Abstract
We assess performance in the structure refinement category in CASP9. Two years after CASP8, the performance of the best groups has not improved. There are few groups that improve any of our assessment scores with statistical significance. Some predictors, however, are able to consistently improve the physicality of the models. Although we cannot identify any clear bottleneck to improving refinement, several points arise: (1) The refinement portion of CASP has too few targets to make many statistically meaningful conclusions. (2) Predictors are usually very conservative, limiting the possibility of large improvements in models. (3) No group is actually able to correctly rank their five submissions—indicating that potentially better models may be discarded. (4) Different sampling strategies work better for different refinement problems; there is no single strategy that works on all targets. In general, conservative strategies do better, while the greatest improvements come from more adventurous sampling–at the cost of consistency. Comparison with experimental data reveals aspects not captured by comparison to a single structure. In particular, we show that improvement in backbone geometry does not always mean better agreement with experimental data. Finally, we demonstrate that even given the current challenges facing refinement, the refined models are useful for solving the crystallographic phase problem through molecular replacement.
1 Introduction
We review here the performance of the 32 research groups who participated in the refinement category in CASP9. This is the third round of the CASP refinement experiment and second time within the main CASP framework (the first being in CASP8 [1]). The refinement experiment is a blind test taking place just after the main structure prediction portion of CASP. For selected targets, the best models submitted during the template-based modeling (TBM) category are assigned as refinement targets. Along with the model and sequence, the predictors are sometimes given a series of hints regarding where problematic regions are and how accurate the starting model is.
Refinement is different from other categories in CASP and what would normally be called refinement in the literature [2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16] in that the refinement structures have already been refined during the TBM portion of CASP. The best TBM predictors are already able to add value, on average, to the single best template [17]. Here, predictors are trying to take this to the next level.
In short, the most substantial improvements from the CASP9 refinement experiment are in improved physicality of the models. Improving the global positioning of the backbone remains a challenge and few groups were successful at this. The small size of the experiment—only 14 targets—leads to very few results that are statistically significant.
We find that no group correctly rank-orders its models. So, even when predictors generate an improved model, they may not know it.
The relative performance of different groups depends on the target protein. There is currently no best strategy that works uniformly across targets.
We also compare the predicted structures directly with experimental data. We find that improvement in the position of the backbone is only partially correlated with the experimental data, which illustrates the difficulty in assessing refinement predictions. Finally, we show that despite all of the challenges facing refinement, the refined models are useful as input for solving the crystallographic phase problem through molecular replacement.
2 Targets used for refinement in CASP9
There were 14 targets available for refinement in CASP9. Targets were selected by the CASP organizers using the following criteria: (1) they should be relatively small domains; (2) the area to be refined should, ideally, be free of crystal or oligomeric contacts; (3) the experimental group who determined the structure is able to offer a further three week extension—in addition to the usual three week prediction window—before the release of the experimental structure. Table 1 shows the targets for refinement in CASP9.
Table 1.
Summary of CASP9 refinement targets.
