Abstract
In this study, we have examined the role of post-translational modification of the myeloid master regulator C/EBPα by small ubiquitin-related modifier (SUMO). We have used transient transfection analysis, oligonucleotide pulldown assays and chromatin immuno-precititation in all-trans retinoic acid (ATRA)-inducible promyelocytic cell lines MPRO and NB4. We demonstrate that sumoylated wild-type p42-C/EBPα is associated with negative regulation of the myeloid specific lactoferrin (LF) gene in early myeloid cells and that a reduction in p42-C/EBPα sumoylation coincides with expression of the LF gene in maturing myeloid cells. In the acute promyelocytic leukemia cell line NB4 however, sumoylated p42 remains persistently bound to the LF promoter following ATRA-induction. This correlates with lack of lactoferrin expression in these cells. Changes in sumoylation status of C/EBPα thus appear to contribute to a switch that regulates transcriptional activity of this master regulator during normal neutrophil development. We also demonstrate that sumoylation of the AML associated dominant negative p30-C/EBPα isoform does not alter transactivation activity of the LF promoter. This may be because the p30 C/EBPα isoform binds to the LF promoter much less efficiently than its full length counterpart. Our data suggest that the activity of p42-C/EBPα in the developing neutrophil is more sensitive to changes in sumoylation than the p30 isoform. This difference may contribute to the leukemogenic potential of p30-C/EBPα.
Key words: myeloid transcription, post-translational modification, sumoylation, leukemogenesis.
Introduction
C/EBPα is the founding member of a family of basic region/leucine zipper (bzip) transcription factors that is a master regulator of granulopoiesis.1 It is expressed at high levels throughout myeloid differentiation and binds to the promoters of multiple myeloid- specific genes at different stages of myeloid maturation. Several groups have reported mutations in the C/EBPα gene in a subset of patients with AML with normal karyotype.2 These mutations can be classified into two main categories. The first includes in-frame mutations clustered in the C-terminus of the C/EBPα protein. This domain harbors the conserved basic-region leucine-zipper responsible for DNA binding, as well as the dimerization domain. Often additional mutations in the second C/EBPα allele accompany these mutations resulting in functional loss of C/EBPα. The second category of mutations involves the N-terminus of C/EBPα. These frame-shift mutations are responsible for premature termination of the full length p42 C/EBPα isoform while keeping the truncated p30C/EBPα protein intact.2 The remaining p42C/EBPα is thought to be rendered inactive by the dominant-negative activity of the p30 isoform by an unknown mechanism.3,4 More recently, mice engineered to express only the p30 isoform from the C/EBPα locus developed AML with complete penetrance.5 This highlights the functional differences between the full length p42 isoform, with its tumor suppressive activity, compared to the truncated p30 C/EBPα, which has leukemogenic potential.5
C/EBPα potently activates myeloid differentiation. However, chromatin immunoprecipitation studies from our laboratory have previously demonstrated that C/EBPα associates with the promoters of late myeloid genes such as lactoferrin (LF) in immature neutrophils where LF is not expressed. Upon induction towards neutrophil maturation C/EBPε binds to the LF promoter which correlates with LF expression. This suggests that C/EBPα associates with a DNA binding complex that negatively regulates LF gene expression.6 In an attempt to understand the mechanism underlying this observation, we hypothesized that the negative regulatory effect of C/EBPα may be due in part, to post-translational modification(s) of the C/EBPα protein. In this regard, C/EBPα was recently shown to be post-translationally modified by small ubiquitin-related modifier (SUMO) at a lysine residue (K159) within a region of the C/EBPα protein that can negatively affect transcriptional activity.7,8 Sumoylation at K159 in the C/EBPα protein is thought to prevent association of the SWI/SNF chromatin remodeling complex with C/EBPα, thereby hampering transactivation. Sumoylation of p42-C/EBPα has been shown to be increased in p30-C/EBPα expressing AMLs via a mechanism that upregulates Ubc9, a Sumo E2 conjugating enzyme essential for Sumoylation, in a p30-C/EBPα dependent manner.8 The authors however do not explain if upregulation of Ubc9 has a functional effect on sumoylation of p30-C/EBPα itself. We therefore chose to examine the impact of sumoylation on the activity of both isoforms of C/EBPα during myeloid differentiation and to ascertain their potential impact on leukemogenesis.
