Abstract
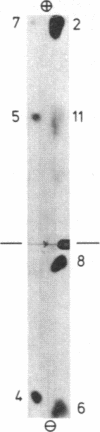
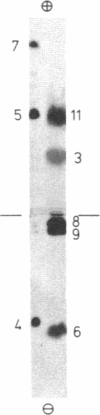
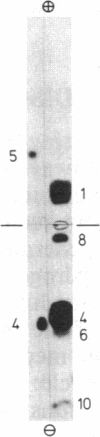
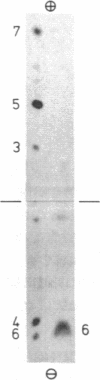
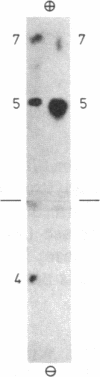
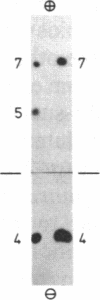
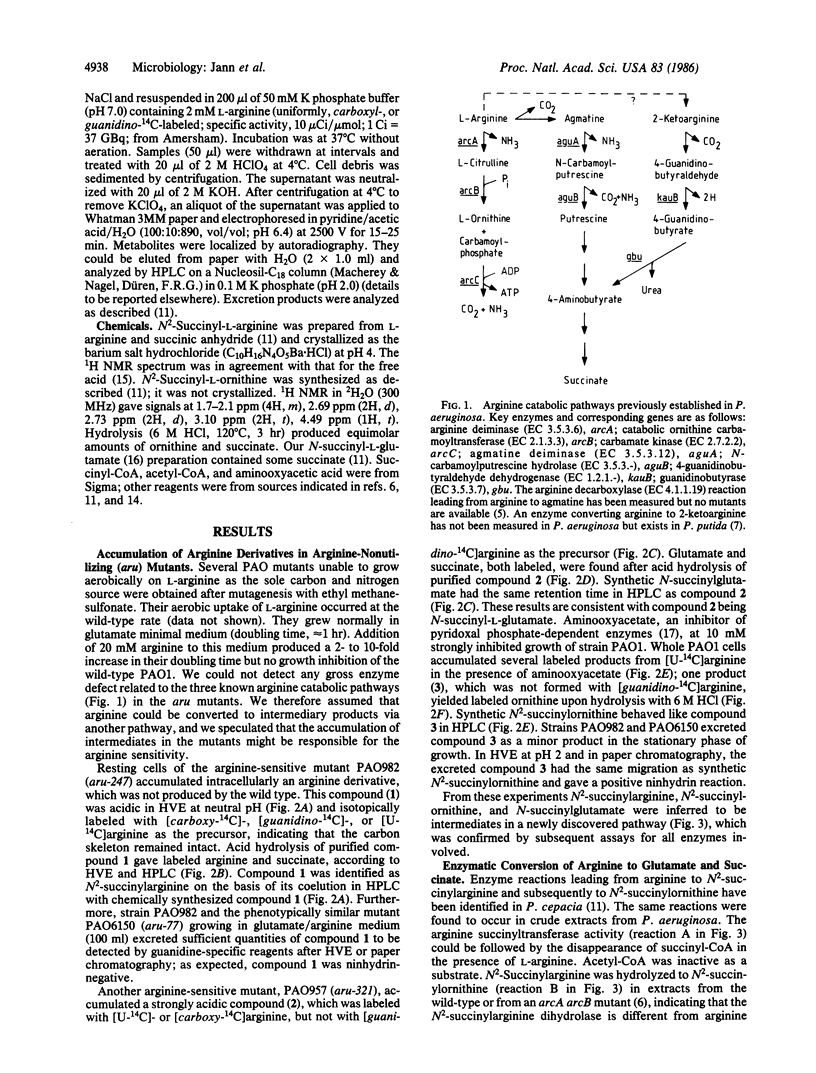
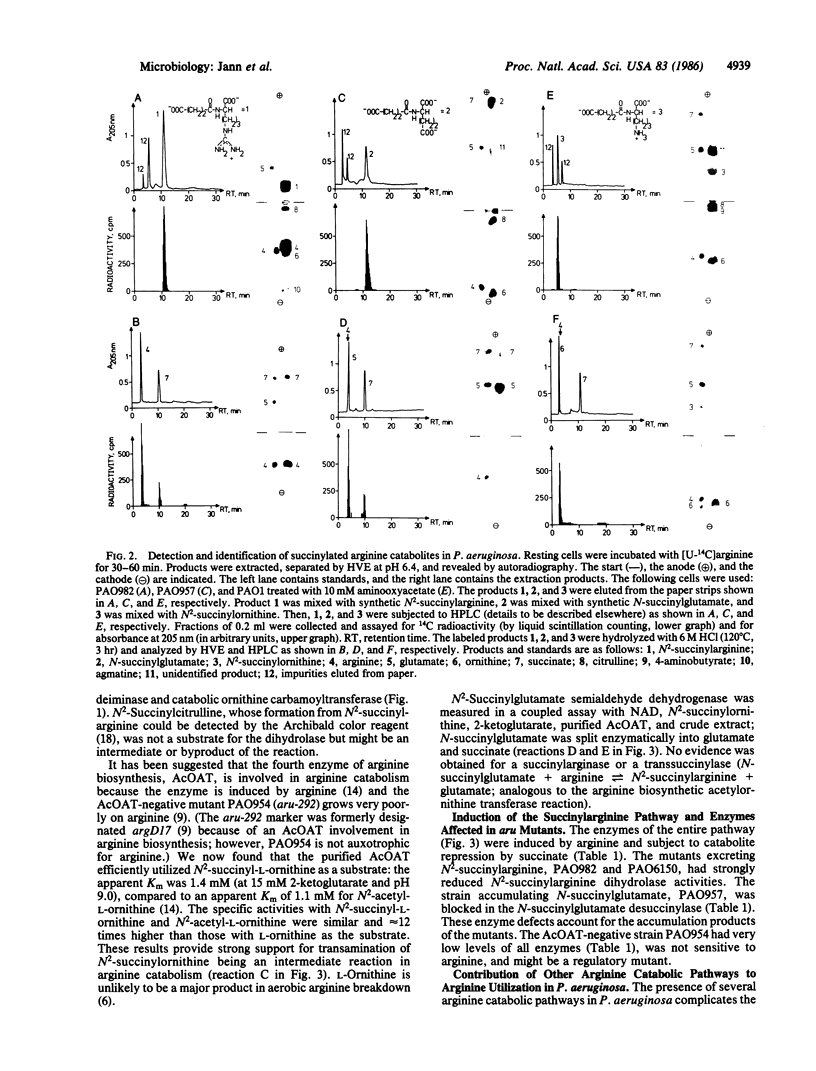
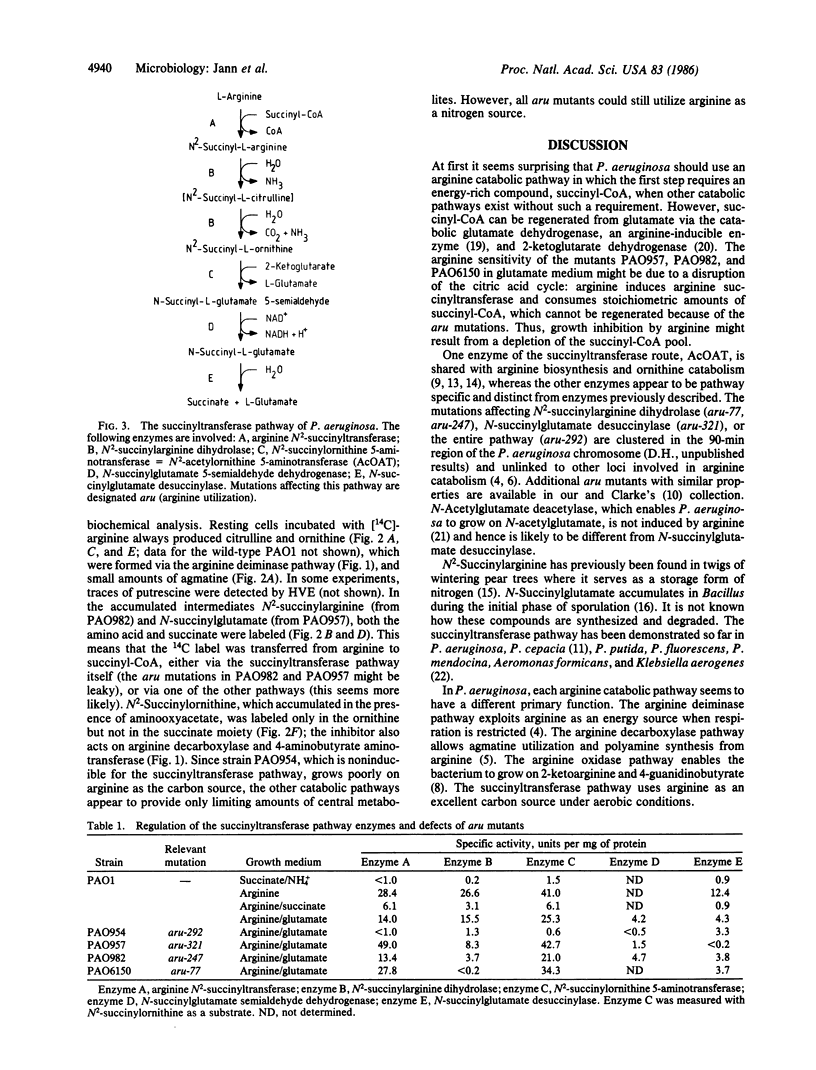
Arginine-nonutilizing (aru) mutants of Pseudomonas aeruginosa strain PAO converted L-arginine to N2-succinylarginine or N-succinylglutamate, which were identified by high-voltage electrophoresis and HPLC. Addition of aminooxyacetate, an inhibitor of pyridoxal phosphate-dependent enzymes, to resting cells of the wild-type PAO1 in arginine medium led to the accumulation of N2-succinylornithine. Enzyme assays with crude P. aeruginosa extracts established the following pathway: L-arginine + succinyl-CoA → N2-succinylarginine → N2-succinylornithine → N_succinylglutamate 5-semialdehyde → N-succinylglutamate → succinate + glutamate. Succinyl-CoA may be regenerated from glutamate via 2-ketoglutarate. L-Arginine induced the enzymes of the pathway, and succinate caused catabolite repression. Purified N2-acetylornithine 5-aminotransferase (N2-acetyl-L-ornithine: 2-oxoglutarate aminotransferase, EC 2.6.1.11), an arginine biosynthetic enzyme, efficiently transaminated N2-succinylornithine; this explains the enzyme's dual role in arginine biosynthesis and catabolism. The succinylarginine pathway enables P. aeruginosa to utilize arginine efficiently as a carbon source under aerobic conditions, whereas the other three arginine catabolic pathways previously established in P. aeruginosa fulfill different functions.
