1988; Klippel et al., 1988; Olorunniji et al., 2008; Rowland et al., 2009; Sarkis et al., 2001)
The only serine recombinase for which crystal structures of both dimeric and tetrameric forms are available is γδ resolvase. γδ is 31% identical to Sin and shares a similar domain structure, with an N-terminal catalytic domain followed by a semi-flexible linker and a C-terminal helix-turn-helix DNA binding domain. Structures of WT γδ resolvase in the apo- and site-I bound forms are dimeric.(Rice and Steitz, 1994a; Yang and Steitz, 1995) Both structures represent a catalytically inactive conformation: some essential residues (e.g. Arg71) point away from the catalytic serines, and in the DNA complex structure those serines are > 13 Å from the scissile phosphates. More recent structures of activated mutants depict the protein in a tetrameric state, and reveal that activation entails large conformational changes.(Kamtekar et al., 2006; Li et al., 2005) The protein-protein interface of the inactive dimer is remodeled, and within each catalytic domain the last helix (helix E) shifts dramatically. This repacking leads to a large, flat central interface with helix E at its center, about which subunit rotation could occur (as cartooned in Figure 1). Three of these activated γδ resolvase mutant structures include site I DNA and provide independent snapshots of the covalent intermediate resulting from DNA cleavage. In these, the two halves of site I have moved over 10Å apart, placing the free 3′ hydroxyl ‘leaving group’ over 13 Å from the phosphoserine linkage: the mechanism for religation thus remains unclear. The resolution of these structures (3.4–3.9Å) renders the detailed placement of side chains uncertain. (Li et al., 2005) However, in maps calculated from the deposited data, two of the highly conserved arginines (8 and 68) were well-ordered and engaged with the phosphoserine, but there was little to no electron density for the side chains of 2 others (45 and 71), implying that the active site was no longer fully assembled. The structure of the isolated catalytic domain of an activated mutant was reported at much higher resolution, but the active site residue Arg68 has been mutated to a histidine, the loop containing the serine nucleophile is poorly ordered, and in two of the subunits the packing of helix E relative to the core is closer to that seen in the inactive dimer than in the DNA-bound tetramer.(Kamtekar et al., 2006) Finally, the WT catalytic domain of a more distantly related serine integrase crystallized as a tetramer even though it was dimeric in solution.(Yuan et al., 2008) In this structure, the internal packing of each subunit also resembles that seen in the inactive dimer. In all five of these tetrameric structures, the relative orientation of the 4 central E-helices is approximately the same, implying that they have all trapped the same rotational substate.
Most recently, a crystal structure of dimeric WT Sin bound to a regulatory site (site II) has transformed our understanding of the architecture of the regulatory apparatus.(Mouw et al., 2008) A plausible structural model of the full synaptosome was constructed entirely from experimentally determined crystal structures, and is consistent with extensive genetic and biochemical data (Rowland et al., 2009; Rowland et al., 2005). This model implies that the synaptosome biases the conformational equilibrium at site I toward the tetramer at least in part by mass action: it brings the two site I’s into close proximity and also aligns them properly. The model also accommodates the evidence for direct contacts between site II- and site I-bound subunits. However, this model used a γδ resolvase tetramer at site I as a substitute for the unknown structure of an activated Sin tetramer.
While the structural studies outlined above brightened our understanding of these complex recombination systems, some areas of mystery remained. For instance, how is the oligomeric state coordinated with catalytic activity at site I? The dimer-to-tetramer change could clearly facilitate subunit rotation after DNA cleavage, but how does it initially activate the catalytic sites? As discussed above, no current structure has offered a glimpse of a fully assembled active site, or of the tetramer that catalyzes strand cleavage. It has also been unclear to what extent the lessons learned from the activated γδ resolvase structures could be applied to other serine recombinases. To address these questions at as high a resolution as possible, we determined the structure of the catalytic domain of an activated Sin variant. The mutation used, Q115R, has the strongest activating effect of any single mutation yet studied, and allows complete bypass of the regulatory mechanism of the synaptosome (Rowland et al., 2009). This variant tetramerizes readily even in the absence of DNA (at high nM concentrations), and efficiently recombines isolated site Is (Rowland et al., 2009). The resulting structure provides a new basis for modeling the cleavage-ready synaptic tetramer, and provides a snapshot of a serine recombinase tetramer in a different rotational substate from that seen previously, lending strong support to the subunit rotation hypothesis.
Results and Discussion
Structure Determination
The catalytic domain of Sin R54E/Q115R, residues 1–128, was purified and crystallized as described in the methods section. Since Q115R greatly reduces the protein’s solubility relative to WT, the R54E mutation was included to reduce aggregation: Arg54 is a surface residue that is important for site I – site II interactions in the full synaptosome, but is irrelevant in the context of recombination between isolated site Is (Figure 1B) (Rowland et al., 2009) The structure was solved through SAD phasing using selenomethionine-containing crystals. The crystals contained 12 protomers in the asymmetric unit, assembled into three nearly-identical tetramers (Figure S1). These tetramers are related to one another by strong pseudotranslational symmetry, as described under “methods”. The final model was refined against native data to 1.86 Å to an Rworking of 20.7% and an Rfree of 24.7%. Diffraction and refinement statistics can be found in Table 1.
Table 1.
Crystallographic statistics.
Selected statistics from data collection, phasing and refinement are shown. Values in parentheses are data from highest resolution shell. Figure of merit values were calculated prior to density modifications.
| Data Collection Statistics | Native | SeMet |
|---|---|---|
| Wavelength (Å) | 0.9794 | 0.9794 |
| Resolution range (Å) | 50 – 1.86 | 50 – 2.07 |
| Unique Reflections | 156340 | 37646 |
| Rmergea | 0.062 (0.734) | 0.082 (0.560) |
| 〈I〉/〈σI〉b | 28.9 (2.4) | 45.7 (2.3) |
| Redundancy | 6.3 (6.3) | 10.4 (4.9) |
| Completeness (%) | 99.9 (100.0) | 99.2 (97.3) |
|
| ||
| Refinement Statistics | ||
|
| ||
| Space group | P65 | P65 |
| Unit cell dimensions | ||
| a = b (Å) | 128.208 | 74.059c |
| c (Å) | 182.015 | 182.275 |
| α = β (°) | 90 | 90 |
| γ (°) | 120 | 120 |
| Solvent content (%) | 52 | 52 |
| Resolution Range (Å) | 50 – 1.80 | |
| Number of atoms | 13151 | |
| Protein | 12141 | |
| Ligand (glycerol, ethylene glycol, sulfate) | 120 | |
| Water | 890 | |
| Rmsds | ||
| Bond length (Å) | 0.007 | |
| Bond angle (°) | 0.936 | |
| Rworking/Rfreed | 20.7/24.6 | |
| Ramachandran analysis (%)e | ||
| Most favored | 91.9 | |
| Additionally allowed | 7.8 | |
| Generously allowed | 0.2 | |
| Disallowed | 0.0 | |
Rmerge is Σj| Ij−〈I〉|, where Ij is the intensity of an individual reflection and 〈I〉 is the mean intensity for multiply recorded reflections.