Materials and Methods
Tissue culture, transient transfection and luciferase assays
HEK293T (293T) cells were a gift from Dr. Ghosh (Yale University School of Medicine, New Haven, CT), MPRO cells obtained from Dr. Schickwann Tsai (University of Utah, Salt Lake City, UT) and NB4 cells were grown as previously described.6,9,10 All cells were maintained at 37°C in a humidified 5% CO2 incubator. HEK293T cells were transfected with the previously described LF89 lactoferrin reporter plasmid11 using FuGene (Roche Applied Sciences) according to the manufacturer’s protocol 0.5 µg pCMVβgal (Clontech, Palo Alto, CA, USA), was cotransfected as an internal control plasmid to monitor transfection efficiency as previously described.12 Transfected cells were incubated at 37°C in 5% CO2 for 16–20 hours. Luciferase activity was determined using a kit from Promega Biotech (Madison, WI) per manufacturer’s instructions. Co-transfection experiments included 1 µm of pCDNA3.1 C/EBPα p42, pCDNA3.1C/EBPαp30, (a gift from Dr. Peter Johnson, NCI-Frederick, MD), or pCMV Flag SUMO1 (a gift form Dr. N. Nucifora, University of Illinois, Chicago, USA). Mutant K159A p42 construct was a gift from Dr. K Miyake (Nayoga University, Japan) while K159A p30 was constructed by mutating a lysine (K) residue to an alanine residue (A) in the p30 cDNA using the QuikChange II method per manufacturers instructions (Stratagene, La Jolla, CA, USA). Luciferase expression levels were normalized to levels of β-galactosidase expression.12
RNA isolation and northern blot analysis
Total RNA was extracted from uninduced and ATRA-induced MPRO and NB4 cells as previously described, and 10 µm analyzed by northern blot analysis. Blots were probed with cDNA probes for mouse or human lactoferrin as described elsewhere.11
Oligonucleotide pulldown assay
This assay was conducted as previously documented.13 Briefly, uninduced and 48hour ATRA-induced MPRO or NB4 cells were lysed in NP-40 lysis buffer (50 mM Tris-Hcl (pH 8.0), 0.5% NP-40, 150 mM NaCl, 0.1 mM EDTA, 1mM DTT, 10% glycerol and the complete protease inhibitor cocktail (Roche). 200 µm of cell lysates or nuclear extracts (NE-PER kit) were incubated with 20 µm of double–stranded biotinylated oligonucleotide probes corresponding to the C/EBP site in the LF promoter. LF/C/EBP WT:5′GTCTATTGGGCAACAGG 3′ or mut LF/C/EBP 5′GTCTAGGTTTACGCAGG3′ DNA-protein complexes were then collected and resolved in a 4–12% gradient NuPage gel (Invitrogen). Western blotting was performed as described below. Antibodies used were: C/EBPε (Santa Cruz Biotech, sc-158, 1:3000), Sumo-1 (Santa Cruz Biotech, 1:1000) and C/EBPα (Santa Cruz Biotech, sc-61, 1:3000) antibodies in TBS-T incubated at 4°C overnight.
Chromatin immunoprecipitation analysis
ChIP analysis was performed essentially as previously described6 using the following oligomers: Human LF C/EBP: FP: TGGCGGGGAGTGGGAGGGAA; RP: AAGCTTGTCGACCGACTTGGCAAACGAAG and HNP: FP: 5′GTCAACTGTGTTAGGAGCCAT3′; RP: 5′CGTGCACAAGTGGACTTC3′ PCR products were subcloned into the pCRII vector (Invitrogen) and sequenced by standard dideoxy sequencing technology to confirm their identity.