| Target | PDB | Residues | Method | GDT-HA | RMSD (Å) | GDC-SC | SG | MP | Missing | Focus |
|---|---|---|---|---|---|---|---|---|---|---|
| TR517 | 3pnx | 159 | X-ray | 54.7 | 4.6 | 30.2 | 55.2 | 1.4 | - | 65–88 |
| TR530 | 3npp | 115 | X-ray | 70.9 | 2.0 | 39.8 | 83.8 | 0.9 | 1–35 | 45–58; 75–79 |
| TR557 | 2kyy | 145 | NMR | 48.6 | 4.1 | 28.8 | 42.4 | 1.5 | 126–145 | 15; 61 |
| TR567 | 3n70 | 145 | X-ray | 59.0 | 3.4 | 38.1 | 80.3 | 1.4 | 1–3 | 63; 103 |
| TR568 | 3n6y | 158 | X-ray | 35.8 | 6.1 | 16.8 | 18.6 | 1.5 | 1–61 | 102; 116 |
| TR569 | 2kyw | 79 | NMR | 52.9 | 3.0 | 35.2 | 45.6 | 0.7 | - | 50–60 |
| TR574 | 3nrf | 126 | X-ray | 39.7 | 3.6 | 21.6 | 44.1 | 3.6 | 1–24 | - |
| TR576 | 3na2 | 172 | X-ray | 46.9 | 6.9 | 17.4 | 50.0 | 3.7 | 1–24; 140–172 | 57–65; 120+ |
| TR592 | 3nhv | 144 | X-ray | 74.0 | 1.3 | 43.4 | 92.4 | 3.5 | 1–16; 122+ | 33; 63 |
| TR594 | 3ni8 | 140 | X-ray | 67.5 | 1.8 | 39.2 | 87.1 | 2.9 | - | 110 |
| TR606 | 3noh | 169 | X-ray | 53.2 | 4.8 | 29.6 | 52.0 | 3.2 | 1–45; 169 | 46–55; 149–168 |
| TR614a | nab | 135 | X-ray | 52.6 | 5.3 | 27.9 | 43.8 | 4.0 | 1; 123–135 | - |
| TR622 | 3nkl | 138 | X-ray | 49.4 | 7.5 | 25.6 | 53.3 | 3.7 | 123–138 | 97+ |
| TR624 | 3nrl | 81 | X-ray | 36.6 | 5.2 | 14.6 | 46.4 | 1.9 | 1–4; 74–81 | 14; 24; 54 |
For target TR614, two starting models were provided and we show the average score.
As of publication, the structure of target TR614 was not available from the PDB.
The predictors were provided with hints. Predictors were told the GDT-TS (see Section 3.2.1 for definition) of the starting model and were told which groups of residues needed refinement (“Focus” column in Table 1). The specificity of the advice varied. Sometimes only the specified residues needed to be changed; in other cases, changes to additional residues were required.
Some of the refinement targets were problematic for two reasons: (1) some starting models are already close enough to the experimental structures that small changes are not clear improvements within the protein’s thermal ensemble, and (2) some of the structures have oligomeric or crystal contacts. We found several examples that we believe may be problematic, but for completeness we include the results for all targets.
In order to determine which starting models may be too close to the crystal structure, we generated a thermal ensemble around the crystal structure and then checked to see if the starting model fell within this ensemble. We generated the thermal ensemble by performing molecular dynamics simulations starting from the crystal structure. We used a current forcefield (parm99sb [18]) and explicit water (TIP3P [19]) in the AMBER10 software package [20]. We performed constant-temperature (300K) molecular dynamics for 10 ns. The first 5 ns was discarded as equilibration and we calculated the RMSD between each snapshot of the final 5 ns and the starting model. For most targets the thermal ensemble is near the experimental structure, far from the starting model, and usually far from most of the predictions. For such targets, it is meaningful to attempt refinement.
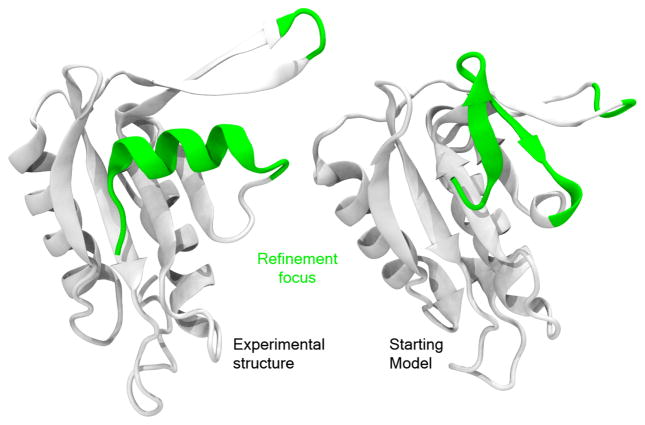
However, for target TR592, we believe it was not meaningful to attempt refinement of the backbone. On average, the thermal ensemble for this target is as far away from the starting model as from the experimental structure, and no further than 2Å in any case. The closest any structure from the ensemble gets to the experimental or starting model structure is also about the same (between 1.2 and 1.8 Å). Thus, we don’t believe there is value added in attempting to refine the backbone of this particular target since it is already within the native thermal ensemble (Figure 1). It is, however, possible that other non-backbone aspects of the structure may be improved through refinement.