〈I〉/〈σI〉 is the mean intensity divided by the mean error.
The unit cell dimensions change only when ignoring the systematically weak refelections.
Rworking is Σ| | Fo| − | Fc| |/Σ| Fo| where Fo is an observed amplitude and Fc is a calculated amplitude; Rfree is the same statistic calculated over a subset of the data that have not been used for refinement.
Ramachandran analysis from PROCHECK.(Laskowski et al., 1993)
Quaternary structure and comparison to the regulatory dimer
The Sin tetramer has approximate 222 symmetry, and one can define three different dimeric pairs within it. The first set comprises the “cutting dimers,” the A/D and B/C pairs, that are expected to cleave the two strands of each DNA duplex in a concerted fashion. (Mouw et al., 2010) The second set comprises the “rotating dimers,” the A/C and B/D pairs, that would maintain their internal interactions throughout the hypothesized 180° subunit rotation during strand exchange (Figure 1A, 1B). The interface between these two dimers is flat and almost exclusively hydrophobic, as seen with the γδ and TP901 tetramers.(Kamtekar et al., 2006; Li et al., 2005; Yuan et al., 2008) The third set comprises the A/B and C/D pairs, which would exchange partners during the 180o rotation to become “religation dimers” – structurally identical to cutting dimers. Previous work on γδ resolvase used different notations for the same concept: the L/R dimer (cutting dimer), the L/L′ dimer (rotating dimer), and the L/R′ dimer (religation dimer).(Li et al., 2005)
The overall fold of the catalytic domain presented here is similar to that seen in other serine recombinase structures.(Li et al., 2005; Rice and Steitz, 1994b; Yang and Steitz, 1995; Yuan et al., 2008): a core comprised of a four- stranded β sheet (strands 1–4) and four α helices (A–D), that are connected by a flexible coil to a long helix, E, that mediates oligomerization. The C-terminal DNA binding domain of Sin, which is not present in this structure, would follow helix E. The region between the core and helix E is a β strand in γδ resolvase, but was more α helical in the inactive Sin dimer (Mouw et al., 2008), and was thus denoted helix D′. However, in the tetrameric Sin structure, this region is variable among protomers and is largely random coil. All 12 protomers in the asymmetric unit superimpose quite well on one another, except in this region and into the beginning of helix E (Ser92 – Ile113), indicative of inherent flexibility relative to the rest of the protein (Figure S1). The active sites and tetrameric architectures are very similar among the three tetramers. Helix D′ was rather poorly ordered in the inactive Sin dimer as well, also indicative of inherent flexibility. As has been previously hypothesized, this region acts as a hinge that likely facilitates relative motions between the more rigid core and the E helix.(Kamtekar et al., 2006)
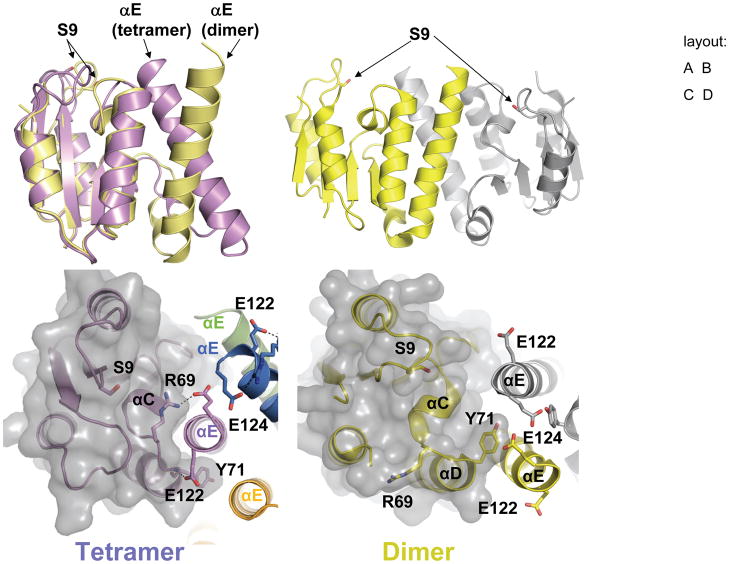
The catalytic domains of the inactive Sin dimer and the active Sin tetramer superimpose well, with a few major exceptions (Figure 2). The most striking is the repacking of helix E. In the dimer, its C-terminus extends away from the core and packs against the opposite protomer (Figure 2B, 2D), whereas in the tetramer it extends towards the core and packs into a small cleft between helix C and the N-terminus of helix D (Figure 2A, 2C). This repacking leads to a new helix-capping hydrogen bond between the N-terminus of helix D and Glu122 from helix E. Among the well-studied serine recombinases, this position (Sin Glu122, γδ resolvase Gln116) is conserved as a hydrophilic residue capable of accepting a hydrogen bond. The flexibility in helix E packing may be conferred in part by an unusually high concentration of methionines, the most flexible of the large hydrophobic residues: there are 5 methionines within the 31-residue segment from Met89 at the end of strand 4 to Met119 in helix E. Tyr 71 also repacks dramatically in the dimer-to-tetramer transition. However, this residue is not highly conserved and the significance of this observation is unclear.
Figure 2. Conformation changes linked to Sin activation.
A. Helix E repacks. One subunit from the inactive WT site II-bound dimer (yellow) and one from the activated tetramer (purple) are superimposed, using the cores as guides. The active site Ser9 nucleophiles are shown as sticks.
B. Inactive dimer. The dimeric catalytic domain from the WT Sin structure is shown in yellow and gray, with the Ser9 nucleophiles shown as sticks, 36.3 Å apart (Cα−Cα). Compare to the cutting dimer in figure 1B in which the serines are 24.7 Å apart.
C. Closeup of helix E packing in the activated Sin Q115R tetramer. The solvent-accessible surface of the left-hand protomer’s core region is shown as a transparent gray surface, and colors are as in figure 1b. Helix E packs in cis against helix C, requiring the movement of Y71. E122 hydrogen bonds to the N-terminus of helix D. R69 swings by ~120° and forms a salt bridge with E124 of the same protomer’s helix E.
D. Closeup of helix E packing in the inactive WT Sin dimer. One protomer is yellow and the other gray. R69 is flipped away from helix E and the active site, and helix E from the opposite protomer packs against helix C.
Although the tertiary structure of the core of the catalytic domain (residues 1 – 92) changes very little in the dimer- to – tetramer transition, at a more detailed level there are several changes in the vicinity of the active site that may explain the enhanced catalytic activity of this conformation. The side chain of Arg69 (corresponding to Arg71 of Tn3 and γδ resolvases), which extends from the helix C–D linker, reorients by roughly 120° and forms salt bridges with a sulfate ion adjacent to the nucleophile S9 and with Glu124 from the repacked helix E (Figure 2C,D and Figure 4). Both Arg69 and Glu124 are highly conserved, and the corresponding γδ resolvase residues were shown to be important for cleavage of the ‘proximal’ DNA strand.(Boocock et al., 1995; Stark and Boocock, 1995) The loop preceeding the highly-conserved Arg43 (aa35-41) was not modeled in the inactive Sin dimer, but is more structured in the active tetramer, where it packs against the linker between helices C and D. Finally, the loop containing the active-site Ser9 itself shifts, as described in more detail under “active site”.