Results
Oligonucleotide pull-down analysis of C/EBPα and C/EBPε during myeloid differentiation of ATRA-induced MPRO cells
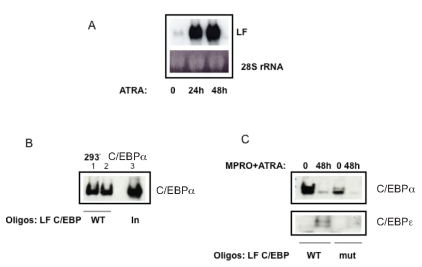
In order to analyze the in vivo protein-DNA interactions at the C/EBP binding site in the lactoferrin promoter during myeloid differentiation, we performed an oligonucleotide pull down assay (OPD), using MPRO cells as a model for neutrophil differentiation. MPRO cells are a factor-dependent promyelocytic cell line harboring a dominant negative RARa gene. These cells undergo neutrophil maturation upon addition of pharmacological levels of ATRA resulting in high levels of LF expression as evidenced in the northern blot (Figure1A). Biotinylated oligomers representing the LF/C/EBP binding site in the LF promoter were used. Nuclear extracts were first prepared from HEK293T cells transfected with a C/EBPα expression plasmid. The DNA-protein complexes formed were recovered using streptavidin–agarose beads and bound proteins resolved by SDS-PAGE and western blot analysis. As shown in Figure 1B (lanes 1 and 2), C/EBPα bound to the LF/C/EBP probe very efficiently and was enriched over input levels (lane 3, In). To assay for binding of endogenous C/EBPα to the probe, the experiments were then repeated using nuclear extracts prepared from uninduced and ATRA-induced MPRO cells, a model for normal in vitro neutrophil maturation. Extracts were incubated with either a wild type (WT) or a mutant (mut) biotinylated LF/C/EBP probe. As shown in Figure 1C, C/EBPα binds to the WT LF/C/EBP probe in uninduced MPPO cells. This binding was significantly reduced in ATRA-induced MPRO cell extracts. As expected, significantly less C/EBPα binding to the mutant LF/C/EBP probe was observed. In contrast, an increase in C/EBPε binding to the LF/C/EBP WT probe was observed upon 48 h ATRA-induction (Figure 1C, WT oligos, lower panel), concomitant with LF expression (Figure 1A, 24 and 48hours in ATRA).
Figure 1.
Binding of C/EBPα to the Lactoferrin (LF) promoter and expression of the LF gene during neutrophil maturation in ATRA-induced MPRO cells. A. Northern blot analysis of uninduced and ATRA-induced MPRO cells demonstrating an increase in LF mRNA expression upon ATRA-treatment of cells for 24 and 48 hours. Ethidium bromidestained 28S rRNA in the lower panel serves as a loading control. B. Oligonucleotide pull down assay using LF C/EBP oligomers and extracts prepared from HEK293T cells. Nuclear extracts prepared from HEK293T cells overexpressing a C/EBPα expression plasmid were incubated with biotinylated wildtype LF/C/EBP oligomers. The DNA-protein complexes were recovered using streptavidin–agarose beads and the bound proteins resolved by SDS-PAGE and western blot analysis. The blot was probed with a C/EBPα antibody (lanes 1 and 2). 10% of the input sample is represented in lane 3 (In). C. Oligonucleotide pulldown assay using LF/ C/EBP oligomers and nuclear extracts from Uninduced and induced MPRO cells. Nuclear extracts prepared from uninduced (0) and ATRA-induced (48h) MPRO cells (a murine myeloid cell line for normal neutrophil maturation) were incubated with wild type (WT) or mutant (mut) biotinylated C/EBP site (LF/C/EBP) oligomers from the LF promoter. The DNA-protein complexes were recovered using streptavidin–agarose beads and the bound proteins resolved by SDS-PAGE and western blot analysis. The blot was probed sequentially with C/EBPα and C/EBPε antibodies.
This experiment suggests that C/EBPα is bound to the LF promoter when the LF gene is not expressed, and that induction of neutrophil maturation is associated with reciprocal changes in the binding of C/EBPα and C/EBPε to the LF/C/EBP binding site, coincident with LF gene expression. We have previously demonstrated this pattern of C/EBP binding to the LF promoter in uninduced and induced MPRO cells by ChIP analysis.6
Sumoylation of C/EBPα limits its ability to transactivate the LF promoter
C/EBPα is an established transcriptional activator. Our observations in Figure1 however, suggest that it negatively regulates the expression LF during myeloid maturation. We postulated that this negative regulation by C/EBPα with respect to LF expression could reflect posttranslational modifications of the C/EBPα protein and/ or changes in associations of C/EBPα with other interacting proteins. As previously noted, human C/EBPα is modified by SUMO-1 at lysine residue 159 within a conserved negative subdomain.14 Sato et al have recently demonstrated that sumoylation of C/EBPα dramatically decreases its ability to transactivate the liver-specific albumin gene. A second study also demonstrated a two-fold reduction in the activity of a generic C/EBP reporter gene in the presence of Sumo-1.8 In order to test whether sumoylation of C/EBPα alters activation of the LF promoter, we co-transfected HEK293 cells with a LF promoter-reporter gene plasmid (LF89) and expression plasmids for C/EBPα and Sumo-1. As shown in Figure 2A, p42 C/EBPα transactivated the LF89 plasmid 7 fold. However, co-transfection with Sumo-1 markedly reduced reporter gene activity (P<0.001). This reduction in transactivation was not observed with a C/EBPα mutant plasmid incapable of being sumoylated (K159A) (Figure 2A). In fact, this mutant increased LF promoter activity two fold compared with the p42 C/EBPα expression plasmid, possibly reflecting loss of repression due to endogenous SUMO activity, even in the absence of the transfected exogenous SUMO-1 expression plasmid. These observations suggest that sumoylation of p42 C/EBPα blocks its ability to transactivate the myeloid-specific LF gene.