Figure 1.

Example of a difficult to refine target: TR592. Difficulties arise from the fact that the starting model is already within the thermal ensemble of the native conformation.
We examined four structures for potential crystal packing problems: TR517, TR567, TR576, and TR592. For target TR517, the residues targeted for refinement are at the interface with another monomer from the asymmetric unit. However, analysis of the predictions shows that several groups were able to improve the structure without knowledge of the interaction partners. Indeed, inspection of the thermal ensemble for this structure in its monomeric form (data not shown) reveals the structure to be very stable, indicating that contacts with neighbor residues may not be needed to hold the structure in place.
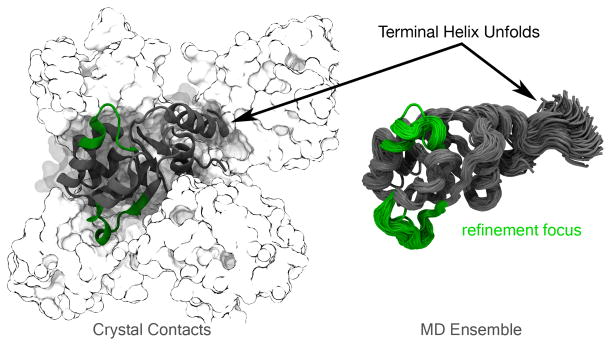
TR567 is a special case, as the presence of crystal contacts stabilize the N-terminus in an alpha helical secondary structure. Without the other monomers in the unit cell, the end of this helix sticks out into empty space and is not stable in the MD ensemble (Figure 2). The conformations of the N- and C-termini represent the largest deviations between the starting model and the native structure—accounting for most of the low starting GDT-HA. The predictors were not told to refine the problematic helical region. Instead, they were asked to refine two other regions that are very similar in the starting model and the native structure. Thus, we feel this target is problematic for two reasons: (1) the regions to refine are already very close to native and within the thermal ensemble; (2) the end of the N-terminal helix is not is not stable and changes in this helix may mask improvements in the focus region.
Figure 2.
Target TR567. Crystal contacts stabilize an alpha helical secondary structure near the N-terminus. However, when those contacts are removed, this helix protrudes into space and is no longer stable..
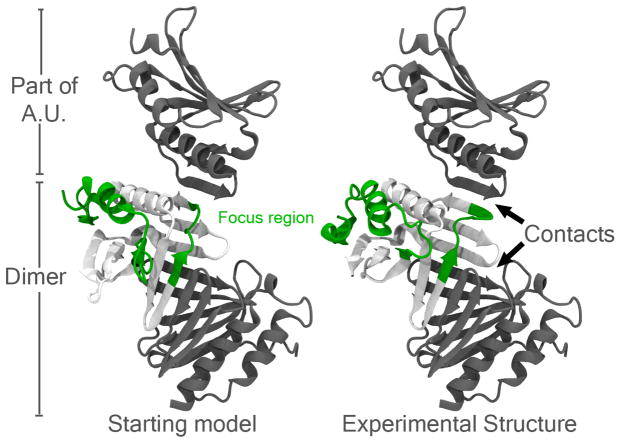
The packing issues appear to be more severe for TR576 (Figure 3) where there are two kinds of interactions: β-sheet stabilization within the biological unit dimer and also β-sheet stabilization between monomers in the asymmetric unit. Both kinds of interactions happen around the same focus area of the protein and determine the position of three β-sheets, thus pulling along the position of other secondary structure elements out of place. Final refinement of this structure is tightly coupled to the identification of these contacts. The fact that this target is the most difficult refinement target to improve (Figure 6) corroborates the importance of these contacts in the determination of the native state.