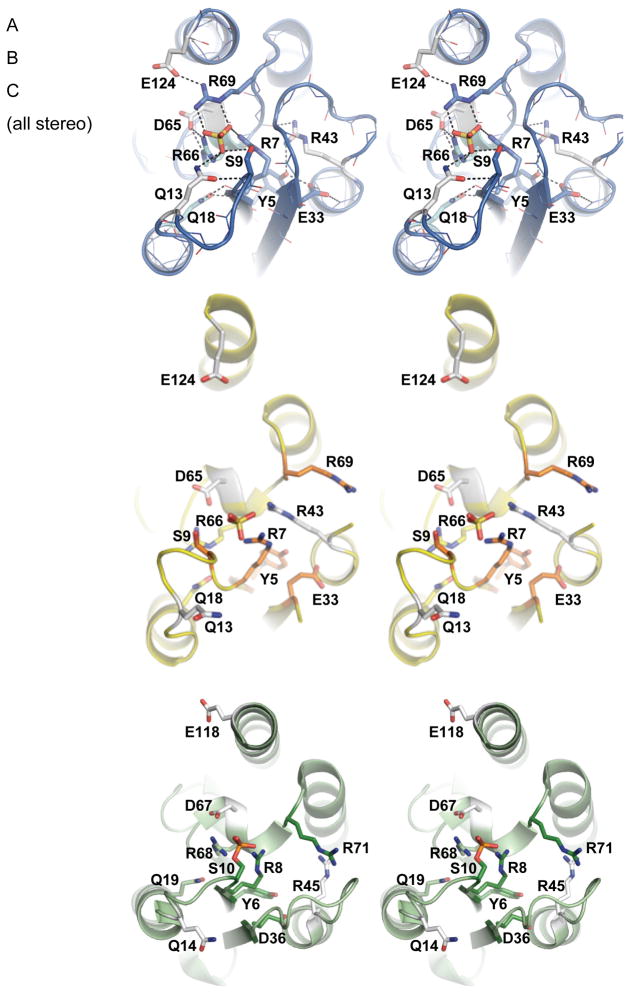
Figure 4. The active site.
A. Stereo view of the fully assembled active site. The critical residues within the active site pocket form a hydrogen bonded network surrounding a sulfate ion from the crystallization buffer. Residues are colored according to the effect of their mutation on DNA cleavage by Tn3 resolvase (Olorunniji and Stark, 2009) : darkest blue for the most deleterious (cleavage rate < = 0.0009x the WT rate) shading to white for the least deleterious (but still < = 0.022x the WT rate).
B. The “inactive site”. The catalytic site in the Sin site II structure is shown in a similar orientation with side chains shaded from orange to yellow to white. A bound sulfate ion roughly marks the active site but makes additional contacts to the protein (not shown) that could not occur in a site I – bound complex. Specific H-bonds are not drawn in panels B and C due to the lower resolution of these structures.
C. The active site from the postcleavage γδ tetramer is shown, with side chains shaded from dark green to white (PDB ID 1ZR4).
A cleavage-competent rotational substate of the tetramer
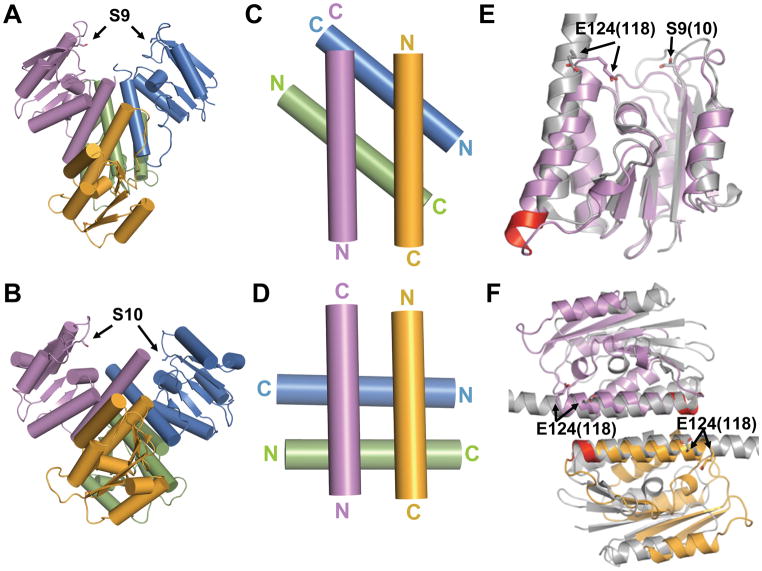
A striking difference between our new structure of Sin and the tetrameric structures of γδ resolvase is the crossing angle of the E helices at the hydrophobic rotational interface (Figure 3). In the γδ resolvase structures as well as the TP901-1 integrase tetramer, the two anti-parallel E helices of one “rotating dimer” cross those of the other rotating dimer at ~85°.(Kamtekar et al., 2006; Li et al., 2005; Yuan et al., 2008) However, in Sin, these pairs of E helices cross each other at ~50°, aligning the two rotating dimer pairs at a different rotational state than is seen in γδ resolvase (Figure 3A–D). The subunit rotation theory for strand exchange predicts that the complex must be able to assume multiple rotational states, at least transiently. Crosslinking experiments on Hin invertase in the presence of ethylene glycol did reflect two distinct rotational stopping points in vitro of ~90° and ~180°.(Dhar et al., 2009a) However, the structure reported here is the first direct visualization of an activated serine recombinase in a different rotational state from those previously observed in other tetrameric serine recombinase structures. Another important difference is that the active-site pockets of the cutting dimers are much closer together in the Sin Q115R tetramer than in the γδ resolvase tetramers: the sulfate ions in the Sin pocket are 18Å apart, vs. 27Å for the scissile phosphates in the post-cleavage γδ resolvase tetramer (Figure 3A,B; Figure 5). As described below, this allows unbroken site I DNA to be modeled onto the cutting dimers without major distortions. In part, the closer spacing in the Sin tetramer is a consequence of the smaller crossing angle between the E helices, but the individual catalytic sites are also closer to their respective E helices in the Sin tetramer. (Figure 3E).
Figure 3. Comparison of the Sin and γδ activated tetramers.
A. Quaternary structure of Sin Q115R, shown as a cartoon with cylindrical helices.
B. Quaternary structure of γδ resolvase. The catalytic domain of a postcleavage structure of an activated mutant of γδ resolvase (1ZR4) is shown from the same vantage point as Sin in part A.
C. E helices of Sin. Only the E helices at the center of the activated tetramer are displayed. The two pairs cross at an angle of ~50°.