Figure 2.
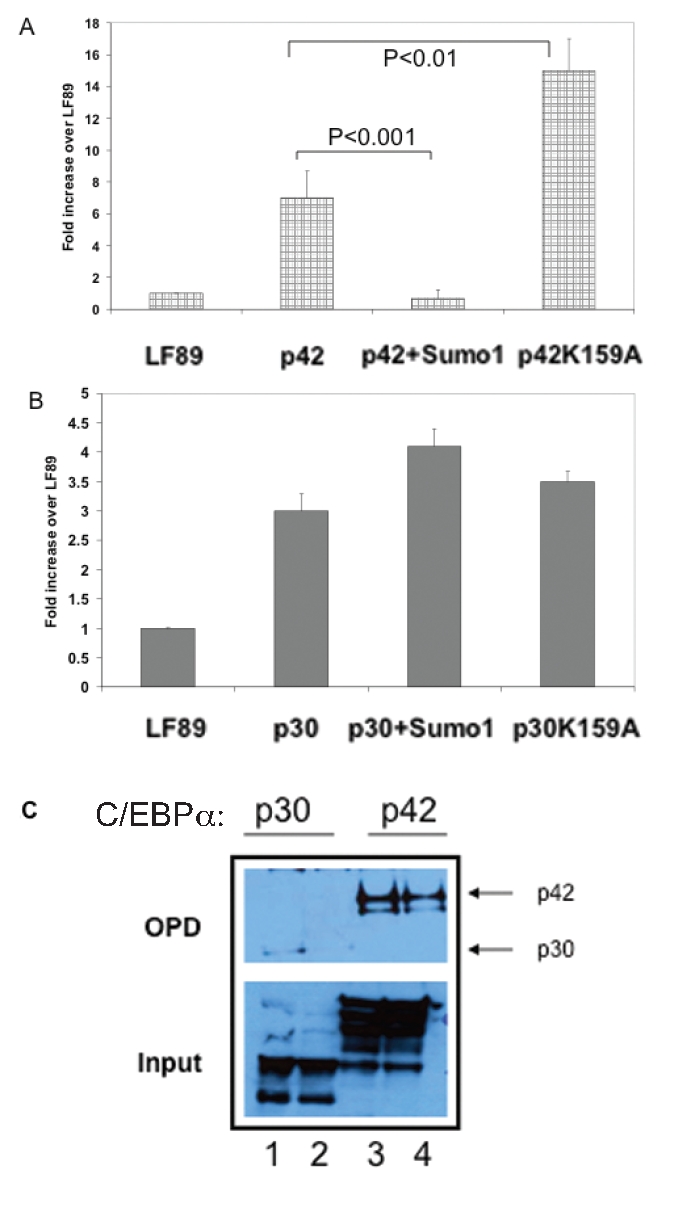
Transient transfection analysis of Lactoferrin promoter (LF89) harboring a C/EBP site in HEK293T cells with expression plasmids for the two C/EBPα isoforms and SUMO-1. A. Transient transfection analysis using a lactoferrin promoter plasmid (LF89) harboring a C/EBP site. HEK293T cells were transiently cotransfected with LF89 and expression plasmids for p42 C/EBPα (wild-type) or p42 K159A (mutant) with and without SUMO-1. Normalized luciferase (to that of β -galactosidase, see Materials and Methods) values have been represented as a fold change over the enzyme activity of LF89 promoter plasmid alone (equal to 1) The figure represents normalized mean+/− s.e. obtained from 3independent experiments, each performed in triplicate. B. Transient transfection analysis of LF89 with expression plasmids for p30C/EBPα (wild-type or p30 K159A mutant) with and without SUMO-1. Analysis was done as indicated above. C. Oligonucleotide pull down assay of LF89 oligomers with extracts prepared from HEK293T cells overexpressing p30C/EBPA (lanes 1 and 2) and p42C/EBPα (lanes 3 and 4). Extracts were incubated with wild type biotinylated C/EBP site (LF/C/EBP) oligomers from the LF promoter. The DNA-protein complexes were recovered using streptavidin–agarose beads and the bound proteins resolved by SDS-PAGE and western blot analysis and probed with a C/EBPα specific antibody. Lower panels represents 5% of each of the input samples.