Figure 3.
Refinement target TR576 which might suffer from crystal packing effects. Contacts are stablished both within the biological dimeric unit and within the asymmetric unit (A.U).
Figure 6.
Summary of scores by target (all models, all groups). The x-axis shows improvement with respect to the starting model. The numeric values are the percentage of the time that the refined model was better or worse than the starting model. The targets are ordered by the fraction of time the GDT-HA score was improved.
Finally, TR592 could have been problematic, but the prediction center identified and trimmed off the problematic portion of the structure.
With the exception of TR576 and TR567, we feel that the refinement challenges posed by the prediction center are relatively unaffected by crystal packing.
3 Data Analysis
3.1 Filtering
Due to our use of all-atom scoring functions and the difficulty in normalizing the scores for predictions of different lengths, we excluded some predictions from our analysis. The predictors were given a model to use as the starting point for their refinement. Accordingly, any prediction missing more than one residue or ten heavy atoms relative to the starting model was excluded from our calculations. In total, 50 of 1615 predictions were excluded. The MIDWAYHUMAN group had the most predictions excluded (25 of 60). Overall, only 3 of 34 groups (including MIDWAYHUMAN) had more than five predictions excluded.
We do not feel that there is a way to fairly evaluate these structures in the context of the all-atom measures we use. The results for these predictions will still be available on the Prediction Center website, but they are not included in any of the analysis in this paper.
3.2 Metrics used for evaluation
We measure the qualities of predicted structures using both global and local measures. We use GDT-HA and RMSD, which are measures of the global positioning of the C-alpha atoms. We use GDC-SC as a global measure of sidechain positioning. We use SphereGrinder as a local all-atom measure of structural similarity. Finally, we use MolProbity as a local measure of the physical reasonableness of the structure. We use a weighted average Z-score to combine these five scores into a single overall score for each prediction.
3.2.1 GDT-HA
GDT-HA is a global measure of the fraction of C-alpha atoms that are positioned correctly [21]. GDT-HA is similar to GDT-TS, which has been a standard assessment tool in previous CASPs. Both GDT-TS and GDT-HA are based on multiple superpositions of the predicted and experimental structure. The difference between the two scores is in the cutoffs used for these superpositions. GDT-HA uses cutoffs of 0.5, 1, 2, and 4 Å, while GDT-TS uses 1, 2, 4, and 8 Å. As a result, GDT-HA is more sensitive to small errors. GDT-HA and GDT-TS are strongly correlated (Figure S1), so we chose to only use GDT-HA in our analysis.
3.2.2 RMSD
We assessed the root-mean-squared deviation (RMSD) of the C-alpha positions. Like GDT-HA, the RMSD is a global measure of the correct positioning of the C-alpha atoms. GDT-HA and RMSD are only weakly correlated (Figure S2) because unlike GDT-HA, RMSD is based on a single superposition and RMSD lacks any kind of distance cutoff. These differences make RMSD more sensitive to large changes in the position of a few atoms.
3.2.3 GDC-SC
We use GDC-SC [22] as a global measure of the correct positioning of the sidechains. GDC-SC is similar to GDT-HA, but uses a single characteristic atom near the end of each sidechain rather than the C-alpha atoms. GDC-SC also uses different weighting of ten different super-positions (rather than four as with GDT-HA), although it is otherwise conceptually similar.
3.2.4 SphereGrinder
SphereGrinder is a recently introduced all-atom score that measures the local environment around each residue. For each residue in the experimental structure, we consider the set of atoms within 6 Å of the C-alpha atom and perform an all-atom RMSD fit between the experimental and predicted structures using only the atoms within this sphere. We then calculate the fraction of atoms inside the sphere that are within 2 Å of their experimental counterparts. This gives a per-residue score, which is then averaged over all residues to obtain the SphereGrinder score for the structure.