D. E helices of γδ resolvase. Only the E helices from the activated γδ resolvase catalytic domain structure (2GM5) are shown. In all the activated γδ resolvase structures, these cross at ~85°.
E. Comparison of activated Sin and γδ protomers. One protomer from each activated tetramer is superimposed, guided by the core region. Sin is in purple and γδ (1ZR4) gray. The active site serines and the conserved glutamate from helix E are shown as sticks (Sin E124 and γδ E118). Helix E of Sin has a 1-turn extension at its N-terminus that is highlighted in red. The angle between the axes of the two E helices is ~20°, and the difference in rotation about the helical axes is ~25 °.
F. Comparison of Sin and γδ rotating dimers. One rotating dimer from each activated tetramer was superimposed by aligning the approximate twofolds between the E-helices. Sin is colored as in part A, and γδ gray (1ZR4). (See also figure S2).
Figure 5. Docking of duplex DNA into the catalytic sites.
A. Uncut duplex DNA taken from the dimeric γδ resolvase – site I complex (1GDT)(Yang and Steitz, 1995) was docked onto the Sin tetramer by superimposing the E-helices of subunit A from each structure (Sin subunit A is shown in lavender). Scissile phosphates from the site I DNA are shown as red spheres; the sulfate ions marking the Sin catalytic sites as yellow spheres, and the nucleophilic serines as red sticks.
B. View of the model in part A rotated ~90° about a horizontal axis.
C. Closeup stereo view of the active site surrounding the scissile phosphate of the model DNA. View is from the upper left background of panel A, and the same side chains are shown as in Fig. 4A.
D. Cartoon of the γδ resolvase – site I tetramer (PDB ID 1ZR4) for comparison to panel b. The phosphoserine linkages are highlighted with a red sphere and the displaced 3′ hydroxyl groups with a black sphere.
E. Model of Sin in the same rotational substate as the γδ structure in panel d, made by changing the relative orientation of the two rotating dimers. The scissile phosphates of uncleaved DNA can still be superimposed on the sulfate ions, but the DNA clashes with the S9-containing loop and helix A would interfere with the binding of the opposite subunit’s helix E in the DNA’s minor groove.
It is not immediately obvious why the Sin tetramer crystallized in a different rotational state from γδ resolvase and TP901-1 integrase. As described below, the Q115R mutation used to stabilize the activated form mediates interactions within rotating dimers, not between them. Although activation requires an overall conformational change from dimer to tetramer, subunit rotation requires that a continuum of rotational substates be accessible within the tetramer, and that the energetic differences among them be small enough to be readily overcome by thermal energy. Thus, different activating mutations might tip the balance slightly more towards one substate than another. We propose that the strongly activating Q115R mutation in Sin stabilizes a substate closer to the cleavage-competent form, whereas the sets of mutations used in the γδ resolvase structures preferentially stabilize a post-cleavage form.
Two additional noteworthy differences exist between the Sin and γδ resolvase tetramers. The primary sequence of Sin contains a small insertion at the N-terminal end of helix E that extends it by one turn (Figure S2). As shown in figure 3E, this doesn’t alter the alignment of conserved residues on helix E with the core region of the same molecule (e.g. Sin E124/γδ E118 vs. Sin S9/γδ S10). However, the relative alignment of the two E-helices within the rotating dimer differs (Figure 3F). When the rotating dimers are aligned by their pseudo-twofold axes, the conserved residues in each Sin subunit’s E helix are translated one helical turn towards the C-terminal end of that helix, relative to their positions in γδ. Because the Sin E helix is one turn longer, the overall contact area between these helices is similar: in both cases, the first 5 ½ turns of each E helix pack against each other. Assuming this different translational register of the rotating dimer’s E helices is a fixed attribute of each system, this implies that for DNA cleavage and religation, the two cutting dimers will be held slightly further apart in the Sin system (ie ~2 α-helical turns). A second difference lies in the intramolecular packing of the E-helices: when superimposing individual protomers from the Sin and γδ tetramers, the core regions align very well, while the C-terminal region of Sin’s E helix lies closer to the active site residues in the core (Figure 3E), bringing Glu124 within H-bonding distance of Arg69, as noted above (Figure 2C). The γδ resolvase helix E nevertheless makes a D helix capping interaction equivalent to that described above for Sin (Figure 2C), and a small additional shift in the position of helix E could potentially bring the conserved active site residues together in a configuration similar to that seen with Sin.
Active Site Architecture
Previous biochemical and structural studies of serine recombinases have highlighted a set of conserved residues surrounding the serine nucleophile that are critical for catalytic function.(Hughes et al., 1990; Olorunniji and Stark, 2009) Rigorous experiments with hyperactive Tn3 resolvase identified six of these residues as crucial for catalysis: Tyr5, Arg7, Ser9, Asp(Glu)33, Arg66, and Arg69 (the residue numbering for Sin is used here). (Olorunniji and Stark, 2009) Mutation of another group of highly conserved residues also inactivated the enzyme in vivo and had strong, although not as drastic, effects on DNA cleavage in vitro: Gln13, Gln18, Arg43, Asp 65 and E124 (Sin residue numbering). These were proposed to play auxiliary roles in organizing the active site.
The Sin tetramer, unlike previous serine recombinase structures, depicts all of these residues in a hydrogen-bonded network that includes the nucleophile Ser9 and a sulfate ion from the crystallization mother liquor that binds at the position of the scissile phosphate group in the γδ resolvase – site I tetramers (Figure 4). The 3 critical arginines (7, 66 and 69) make bidentate salt bridges to the sulfate ion. Two of these also interact with conserved carboxylates: Arg66 with Asp65, and Arg69 with Glu124. The third, Arg7, is close to Ser9: Arg NH1- Ser OH is 3.5–4Å in two monomers, and would be that close on the other two if the serine adopted a different rotamer. Arg43, Tyr5 and Glu33 form a hydrogen bonding chain that extends from the backbone carbonyls at the C-terminus of helix C (containing Arg66 and Arg69) to the backbone amides of helix B. These residues may be primarily important for holding the tertiary structure together, but they might also participate more directly in catalysis, as discussed below. Finally, Gln13 and Gln18 appear to buttress the position of Ser9 by hydrogen-bonding to the peptide backbone of residues 7-9. However, some details of the interactions may vary in the ground and/or transition states of the DNA-bound complex, because a sulfate ion does not perfectly mimic the scissile phosphate: it contains two negative charges whereas a phosphate group within the DNA backbone would only have one. The transition state of the phosphotransfer reaction would be somewhat more charged, but its geometry would differ from that of the sulfate ion.
Although many of these residues undergo some movement when coalescing to form the active site in the tetrameric form (relative to the dimer), the most striking changes are in Arg69 and Glu124 (Figures 4 and 2C,D). The guanidium group of Arg69 swings over 11Å to join the other critical arginines in forming the sulfate-binding pocket. The repacking of helix E positions Glu124 so that it can form a salt bridge with this guanidinium group as well. This interaction was not seen in previous serine recombinase structures and provides a new explanation for why this residue is so highly conserved. Additionally, the loop beginning with Ser9 shifts position, and the serine moves towards the rest of the active site.