Sumoylation of p30 C/EBPα does not suppresses transactivation of the LF promoter
As a result of the differential utilization of alternate translational start codons, the C/EBPα gene yields two proteins, p42kD (full length) and p30kD (truncated) that differ from one another at the N-terminus wherein lie potent transactivation domains. The p30 isoform is a less efficient transactivator than its full length counterpart and is considered a dominant negative form of C/EBPα.3 P30 has been shown to be overexpressed in most cases of acute myeloid leukemia (AML).2
To determine whether sumoylation has an impact on the ability of p30 C/EBPα to transactivate the lactoferrin promoter LF89, we performed transient cotransfection analysis in HEK293 cells. The p30 isoform transactivated LF89 about 2.5- fold less efficiently than the p42 isoform (compare Figures 2A and 2B).
P30 C/EBPα transactivated the LF89 promoter reporter 3-fold above LF89 alone. However, unlike the p42 C/EBPα plasmid, cotransfection of p30 C/EBPα with a Sumo-1 expression plasmid did not significantly change LF89 activity, nor did the sumoylation- blocking mutant p30 C/EBPα K159A (Figure 2B). This suggests that p30 C/EBPα is much less sensitive to sumoylation than p42 C/EBPα.
To confirm binding of the C/EBPα isoforms to the previously defined LF C/EBP cis element, we performed an OPD assay with the LF/C/EBP probe and nuclear extracts prepared from both C/EBPα p42- and p30- overexpressing HEK293T cells. Despite abundant expression of both C/EBP isoforms in the respective HEK293T cells (Figure 2C, lower panel), the p30 isoform demonstrated negligible binding to the LF/C/EBP probe (Figure 2C, lanes 1 and 2) as compared to the p42 isoform (Figure 2C, lanes 3 and 4). This reduced DNA binding of p30 has been demonstrated with other C/EBPα specific downstream targets such as the CSF3 receptor promoter.2
Taken together, these data suggest that sumoylation controls the transactivition activity of the p42C/EBPα isoform but not that of the dominant negative p30C/EBPα isoform.
Binding of sumoylated C/EBPα to the lactoferrin promoter decreases upon neutrophil differentiation in ATRA-induced MPRO cells
Our finding that sumoylation of p42C/EBPα reduces its ability to transactivate LF89 suggested a possible role for the sumoylated form of the protein in negative regulation of the LF gene early in neutrophil development. To test this hypothesis, it was necessary to demonstrate an inverse relationship between the presence of sumoylated C/EBPα and the activity of the LF promoter in a physiologic context. An OPD assay was performed on uninduced and induced MPRO cells, with western blot analysis for sumoylated C/EBPα. The data demonstrate that SUMO-C/EBPα is bound to the LF/C/EBP probe in uninduced MPRO cells (Figure 3, lane 3), and that the binding is significantly decreased by the induction of neutrophil maturation (Figure 3, lane 4). Of note, while the total amount of SUMO-C/EBPα remains about the same during neutrophil development in the MPRO cells, (Figure 3, lanes 1 and 2), the amount of SUMO-C/EBPa bound to the LF/C/EBP probe decreases with maturation, confirming a reciprocal relationship to LF expression during neutrophil development.
Figure 3.
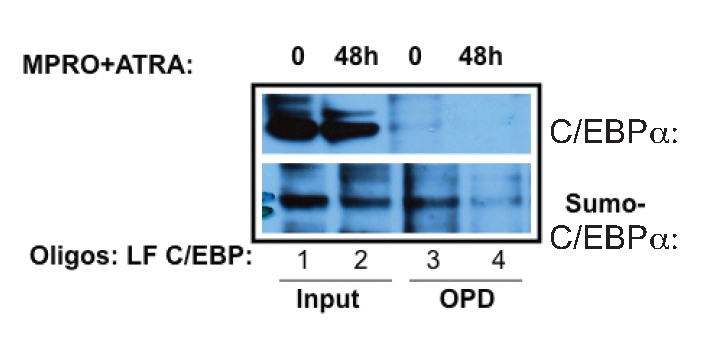
Oligonucleotide pulldown analysis of C/EBPα and sumoylated-C/EBPα during ATRA-induction of MPRO cells. Nuclear extracts prepared from uninduced (0) and ATRA-induced (48h) MPRO cells were incubated with biotinylated C/EBP site (LF/C/EBP) oligomers from the LF promoter and OPD conducted as above. The blot was probed sequentially with anti-C/EBPα (lanes 3 and 4, top panel) and anti-SUMO-1 (lanes 3 and 4, bottom panel) antibodies. Input samples are represented in lanes 1 and 2 of the top (C/EBPα) and bottom (SUMO-1) panels.