3.2.5 MolProbity
MolProbity is an all-atom measure of the physical correctness of a structure based on statistical analysis of high-resolution protein crystal structures [23]. MolProbity is sensitive to steric clashes, rotamer outliers, and Ramachandran outliers. High-resolution protein crystal structures contain very few defects of these types. We use a combination of these terms, defined as the MPscore [22, 23]. Unlike the other scoring measures we use, MolProbity is not native-centric, i.e. the native structure is not required to calculate it. As such, MolProbity could be used by groups in the prediction stage to improve the physicality of their models. We believe that having physically believable models is important, particularly at the high accuracy end of structure prediction.
3.3 Normalizing and combining scores
Better values of GDT-HA, GDC-SC, and SphereGrinder are indicated by higher scores. Better values of RMSD and MolProbity are indicated by lower scores. We report improvement in these scores rather than raw values, so that more positive scores are always better.
In order to efficiently compare between different groups, targets, and scores, a normalized measure is needed. Z-scores are often used for this purpose; However, the presence of outliers can skew the results. To minimize the influence of outliers, we use a robust version of the Z-score based on the median and median absolute deviation (MAD) of the median, rather than the mean and standard deviation (Equations 1 and 2).
For a given target, we calculate the median absolute deviation using:
| (1) |
Here, xi is the set of all scores of one type (e.g. GDT-HA) for a particular target. The notation mediani (xi) denotes the median of this set of scores and | · |is the absolute value. The robust Z-scores are then calculated as:
| (2) |
The factor of 1.4826 scales the MAD to be the same as the standard deviation for a normal distribution.
To combine the results for all five scores into a single overall score, we use a weighted average of the five scores, where GDT-HA is given a weight of 4 (Equation 3). GDT-HA makes up half of the overall score, while the other four measures make up the remaining half. As we noted in our assessment of refinement in CASP8 [1], such a weighting is arbitrary and changing the weighting factors would emphasize different aspects of refinement and will change the rankings of different groups.
| (3) |
3.4 The “Null” group
As a control, we created an additional prediction group—the “Null” group—where we did nothing to the starting model. Groups that perform worse than the Null group have made the starting model worse rather than better.
There were two starting models for target TR614. In this case, our model for the Null group was to randomly choose the two starting models with equal probability. Thus, the expected value for each of refinement measure is simply the mean of the values for the two starting models.
The performance of each group was compared to the Null group using the Wilcoxon signed-rank test using p = 0.05. The Wilcoxon test is similar to a paired t-test, but it does make any underlying assumption of the normality of the distributions being compared. Figure 5 shows that due to the small number of targets, the distributions for the individual groups differ substantially from normal distributions.
Figure 5.
Summary of scores for the ten groups (all models, top ten groups as judged by cherry-picked overall score). The x-axis shows improvement with respect to the starting model. The numeric values are the percentage of the time that the refined model was better or worse than the starting model. The groups are ordered by the overall performance considering the best model for each target (see Figure 8).
3.5 Selection of predictions
Each group was asked to submit up to five predictions rank ordered from best to worst. We perform one set of analysis using only Model 1, which is the model that the predictor believes is the best. Due to the inability of groups to correctly rank their structures, we performed a second set of analysis, where for each target we select the best model (as judged by overall score) from each group. We refer to this as “cherry-picking”. The cherry-picked results may or may not be useful depending on the specific application of refinement. For example, as we discuss later, when using structure predictions as models for molecular replacement[24, 25], it is possible to try many different models and use the one that agrees best with experimental data. In this case, the ability to rank order the models is desirable—but not required. Other applications, such as a very expensive physics based free-energy calculations [26], may be too computationally demanding to perform on more than one model.
Ideally, our assessment would use Model 1 exclusively, but since predictors cannot correctly identify their best models, we feel it is reasonable to evaluate the best models. Unfortunately, this rewards predictors who employ a “safety-net” strategy of always submitting at least one model that is very close to the starting model. The assessors view this as an undesirable strategy, but we note that we do not see evidence of the wide-spread adoption of this strategy. In order to avoid bias when measuring improvement between CASP experiments or deciding who “won” refinement in CASP9, we feel it is important to use only the Model 1 results. This is the fairest measure of comparison and does not reward safety-net strategies.