Previous hypotheses on the roles of the active site arginines are consistent with the structural information presented here (Boocock et al., 1995; Droge et al., 1990; Grindley et al., 2006; Rice and Steitz, 1994b)(Olorunniji and Stark, 2009). The active sites of serine recombinases are devoid of residues such as histidine that are generally thought of as good general acid/base catalysts. However, Arg7 was proposed to be the general base that abstracts a proton from Ser9 in the cleavage reaction, and Arg69 is a candidate for the general acid that protonates the leaving 3′ hydroxyl (Kent Mouw & PAR, unpublished data), and in our structure, they are nicely positioned for such roles. Two other residues that (in Tn3 resolvase) are critical for catalysis, Glu33 and Tyr5, do not directly contact the sulfate. However, they do form a hydrogen bond network with Arg43 and the polypeptide backbone, which may help to stabilize the local structure. This triad might also act as a proton shuttle to aid in deprotonating another arginine residue in close proximity (e.g. Arg7). Normally, the high pKa of arginine residues (~12.5) renders them unsuitable for general acid/base catalysis, but are precedents for arginine to function as a strong general base.(Charnock et al., 2002; Guillen Schlippe and Hedstrom, 2005a, b; Guillen Schlippe et al., 2004) The pKas of the arginines in Sin’s active site might be lowered in part by their proximity, which would electrostatically destabilize the positively charged form of each individual arginine. Additionally, strong hydrogen-bonding interactions may lower the pKa of arginine by straining the planar conformation of the guanidinium, decreasing the resonance that stabilizes the charged form.(Guillen Schlippe and Hedstrom, 2005b) Asp65, Glu124, and the carbonyl groups at the end of helix C may play this role, although they also may simply localize the arginine side chains. Unfortunately, even at 1.86Å resolution, a subtle twist in the side chain’s tip would not be apparent. Arginines 7, 66 and 69 may also be important in orienting the scissile phosphate and in stabilizing the geometry and additional negative charge of the transition state. Finally, Arg 125 is strongly conserved but has not been implicated directly in catalysis. It could not be modeled in this crystal structure, but small bits of electron density suggest it points towards the sulfate in the active site. While its role is unclear, it may help to stabilize the negative charge of the DNA backbone upon binding.
The activating mutation Q115R
The critical mutation that forces the apo protein into its tetrameric form is Q115R, located on the E helix. This mutation, which was originally selected for its activating effect on catalysis, may simultaneously destabilize the dimeric and stabilize the tetrameric forms. In the inactive dimer, Gln115 is mainly solvent-exposed, but on one side of this slightly asymmetric structure it could form hydrogen bonds across the dimer interface with the N-terminus of helix C and with the β4-αD′ turn. However, in the activated structure, the guanidinium groups of the two Arg115s within each rotating dimer form Pi-Pi stacks with one another (Figure 1C). Each Arg115 also interacts with Asp111, which is located one helical turn away, and presumably helps mitigate the charge-charge repulsion between the arginines. In the crystal structure each pair of Arg115s also interacts with a negatively charged sulfate ion that was present in the crystallization buffer but is not necessary for activity in vitro (Figure 1C, and data not shown). Although two positively charged moieties would normally repel each other, this type of stacking between arginine side chains has been observed before at protein interface hotspots as well as implicated in high affinity protein-protein interactions such as a receptor engaging its ligand.(Hughes et al., 1990; Persson et al., 2009) Arg-Arg pairs have been shown to defy electrostatics and confer thermodynamic stability.(Pednekar et al., 2009) The relative orientation of the guanidinium groups makes Pi-Pi stacking favorable while still allowing the formation of hydrogen bonds, as with the aspartates and sulfate group in this structure. A simple model made by substitution of the arginine with the WT glutamine shows a very plausible hydrogen bonding interaction that likely exists within the WT synaptic complex (Figure 1D).
How stabilizing could the Arg-Arg interaction be? Previous work in our labs showed that the Q115R mutation strongly stabilizes the tetrameric over the dimeric form of Sin. From analytical ultracentrifugation experiments, Kd for the dimer-tetramer equilibrium of Sin Q115R/R54E was determined to be ~400–450 nM in DNA-bound dimers.(Mouw et al., 2010) Given that the WT protein was dimeric even at 50μM, we estimate that the Q115R mutation decreases the Kd for tetramerization by >100 fold. Computational simulations predict that two guanidinium moieties of an arginine side chain in water interact with a ΔΔG = ~−2.1 kcal/mol. (Vondrasek et al., 2009) Since each Q115R tetramer contains two R115-R115 interactions, one can estimate ΔΔG = ~−4.2 kcal/mol (~−17.4kJ/mol) per tetramer: this would be sufficient to account for a ~1000-fold change in the Kd for tetramerization.
Although the Arg-Arg stacking may theoretically be strong enough, it is probably not the complete explanation for the activating effect of Q115R. Certain other substitutions at position 115 also led to markedly increased activity over WT Sin, although their effect is not as strong as that of Q115R.(Rowland et al., 2009) These were, in, order of activity, C > Y > K (while the R115E change was inhibitory). Modeling indicates that tyrosines at position 115 could also make favorable pi-pi stacking interactions, as seen in many protein interfaces (Flocco and Mowbray, 1994; Ma et al., 2003; Rajamani et al., 2004; Rice and Steitz, 1994b). It is less obvious how a cysteine or lysine at position 115 could stabilize the tetramer (simulations predict a ΔΔG = ~+7.4 kcal/mol for the association of two NH4+ moieties on lysine residues). Perhaps destabilization of the dimer is the main factor here.
Activating mutations in other serine recombinases have also been suggested to alter the equilibrium between dimers and tetramers, by destabilizing the former and/or stabilizing the latter. In support of this, they generally lie in the region that must repack during the transition, (Li et al., 2005; Olorunniji et al., 2008; Rowland et al., 2009) and similar activating mutations in γδ resolvase force the catalytic domain into a tetramer in the absence of DNA.(Kamtekar et al., 2006) Interestingly, the catalytic domain of TP901-1 integrase, a member of the large serine recombinase family, crystallized as a tetramer despite being dimeric in solution.(Yuan et al., 2008) This tetramer may be stabilized in part by a non-native residue from the C-terminal tag, Glu137, that extends from helix E to cap helix D, similar to the WT Glu122 of Sin (Figure 2).
Sin site I model
The uncleaved duplex DNA from the inactive, WT γδ resolvase – site I structure fits remarkably well into the catalytic sites of the Sin tetramer (Figure 5). The DNA was docked by rigid-body superposition of subunit A from the γδ resolvase structure (which has a slightly kinked E helix) onto subunit A of Sin, using the E helices as guides. With no manual adjustment, one scissile phosphate fits almost exactly onto the sulfate ion that marks the opposite Sin subunit’s active site, and the other phosphate-sulfate pair are nearly aligned. Thus, even though the DNA is far from the catalytic sites in the inactive dimer, the bending and widening of the minor groove induced by its interactions with the C-terminal segments of the E-helices appear to be preset for interactions in an active tetramer.