Sumoylated C/EBPα remains bound to the LF/C/EBP site in the acute promyelocytic cell line NB4
NB4 is an acute promyelocytic leukemia cell line that contains the t(15;17) PML-RARa translocation and undergoes neutrophil maturation with addition of ATRA. We have previously demonstrated that following ATRA-induction, NB4 cells uniformly fail to express not only LF, but all the secondary granule protein genes.9 We further demonstrated that this failure of expression occurs despite appropriate binding of C/EBP factors known to upregulate LF expression. We therefore investigated whether sumoylation of the abundantly available C/EBPα in NB4 cells contributed to the failure of LF gene expression. In order to test this hypothesis, we first performed ChIP analysis using nuclear extracts prepared from uninduced and 48 hour ATRA-induced NB4 cells. As shown in Figure 4A (upper panel), ChIP analysis using a C/EBPα antibody indicated binding of C/EBPα to the control HNP (human neutrophil peptide also known as a-defensin) promoter in uninduced but not ATRA-induced NB4 cells. We have previously shown that the HNP gene, encoding a late primary granule protein, is in fact expressed in ATRA-induced NB4 cells.6 Binding of C/EBPα to the LF promoter, on the other hand, persisted in both uninduced and ATRA-induced NB4 cells corresponding to the absence of LF expression in ATRA-induced NB4 cells. Thus, loss of C/EBPα- binding to myeloid promoters (HNP) in the maturing neutrophil is concomitant with expression of these genes, while persistent binding parallels nonexpression, as is the case for LF.
Figure 4.
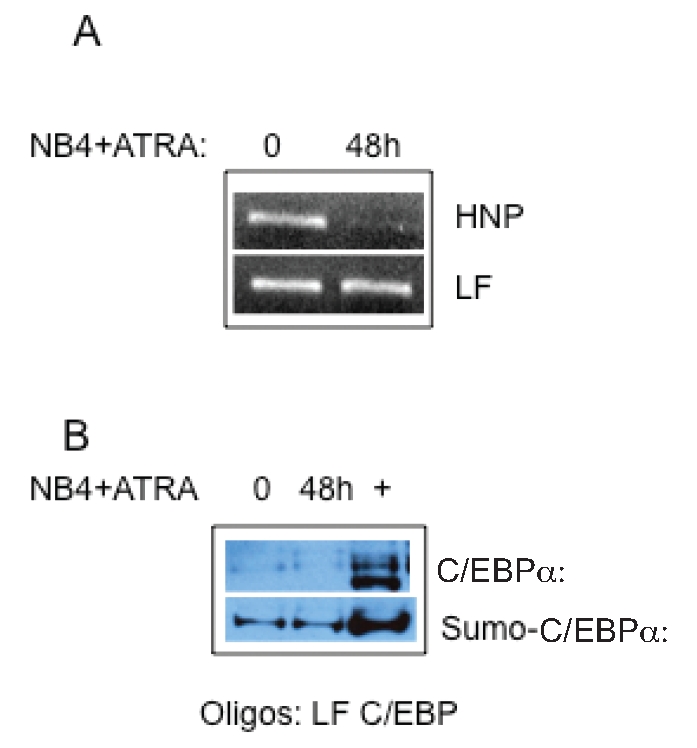
Binding of Sumoylated-C/EBPA to the LF C/EBPα oligomers in ATRA-induced NB4 leukemic cells: A. Chromatin immunoprecipitation was performed from uninduced (0) and ATRA-induced (48h) NB4 cells using antibodies specific for C/EBPα . The precipitated chromatin was analyzed using primers specific for the human LF (LF) promoter (bottom panel) and the HNP promoter (top panel). This experiment was repeated three times. B. Oligonucleotide pulldown assay of C/EBPα and sumoylated-C/EBPα during ATRA-induction of NB4 cells. Nuclear extracts prepared from uninduced (0) and 48h ATRA-induced NB4 cells were incubated with biotinylated C/EBP site (LF/C/EBP) oligomers from the LF promoter and oligonucleotide pulldown assay conducted as above. The blot was probed sequentially with anti-C/EBPα (top panel) and anti-SUMO-1 (bottom panel) antibodies. + represents a positive control (uninduced MPRO extracts).
