Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder with a specific genetic abnormality, the Philadelphia chromosome (Ph) translocation t(9;22)(q34;q11) generating the BCR-ABL1 gene fusion. The role of the Philadelpia chromosome translocation t(9;22)(q34;q11) in CML development is well documented and early in the chronic phase of the disease it is the sole chromosome anomaly in the majority of patients with CML. By contrast, disruption of the mixed lineage leukemia (MLL) gene located on 11q23 is a recurrent genomic change in acute leukemia and is particularly prevalent in secondary, acute myeloblastic leukaemia (AML), arising following cytotoxic treatment.While the dual combination of 11q23 rearrangement and the Ph translocation has been described in a several CML patients, the rearrangement of MLL gene was confirmed only in rare cases.1–4 We report a case of combination of the Philadelphia translocation t(9;22)(q34;q11) and t(11;19) (q23;p13.1) in a CML patient who developed accelerated phase of the disease while on imatinib. The patient, a 28-years old female was diagnosed with Ph-positive CML in December 2003. She was treated with hydroxyurea until January 2006 when she was pregnant 26 weeks. Treatment with interferon-α (IFN) was started and she delivered a normal baby, but failed to achieve complete cytogenetic response. From September 2006, she switched to imatinib with an initial dose of 400 mg/day resulting in a complete hematological and cytogenetic remission 6 months later. She maintained her complete response until March 2008, when 25% of the examined metaphases showed the Philadelphia translocation. The initial dose was increased to 600 mg/day, but 6 month later she developed accelerated phase of the disease. A complete blood count revealed: hemoglobin, 8.9. g/dL, leukocyte count 11.6×109/L (neutrophils 38%, lymphocytes 26%, monocytes 23%, myelocytes 3%, metamyelocytes 6%; blasts 4%) and platelets 125×109/L. Bone marrow aspirate showed hypercellularity with a predominance of myeloid cells including including 17% myeloblasts (promyelocytes 1.5%, myelocytes 22%, metamyelocytes 27%, lymphocytes 7.5%, poly 12%, eryhroblasts 13%). Chromosome analysis performed from bone marrow aspiration revealed 100% Ph-positive metaphases and a new cytogenetic finding, a t(11;19)(q23;p13) in 18 out of the 20 examined metaphases. FISH analysis showed 95% BCR-ABL1 positive cells. MLL rearrangement was identified using the LSI MLL (11q23) Dual Color Break Apart Rearrangement probe (Vysis) revealing 80% of cells with MLL rearrangement. The disruption of MLL was confirmed in metaphases demonstrating that the distal part of the MLL was juxtaposed to the der(19) chromosome, adjacent to the centromeric region (Figure 1). Treatment with Tasigna (400 mg/day) was started, but 2 months later her leukocyte count increased to 100×109/L with 19% blasts. Serum biochemistry was remarkable for lactatdehydrogenase 1014 U/L and GGT 70 U/L. One month later, she developed fever, progressive dyspnoea and tachycardia and she ultimately died one month later.
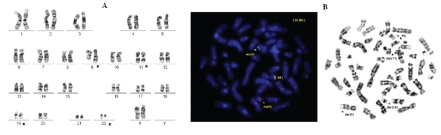
Figure 1.
Karyotype of the patient showing simultaneous occurrence of t(9;22)(q34;q11) and t(11;19)(q23;p13.1) (A). FISH using a MLL dual-color, break-apart probe revealing the juxtaposition of the distal part of the MLL gene from chromosome 11 to the der(19) chromosome, adjacent to the centromeric region (red signal) (B).
While CML progression is frequently accompanied by nonrandom secondary changes, the mechanism(s) responsible for severe block in differentiation and apoptosis remains poorly understood. The acquisition of t(11;19) (q23;p13.1) and its underlying MLL rearrangement in our patient is an unusual finding and raises several questions. Firstly, the t(11;19)(q23;p13.1) fusing the MLL gene on chromosome 11q23 with the ELL (MEN) gene at 19p13.1 is found in a large proportion of AML patients who have been treated by chemotherapy,5,6 raising the possibility that the acquisition of MLL rearrangement in our patient is the direct consequence of toxic treatment. However; in our case hydroxyurea and interferon was administrated, followed by imatinib therapy for about 2 years, arguing against a direct genotoxic effect of imatinib. In addition, the aberration is not limited to chemotherapy-related mutagenesis and has also been identified in association with de novo myeloid leukemias. Therefore, it is more likely that the cytogenetic evolution observed in our case may represent the changing natural history of CML.