The precise fit of the DNA duplex onto the Sin tetramer implies that this rotational substate is consistent with a cleavage-competent state of the tetramer. Further rotation of the subunits relative to each other would require DNA cleavage, as shown by rotating the Sin subunits about the flat interface so that their E-helices cross at the angle seen for γδ resolvase (Figure 5D,E). The serines remain closer together than in the γδ tetramer, due to the different orientation of each catalytic core relative to its own helix E (see Fig. 3E). However, docking uncleaved DNA to this tetramer model so that relevant phosphates and sulfates align requires that the DNA cross the catalytic domains at a different angle, which creates steric clashes and would be incompatible with the extension of helix E binding in the minor groove.
Although DNA binding may lead to additional conformational changes, the ease of modeling DNA onto the Q115R Sin tetramer implies that such changes may be quite small. We are hesitant to predict detailed interactions from the model. However, the position of the highly conserved Gln13 suggests that it stabilizes the backbone of the 2 nt 3′ overhang before religation. Modeling of the extension of the E helix not included in our tetramer structure implies that the highly conserved Arg131 does so as well, but would be located near the active site of the opposite subunit in the cutting dimer. More detailed hypotheses would require a structure of the activated Sin tetramer with site I DNA.
Conclusions
This structure offers the first high-resolution snapshot of the catalytic site of a serine recombinase in what appears to be a fully assembled state, with a sulfate ion bound at the anticipated location of the scissile phosphodiester bond (Figure 4). The ease with which uncleaved DNA could be modeled onto this structure implies that the quarternary structure is also poised for DNA cleavage, and that the two cleavage events on a single duplex could be simultaneous (Dhar et al., 2009a; Kamtekar et al., 2006; Li et al., 2005; Smith et al., 2010), rather than sequential as has occasionally been postulated (Figure 5).(Yang and Steitz, 1995)
This structure strongly reinforces the subunit rotation model for strand exchange: a 35 degree rotation of one ‘rotating dimer’ of the Sin tetramer reported here would create a configuration resembling the post-cleavage tetramers previously reported for γδ resolvase (Fig 3). The fact that both tertiary as well as quarternary differences exist between these tetramers also supports the idea that the full reaction pathway proceeds through a number of different substates of the tetramer.
Many questions remain. Deeper analyses of the active site are needed to elucidate the precise function of each residue in catalysis. The functions of the regulatory apparatus (Sin site II and HU) need to be more closely examined: it remains unknown whether the regulatory apparatus functions through an allosteric mechanism to activate tetramerization at site I, or whether it does so simply by aligning two site I-bound dimers at a high local concentration and in the correct relative orientation. Future experiments will focus on these two areas to gain a more complete understanding of the structure and function of the Sin synaptosome at the molecular level.
Materials and Methods
Native Protein Purification
The plasmid pSA1162 encoding the 14 kDa N-terminal domain (NTD) of pI9789 Sin was derived from pSA1122 by introducing a stop codon following residue 128 and by introducing the R54E and Q115R mutations. Rosetta (DE3)[pLysS] cells (Novagen) were transformed with pSA1162 and grown at 225 RPM and 37°C in Luria-Bertani medium containing 50 μg/ml kanamycin and 33 μg/ml chloramphenicol. The cells were grown to an OD600 of ~0.6 and induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Following induction, the cells were grown for an additional four hours and harvested by centrifugation. The cell pellet from 2 liters of culture(~10g ) was frozen at −80°C for later use. The pellet was thawed on ice, resuspended in 50 ml Lysis Buffer (25 mM TRIS [pH 8.0], 10 mM NaCl, 1mM EDTA and protease inhibitor cocktail [Roche Complete]) sonicated, and centrifuged at 18,000 rpm in an SS34 rotor for 30 minutes. Sin remained in the pellet and was resuspended in Buffer A (25 mM TRIS [pH 8.0], 10 mM NaCl, 1mM EDTA, and 3M urea) followed by centrifugation at 18,000 rpm for 30 minutes. Soluble denatured Sin was filtered with a .22 μm filter, loaded onto a Q FF anion exchange column (Amersham), and eluted with Buffer B (Buffer A except 1M NaCl). Pooled fractions were stepwise dialyzed into Buffer C (25 mM TRIS [pH 8.0], 2 M NaCl, 1mM EDTA, and 2M urea), Buffer D (Buffer C except 1M urea), Buffer E (25 mM TRIS [pH 8.0], 2 M NaCl, and 1mM EDTA), Buffer F (25 mM TRIS [pH 8.0], 1 M (NH4)2SO4, and 1mM EDTA) and finally Buffer G (25 mM TRIS [pH 8.0], 500 mM (NH4)2SO4, 1mM EDTA, and 5% glycerol). The protein was concentrated to ~8 mg/ml in centrifugal filters (MWCO 3000, Amicon) and stored in 50 μL aliquots at −80°C. The final yield was ~50 mg of protein.
Selenomethionyl Protein Purification
Rosetta (DE3)[pLysS] cells (Novagen) were transformed with pSA1162 and grown overnight at 37°C in ml Luria-Bertani medium containing 50 μg/ml kanamycin and 33 μg/ml chloramphenicol. The cells were spun down and resuspended in M9 media, while flasks containing M9 media plus additives (0.4% Glucose, 10 mM NaCl, 0.1 mM CaCl2, 2 mM MgSO4) were inoculated with the overnight culture. The cells were grown to an OD600 of ~0.5 and an amino acid cocktail containing equal amounts each of L-isoleucine, L-leucine, L-lysine, L-phenylalanine, L-threonine, and L-valine was added to a final concentration of 100 mg/L. Selenomethionine was added to a final concentration of 60 mg/L and the culture was grown for another 15 minutes, at which point the cells were induced with 1 mM IPTG, grown for 4 hours and harvested via centifugation. The protein was purified, concentrated, and stored exactly as the native protein except all buffers contained 10 mM dithiothreitol (DTT).
Crystallization
Crystals were grown by the hanging drop vapor diffusion method. The protein was mixed in a 1:1 ratio with well solutions containing 100 mM HEPES (pH 7.0), 1.8 M (NH4)2SO4, 20 % ethylene glycol, and 5% glycerol. Both native and selenomethionyl (SeMet) crystals grew to full size in ~3 days and were cryoprotected in well solutions containing 10% glycerol and frozen in liquid N2. Similar crystals were grown in 20% PEG 3350 and 0.2M ammonium acetate yielding nearly-identical diffraction quality and space group parameters, but were not used for structure determination due to twinning.