Keywords: aru mutants, N2-succinylarginine, N2-succinylornithine, metabolic versatility
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AUBERT J. P., MILLET J., PINEAU E., MILHAUD G. [N-Succinyl-L-glutamic acid in Bacillus megaterium during sporulation]. Biochim Biophys Acta. 1961 Aug 19;51:529–537. doi: 10.1016/0006-3002(61)90610-2. [DOI] [PubMed] [Google Scholar]
- Früh H., Leisinger T. Properties and localization of N-acetylglutamate deacetylase from Pseudomonas aeruginosa. J Gen Microbiol. 1981 Jul;125(1):1–10. doi: 10.1099/00221287-125-1-1. [DOI] [PubMed] [Google Scholar]
- Früh R., Haas D., Leisinger T. Altered control of glutamate dehydrogenases in ornithine utilization mutants of Pseudomonas aeruginosa. Arch Microbiol. 1985 Mar;141(2):170–176. doi: 10.1007/BF00423280. [DOI] [PubMed] [Google Scholar]
- Haas D. Genetic aspects of biodegradation by pseudomonads. Experientia. 1983 Nov 15;39(11):1199–1213. doi: 10.1007/BF01990357. [DOI] [PubMed] [Google Scholar]
- Haas D., Matsumoto H., Moretti P., Stalon V., Mercenier A. Arginine degradation in Pseudomonas aeruginosa mutants blocked in two arginine catabolic pathways. Mol Gen Genet. 1984;193(3):437–444. doi: 10.1007/BF00382081. [DOI] [PubMed] [Google Scholar]
- Holloway B. W. Genetics of Pseudomonas. Bacteriol Rev. 1969 Sep;33(3):419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercenier A., Simon J. P., Haas D., Stalon V. Catabolism of L-arginine by Pseudomonas aeruginosa. J Gen Microbiol. 1980 Feb;116(2):381–389. doi: 10.1099/00221287-116-2-381. [DOI] [PubMed] [Google Scholar]
- Mercenier A., Simon J. P., Vander Wauven C., Haas D., Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol. 1980 Oct;144(1):159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C. G., Dawes E. A. The role of oxygen in the regulation of glucose metabolism, transport and the tricarboxylic acid cycle in Pseudomonas aeruginosa. J Gen Microbiol. 1982 Jan;128(1):49–59. doi: 10.1099/00221287-128-1-49. [DOI] [PubMed] [Google Scholar]
- Rahman M., Laverack P. D., Clarke P. H. The catabolism of arginine by Pseudomonas aeruginosa. J Gen Microbiol. 1980 Feb;116(2):371–380. doi: 10.1099/00221287-116-2-371. [DOI] [PubMed] [Google Scholar]
- Stalon V., Mercenier A. L-arginine utilization by Pseudomonas species. J Gen Microbiol. 1984 Jan;130(1):69–76. doi: 10.1099/00221287-130-1-69. [DOI] [PubMed] [Google Scholar]
- Vander Wauven C., Piérard A., Kley-Raymann M., Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984 Dec;160(3):928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C., Stalon V. Occurrence of succinyl derivatives in the catabolism of arginine in Pseudomonas cepacia. J Bacteriol. 1985 Nov;164(2):882–886. doi: 10.1128/jb.164.2.882-886.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderbilt A. S., Gaby N. S., Rodwell V. W. Intermediates and enzymes between alpha-ketoarginine and gamma-guanidinobutyrate in the L-arginine catabolic pathway of Pseudomonas putida. J Biol Chem. 1975 Jul 25;250(14):5322–5329. [PubMed] [Google Scholar]
- Voellmy R., Leisinger T. Dual role for N-2-acetylornithine 5-aminotransferase from Pseudomonas aeruginosa in arginine biosynthesis and arginine catabolism. J Bacteriol. 1975 Jun;122(3):799–809. doi: 10.1128/jb.122.3.799-809.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellym R., Leisinger T. Role of 4-aminobutyrate aminotransferase in the arginine metabolism of Pseudomonas aeruginosa. J Bacteriol. 1976 Dec;128(3):722–729. doi: 10.1128/jb.128.3.722-729.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson O. H., Holden J. T. Arginine transport and metabolism in osmotically shocked and unshocked cells of Escherichia coli W. J Biol Chem. 1969 May 25;244(10):2737–2742. [PubMed] [Google Scholar]