In order to determine whether C/EBPα bound to the LF promoter in NB4 cells is sumoylated, we next performed an OPD assay using the LF/C/EBP oligonucleotide and extracts prepared from uninduced and ATRA-induced NB4 cells. As shown in Figure 4B (top panel), C/EBPα binds to the LF/C/EBP site in both uninduced and ATRA-induced NB4 cells. In addition, the bound C/EBPα appears to be sumoylated in both uninduced and ATRA-induced cells (Figure 4B, bottom panel). This contrasts with the loss of C/EBPα binding upon induction observed in the ATRA-responsive LF- expressing MPRO cell line (Figure 1B). Thus, persistent association of sumoylated C/EBPα with the LF promoter in NB4 cells may contribute to the lack of expression of this gene in leukemic cells.
Discussion
In this study we have demonstrated that the sumoylation status of the C/EBPα protein changes during normal myeloid maturation, and that this is accompanied by changes in LF gene expression. We postulate that changes in the sumoylation status of C/EBPα contribute to a switch that regulates its transcriptional activity during normal neutrophil development.
Sumoylation or Sumo modification is a post translational modification wherein SUMO (small ubiquitin – like modifier) is covalently and reversibly conjugated to its target proteins by a series of sequential and evolutionarily conserved enzymatic reactions. This results in alteration of target protein function and cellular fate. 1,15 The list of sumoylated proteins is rapidly increasing and includes several that play pivotal roles in processes as diverse as transcriptional regulation, nuclear targeting, sub-nuclear targeting, chromatin structure and genome stability.
We demonstrate here that the C/EBPα p42 isoform transactivates the myeloid-specific lactoferrin gene promoter approximately 3 fold more efficiently than p30 C/EBPα. Cotransfection of p42 with an expression plasmid for SUMO-1 however, significantly reduces LF89 driven luciferase activity compared to transfection of p42 alone. In comparison, SUMO-1 has little or no effect on the ability of p30C/EBPα to drive LF89 luciferase activity. This suggests that sumoylation alters the transcriptional activity of p42 but not p30 during myeloid differentiation and further indicates that sumoylation of the two C/EBPα isoforms may have functionally disparate effects, possibly due to differential protein-protein binding properties of each sumoylated isoform. In this context, in a recent study, Geletu et al.8 showed that overexpression of p30 C/EBPα in an erythroleukemia cell line resulted in upregulation of Ubc9, a sumo E2 conjugating enzyme, which is essential for sumoylation.1 The authors demonstrate that upregulation of Ubc9 sumoylates the p42 isoform, thereby rendering it inactive with respect to transactivation of a C/EBP- responsive reporter plasmid in transactivation assays. The authors further suggest that sumoylation of p42 C/EBPα in AML may be the result of increased expression of the p30 isoform. Our data extend this finding by showing that sumoylated p42 remains persistently bound to the LF promoter in the acute promyelocytic leukemia cell line NB4 following ATRA-induction. This observation is correlated with lack of lactoferrin expression in these cells. Whether binding of sumoylated p42 C/EBPα to the LF promoter persists due to aberrant activity of the desumoylating SENP enzymes or whether an increase of p30 C/EBPα expression in these leukemic cells causes increased rates of p42 sumoylation remains to be clarified. Approximately 10–15% of AML patients have mutations in C/EBPα2,16,17 that often result in an increase in expression of p30 C/EBPα. In fact, a recent study has shown that mice homozygously expressing p30 from the C/EBPα locus develop AML with complete penetrance.5 Changes in the ratio of p42:p30 have been found to contribute to tipping the scales from normal myelopoiesis to pre-leukemic or even leukemic myelopoiesis.2 While the p30 isoform is thought to exert a dominant negative effect on its full length counterpart (p42), the mechanisms underlying this effect remain unclear. Our data demonstrate that unlike p42, the p30 C/EBPα isoform remains unresponsive to sumoylation and binds poorly to the LF promoter in a C/EBP binding site pulldown assay. The limited ability of p30 to bind to other myeloid specific promoters such as the CSF3 receptor promoter, has also been previously reported.2 This evidence strongly suggests that differences in activity between the two C/EBPα isoforms play a role in the shift from normal myeloid development to leukemogenesis.