Structure Determination
Native diffraction data were collected at the SBC beamline 19-ID at the Advanced Photon Source (Argonne, IL) while SeMet crystal diffraction data were collected at the LS-CAT beamline 21-ID. Both data sets were processed with the HKL3000 suite.(Minor et al., 2006) They were isotropic, with resolution extending to 2.07 Å (〈I〉/〈σI〉 > 2 in the outermost shell) for the SeMet set and 1.86 Å for the native. All crystals belonged to space group P65 and contained a strong pseudotranslational symmetry vector of 〈0.6667, 0.3333, 0.0088〉 (in fractions of the unit cell axes) as determined by the native patterson map with a peak ~66% the height of the origin. Reflecting this, every 3rd reflection (along h and k) was particularly intense. The dimensions of the true unit cell were a=b=128.21 Å, c=182.02 Å. However, ignoring the faint reflections forces the data into a unit cell of the same space group but 1/3 the volume, with dimensions (Å) a/√3=b/√3=74.059, c=182.275, and with the h and k axes shifted by −30°. The true unit cell contains 12 protomers per asymmetric unit, whereas the smaller cell contains 4 (Figure S1).
To simplify the search for Se positions (7/monomer), the structure was initially solved in the smaller unit cell via SAD phasing using the program SOLVE (Terwilliger and Berendzen, 1999), which located all 28 Se sites. The maps were improved by solvent modification with automated phase extension using the program DM (Figure 6A). (Vellieux, 1998) An initial model of one tetramer was built into this map, but residues 97 – 102 were disordered. To take advantage of the information contained in the faint reflections that were not included in this original solution, the larger unit cell was solved via molecular replacement with MolRep(Vagin and Teplyakov, 2010) using the tetramer as a search model, yielding a solution with three tetramers per asymmetric unit. Residues 97-102 were better ordered in the resulting maps, but their conformations differ somewhat among the 12 monomers (Figure 1). Presumably, this region could not be modeled in the small unit cell because the exclusion of the faint 2/3 of the reflections forces the 3 subtly-different tetramers to be identical. In one monomer, residues 95 - 97 remained to poorly ordered to model, as did residues 125 – 128 in all the monomers. The structure was refined against all data to 1.80 Å using REFMAC(Murshudov et al., 1997) in the CCP4 program suite(1994) to an Rworking of 20.7% and an Rfree of 24.7% with very good stereochemistry (the reported nominal resolution of 1.86Å was defined by the highest-resolution shell in which 〈I〉/〈σI〉 is > 2). All models were built with Coot(Emsley and Cowtan, 2004) and figures displayed with Pymol, made by Delano Scientific. Experimental and post-refinement electron density maps are shown in figure 6. Diffraction and refinement statistics can be found in Table 1.
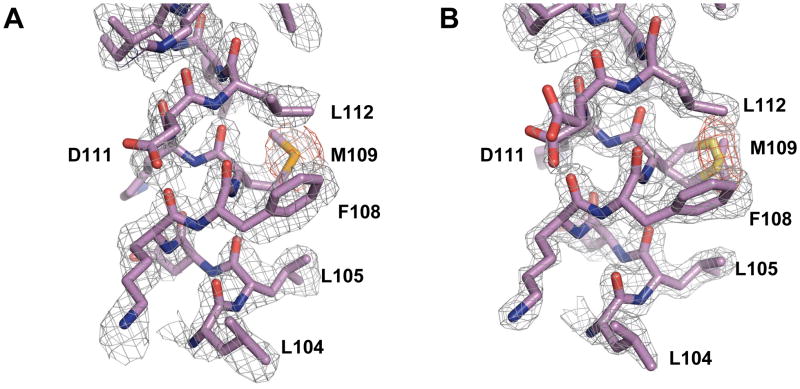
Figure 6. Electron density maps.
A. Experimental electron density. The SAD-phased map, calculated in the small unit cell, was subject to one round of density modification with automated phase extension. The map is contoured at 1.5 σ. An anomalous difference map calculated from the Se SAD data, shown in red, is contoured at 3.5 σ. Both maps contain only experimental phase information without any model bias. The maps are centered on a portion of the E helix containing the residues 104-112, and for clarity, “carved” to remove density > 2Å from atoms. B. Weighted final 2Fo−Fc electron density for the same region as shown in part A, calculated using phases from the refined model in the large unit cell. The map is contoured at 1.5 σ. An anomalous difference map calculated from an Se SAD data set is superimposed on it, contoured at 3.5 σ (red). Due to the high resolution, multiple conformers could be fit for certain side chains, such as Met 109 and Asp111
Supplementary Material
Highlights.
We report the 1.86Å structure of an activated mutant of the Sin catalytic domain
The active site appears to be fully assembled, primed for DNA cleavage
Uncleaved DNA can be modeled into the active sites
Observation of a new rotational substate supports the subunit rotation hypothesis.
Acknowledgments
We thank the staff at the SBC-CAT (19-ID) and LS-CAT (21-ID) beamlines at the Advance Photon Source for help with data collection and processing. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). This work was supported by NIH grant R01-GM086826 (to P.A.R.) and Wellcome Trust Project grant 072552 (to W.M.S, S.J.R and M.R.B). R.A.K was supported in part by the NIH Training Grant 5T32GM007183-35. R.A.K solved the crystal structure and wrote the initial manuscript with training and advice from P.A.R.; P.A.R., W.M.S, S.J.R and M.R.B planned the project, and W.M.S, S.J.R and M.R.B provided clones, and biochemical advice and data. All authors contributed to interpreting the structure and writing the final manuscript. We thank Kent Mouw for helpful discussions on Sin and the manuscript, and Sherwin Montaño, Adrianna Zhang, Ying Zhang Pigli, and Xiaojing Yang for protein purification advice and training in crystallographic methods.