The mechanisms is unclear, however considering MLL's central role as a major player in leukemia, it is likely that the t(11;19) (q23;p13.1) observed in our patient is a form of cytogenetic evolution that play a direct role in CML progression in the same way that results in acute leukemia de novo. The critical contribution of MLL-ELL fusion protein on the proliferative potential of myeloid progenitors as well as its causal role in the genesis of acute myeloid leukemias was demonstrated using transgenic animal models,7,8 though the precise mechanism remains elusive. Further question may be raised regarding the direct or indirect role of BCR-ABL1 in transition, suggesting that the acquisition of MLL rearrangement may be the consequence of genetic instability promoted by BCR-ABL1. Noteworthy, our patient received treatment with hydroxyurea only for about 2 years. While on imatinib therapy, she maintained her complete cytogenetic response for only 18 months. She remained BCR-ABL1 positive by molecular methods during the course of the disease. It is unclear, if and how incomplete supression of BCR-ABL1 tyrosine kinase activity facilitated the development of MLL disruption. As MLL fusion proteins are dependent on hyperactive signalling from stem cell receptor tyrosine kinases, it can be hypothesed that kinase activity in our case could be of crucial importance. In addition, it has been shown, that the loss of ELL function and overexpression of the MLL-ELL have anti-apoptotic effects. Therefore, the acquisition of t(11;19) could result in survival of cells that otherwise may have been destined to die, which could contribute to the development and maintenance of imatinib resistance in our patient. A case similar to ours was reported by Suzuki and all,9 describing a 33-years old CML patient who was initially treated with interferon-α and hydroxyurea but failed to achieve complete cytogenetic response. One year later, the patient developed blastic phase and a secondary karyotypic change of t(11;19) (q23;p13.3). Treatment with chemo therapy and imatinib was administrated, but the patient failed to achieve remission and died five months later. Similar to our case, the clinical course of this patient was aggressive and unresponsive to imatinib treatment.
The dual combination of 11q23 aberration and Ph is an infrequent phenomenon and has only rarely been reported in CML. These cases have been reported almost exclusively in association with disease acceleration or onset of blast crisis, suggesting the direct role of MLL in transformation. As the chromosomal translocation t(11;19)(q23;p13) is a subtle genetic anomaly and may be easily overlooked by standard cytogenetic analysis, our case further reinforce the importance of FISH in revealing cryptic balanced translocations involved in CML progression.10
References
- 1.Barbouti A, Johansson B, Höglund M, Mauritzson N, et al. Multicolor COBRA-FISH analysis of chronic myeloid leukemia reveals novel cryptic balanced translocations during disease progression. Genes Chromosomes Cancer. 2002;35:127–137. doi: 10.1002/gcc.10099. [DOI] [PubMed] [Google Scholar]
- 2.Nishii K, Usui E, Sakakura M, Miyata E, et al. Additional t(11;17)(q23;q21) in a patient with Philadelphia-positive mixed lineage antigen-expressing leukemia. Cancer Genet Cytogenet. 2001;126(1):8–12. doi: 10.1016/s0165-4608(00)00382-4. [DOI] [PubMed] [Google Scholar]
- 3.Otero L, Moellmann AC, Pombo-de-Oliveira MS, Ornellas MH, et al. Additional t(1;11)(q21;q23) with mixed lineage leukemia rearrangement in T-blastic crisis of a Ph-positive chronic myeloid leukemia. Eur J Haematol. 2007;79:179–181. doi: 10.1111/j.1600-0609.2007.00884.x. [DOI] [PubMed] [Google Scholar]
- 4.Royer-Pokora B, Hildebrandt B, Redmann A, Herold C, et al. Simultaneous occurrence of a t(9;22) (Ph) with a t(2;11) in a patient with CML and emergence of a new clone with the t(2;11) alone after imatinib mesylate treatment. Leukemia. 2003;17:807–810. doi: 10.1038/sj.leu.2402877. [DOI] [PubMed] [Google Scholar]
- 5.Moorman AV, Hagemeijer A, Charrin C, Rieder H, Secker-Walker LM. The translocations, t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3): a cytogenetic and clinical profile of 53 patients. Leukemia. 1998;12:805–10. doi: 10.1038/sj.leu.2401016. [DOI] [PubMed] [Google Scholar]
- 6.Huret JL. t(11;19)(q23;p13.1) Atlas Genet Cytogenet Oncol Haematol. 1997 Dec; URL: http://AtlasGeneticsOncology.org/Anomalies/t1119ELL.html.
- 7.DiMartino JF, Miller T, Ayton PM, Landewe T, et al. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood. 2000;96:3887–93. [PubMed] [Google Scholar]
- 8.Lavau C, Luo RT, Du Ch, Thirman MJ. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci USA. 2000;26(97):10984–9. doi: 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki K, Sugawara T, Kowata S, Utsugizawa T, et al. Uncommon karyotypic abnormality, t(11;19)(q23;p13.3), in a patient with blastic phase of chronic myeloid leukemia. Cancer Genet Cytogenet. 2004;150:159–63. doi: 10.1016/j.cancergencyto.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer Lisa G., et al. ISCN 2009; an international system for human cytogenetic nomenclature (2009) S. Karger AG. 2009 [Google Scholar]