Acknowledgements:
the authors would like to thank Drs Peter Johnson, G. Nucifora and K Miyake for reagents used in this study and to members of the Berliner Lab for helpful discussions. This work was supported by NIH PO1-HL63357.
References
- 1.Khanna-Gupta A. Sumoylation and the function of CCAAT enhancer binding protein alpha (C/EBP alpha) Blood Cell Mol Dis. 2008;41:77–81. doi: 10.1016/j.bcmd.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pabst T, Muller B, Zhang P. Dominant negative mutations of CEBPA encodong CCAAT/enhancer binding protein-a (C/EBPa), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 3.Muller B, Pabst T. C/EBPa and the patho-physiology of acute myeloid leukemia. Curr Opin Hematol. 2006;13:7–14. doi: 10.1097/01.moh.0000190110.08156.96. [DOI] [PubMed] [Google Scholar]
- 4.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27:619–28. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirstetter P., Schuster MB, Bereshchenko O, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Khanna-Gupta A, Zibello T, Sun H, et al. Chromatin immunoprecipitation (ChIP) studies indicate a role for CCAAT enhamcer binding proteins alpha and epsilon (C/EBPa and C/EBPε) and CDP/cut in myeloid maturation induced lactoferrin gene expression. Blood. 2003;101:3460–68. doi: 10.1182/blood-2002-09-2767. [DOI] [PubMed] [Google Scholar]
- 7.Sato K Y., Miyake H, Kaneoka H, Iijima S. Sumoylation of CCAAT/enhancer binding protein a and its functional roles in hepatocyte differentiation. J Biol Chem. 2006;281:21629–39. doi: 10.1074/jbc.M600852200. [DOI] [PubMed] [Google Scholar]
- 8.Geletu M, Balkhi M, Peer Zada A, et al. Behre, Target proteins of C/EBPalphap30 in AML: C/EBPalphap30 enhances sumoylation of C/EBPalphap42 via up-regulation of Ubc9. Blood. 2007;110:3301–9. doi: 10.1182/blood-2007-01-071035. [DOI] [PubMed] [Google Scholar]
- 9.Khanna-Gupta A, Kolibaba K, Zibello TA, Berliner N. NB4 cells show bilineage potential and an aberrant pattern of neutrophil secondary granule protein gene expression. Blood. 1994;84:294–302. [PubMed] [Google Scholar]
- 10.Lawson ND, Krause DS, Berliner N. Normal neutrophil differentiation and secondary granule gene expression in the EML and MPRO cell lines. Exp Hematol. 1998;26:1178–85. [PubMed] [Google Scholar]
- 11.Khanna-Gupta A, Zibello TA, Simkevich C, et al. Sp1 and C/EBP are necessary to activate the lactoferrin gene promoter during myeloid differentiation. Blood. 2000;95:3734–41. [PubMed] [Google Scholar]
- 12.Khanna-Gupta A, Zibello T, Kolla S, et al. CCAAT displacement protein (CDP/cut) recognizes a silencer element within the lactoferrin gene promoter. Blood. 1997;90:2784–95. [PubMed] [Google Scholar]
- 13.Khanna-Gupta A, Zibello T, Idone V, et al. Human neutrophil collagenase expression is C/EBP-dependent during myeloid development. Exp Hematol. 2005;33:42–52. doi: 10.1016/j.exphem.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian L, Benson M, Iniguez-Lluhi J. A synegy control motif within the attenuator domain of CCAAT/enhancer binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or Sumo-3. J Biol Chem. 2003;278:9134–41. doi: 10.1074/jbc.M210440200. [DOI] [PubMed] [Google Scholar]
- 15.Fu CT, Zhu KY, Mi JQ, et al. An evolutionarily conserved PTEN-C/EBPα-CTNNA1 axis controls myeloid development and transformation. Blood. 2010;115:4715–24. doi: 10.1182/blood-2009-11-255778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gombart A, Hofmann W, Kawano S, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–40. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- 17.Green C, Koo KK, Hills RK, et al. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol. 2010;28:2739–47. doi: 10.1200/JCO.2009.26.2501. [DOI] [PubMed] [Google Scholar]