Footnotes
Accession Numbers
Crystallographic coordinates and data have been deposited in the Protein Data Bank under 3PKZ.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Arnold PH, Blake DG, Grindley NDF, Boocock MR, Stark WM. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. Embo J. 1999;18:1407–1414. doi: 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock MR, Zhu X, Grindley NDF. Catalytic residues of γδ resolvase act in cis. Embo J. 1995;14:5129–5140. doi: 10.1002/j.1460-2075.1995.tb00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke ME, Arnold PH, He J, Wenwieser SVCT, Rowland SJ, Boocock MR, Stark WM. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol Microbiol. 2004;51:937–948. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- Charnock SJ, Brown IE, Turkenburg JP, Black GW, Davies GJ. Convergent evolution sheds light on the anti-beta -elimination mechanism common to family 1 and 10 polysaccharide lyases. Proc Natl Acad Sci U S A. 2002;99:12067–12072. doi: 10.1073/pnas.182431199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbise A, Dyke KG, el Solh N. Rearrangements in the staphylococcal beta-lactamase-encoding plasmid, pIP1066, including a DNA inversion that generates two alternative transposons. Mol Microbiol. 1995;17:769–779. doi: 10.1111/j.1365-2958.1995.mmi_17040769.x. [DOI] [PubMed] [Google Scholar]
- Dhar G, Heiss JK, Johnson RC. Mechanical constraints on Hin subunit rotation imposed by the Fis/enhancer system and DNA supercoiling during site-specific recombination. Mol Cell. 2009a;34:746–759. doi: 10.1016/j.molcel.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar G, McLean MM, Heiss JK, Johnson RC. The Hin recombinase assembles a tetrameric protein swivel that exchanges DNA strands. Nucleic Acids Res. 2009b;37:4743–4756. doi: 10.1093/nar/gkp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droge P, Hatfull GF, Grindley NDF, Cozzarelli NR. The two functional domains of γδ resolvase act on the same recombination site: implications for the mechanism of strand exchange. Proc Natl Acad Sci U S A. 1990;87:5336–5340. doi: 10.1073/pnas.87.14.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flocco MM, Mowbray SL. Planar stacking interactions of arginine and aromatic side-chains in proteins. J Mol Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Guillen Schlippe YV, Hedstrom L. Is Arg418 the catalytic base required for the hydrolysis step of the IMP dehydrogenase reaction? Biochemistry. 2005a;44:11700–11707. doi: 10.1021/bi048342v. [DOI] [PubMed] [Google Scholar]
- Guillen Schlippe YV, Hedstrom L. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch Biochem Biophys. 2005b;433:266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Guillen Schlippe YV, Riera TV, Seyedsayamdost MR, Hedstrom L. Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase reaction. Biochemistry. 2004;43:4511–4521. doi: 10.1021/bi035823q. [DOI] [PubMed] [Google Scholar]
- Haffter P, Bickle TA. Enhancer-independent mutants of the Cin recombinase have a relaxed topological specificity. Embo J. 1988;7:3991–3996. doi: 10.1002/j.1460-2075.1988.tb03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlander SK, Hulten KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, et al. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes RE, Hatfull GF, Rice P, Steitz TA, Grindley NDF. Cooperativity mutants of the γδ resolvase identify an essential interdimer interaction. Cell. 1990;63:1331–1338. doi: 10.1016/0092-8674(90)90428-h. [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SVCT, Boocock MR, Grindley NDF, Steitz TA. Implications of structures of synaptic tetramers of γδ resolvase for the mechanism of recombination. Proc Natl Acad Sci U S A. 2006;103:10642–10647. doi: 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A, Cloppenborg K, Kahmann R. Isolation and characterization of unusual gin mutants. Embo J. 1988;7:3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley NDF, Steitz TA. Structure of a synaptic γδ resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci U S A. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Mouw KW, Rowland SJ, Gajjar MM, Boocock MR, Stark WM, Rice PA. Architecture of a serine recombinase-DNA regulatory complex. Mol Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw KW, Steiner AM, Ghirlando R, Li NS, Rowland SJ, Boocock MR, Stark WM, Piccirilli JA, Rice PA. Sin Resolvase Catalytic Activity and Oligomerization State are Tightly Coupled. J Mol Biol. 2010;404:16–33. doi: 10.1016/j.jmb.2010.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Olorunniji FJ, He J, Wenwieser SVCT, Boocock MR, Stark WM. Synapsis and catalysis by activated Tn3 resolvase mutants. Nucleic Acids Res. 2008;36:7181–7191. doi: 10.1093/nar/gkn885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olorunniji FJ, Stark WM. The catalytic residues of Tn3 resolvase. Nucleic Acids Res. 2009;37:7590–7602. doi: 10.1093/nar/gkp797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Gillespie MT, Littlejohn TG, Hanvivatvong O, Rowland SJ, Dyke KG, Skurray RA. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene. 1994;141:109–114. doi: 10.1016/0378-1119(94)90136-8. [DOI] [PubMed] [Google Scholar]
- Pednekar D, Tendulkar A, Durani S. Electrostatics-defying interaction between arginine termini as a thermodynamic driving force in protein-protein interaction. Proteins. 2009;74:155–163. doi: 10.1002/prot.22142. [DOI] [PubMed] [Google Scholar]
- Persson BD, Muller S, Reiter DM, Schmitt BBT, Marttila M, Sumowski CV, Schweizer S, Scheu U, Ochsenfeld C, Arnberg N, et al. An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46. J Virol. 2009;83:673–686. doi: 10.1128/JVI.01967-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc Natl Acad Sci U S A. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Steitz TA. Model for a DNA-mediated synaptic complex suggested by crystal packing of γδ resolvase subunits. Embo J. 1994a;13:1514–1524. doi: 10.2210/pdb1gdr/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Steitz TA. Refinement of γδ resolvase reveals a strikingly flexible molecule. Structure. 1994b;2:371–384. doi: 10.1016/s0969-2126(00)00039-3. [DOI] [PubMed] [Google Scholar]
- Rowland SJ, Boocock MR, McPherson AL, Mouw KW, Rice PA, Stark WM. Regulatory mutations in Sin recombinase support a structure-based model of the synaptosome. Mol Microbiol. 2009;74:282–298. doi: 10.1111/j.1365-2958.2009.06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland SJ, Stark WM, Boocock MR. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol Microbiol. 2002;44:607–619. doi: 10.1046/j.1365-2958.2002.02897.x. [DOI] [PubMed] [Google Scholar]
- Rowland SJ, Boocock MR, Stark WM. Regulation of Sin recombinase by accessory proteins. Mol Microbiol. 2005;56:371–382. doi: 10.1111/j.1365-2958.2004.04550.x. [DOI] [PubMed] [Google Scholar]
- Rowland SJ, Dyke KG. Characterization of the staphylococcal beta-lactamase transposon Tn552. Embo J. 1989;8:2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis GJ, Murley LL, Leschziner AE, Boocock MR, Stark WM, Grindley NDF. A model for the γδ resolvase synaptic complex. Mol Cell. 2001;8:623–631. doi: 10.1016/s1097-2765(01)00334-3. [DOI] [PubMed] [Google Scholar]
- Smith MC, Brown WR, McEwan AR, Rowley PA. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans. 2010;38:388–394. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- Stark WM, Boocock MR. Gatecrashers at the catalytic party. Trends Genet. 1995;11:121–123. doi: 10.1016/s0168-9525(00)89016-2. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Vellieux FM. A comparison of two algorithms for electron-density map improvement by introduction of atomicity: skeletonization, and map sorting followed by refinement. Acta Crystallogr D Biol Crystallogr. 1998;54:81–85. doi: 10.1107/s0907444997008081. [DOI] [PubMed] [Google Scholar]
- Vondrasek J, Mason PE, Heyda J, Collins KD, Jungwirth P. The molecular origin of like-charge arginine-arginine pairing in water. J Phys Chem B. 2009;113:9041–9045. doi: 10.1021/jp902377q. [DOI] [PubMed] [Google Scholar]
- Yang W, Steitz TA. Crystal structure of the site-specific recombinase γδ resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- Yuan P, Gupta K, Van Duyne GD. Tetrameric structure of a serine integrase catalytic domain. Structure. 2008;16:1275–1286. doi: 10.1016/j.str.2008.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.