4 Results and discussion
4.1 Overall results
Figure 4 shows the overall performance of refinement in CASP9 across all targets and all models from all groups. Some scores are improved more frequently than others. For example, improvement in the global position of the backbone atoms—as judged by GDT-HA—is observed in less than 25 percent of all predictions. Even in cases where GDT-HA is improved, the improvement is typically modest. In contrast, the physicality of the models—as judged by MolProbity—is improved in nearly 50 percent of all predictions. Clearly, the predictors are better at improving physicality than at improving the backbone positioning. This suggests that improvements can be made in the future by focusing method development on global search techniques that can improve the overall fold, rather than on local search techniques that can improve the physicality of the models.
Figure 4.
Summary of aggregate (all models, all groups) results by score. The numeric values are the percentage of time the structure was made better or worse for each metric.
The distributions in Figure 4 are multimodal, which indicates that not all targets are equally easy to refine and that not all groups are equally good at refinement. It is also clear that the best improvements are not large enough. The worst starting GDT-HA is 35, so the largest possible improvement would be 65. The largest improvement we actually observe, however, is only about 15. We will show later that the predictors are too conservative overall and that for most groups there is little chance of such a large GDT-HA improvement.
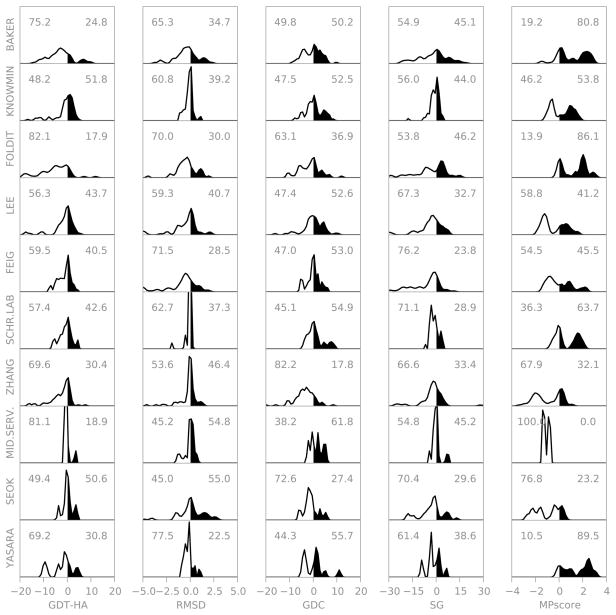
If we focus on the top ten groups (Figure 5), we observe that different groups perform better at different aspects of refinement. The distributions for each group are multimodal due to variations between targets. We see interesting variations between groups. For example, compared to the BAKER group, several groups (e.g. KNOWMIN) have higher average GDT-HA and more frequently improve GDT-HA. However, when the BAKER group improves GDT-HA it is often by a larger amount.
Figure 5 also illustrates the difficulty of formulating an overall score. Different groups excel at different aspects of refinement and changing the relative importance of the different scores will clearly change the ordering of the groups.
Different aspects of refinement are easier in some targets than in others (Figure 6). Different targets will be easier or harder for different groups depending on what aspect needs to be improved. We believe that one way to improve refinement in the future is to combine algorithms with different specialties in a smart way, so that the composite algorithm is better than its individual constituents.
We have spent considerable effort trying to understand why a particular target is easy or hard, but we have been unable to reach any compelling conclusions. After assessing both CASP8 and CASP9, we cannot predict whether one particular target will be easy or hard. For example, target TR592 has the highest starting GDT-HA (74.1), while target TR574 has one of the lowest (39.7). We expected to see some difference in how often and by how much these initial models were improved. However, Figure 6 shows that these two structures are approximately in the middle of the range of improvement. In our view, this inability to predict target difficulty is one of the most pressing questions in the field at the moment. If we do not know what is hard about the problem, how can we hope to improve? When we examine the relationship between the starting value of any of the scores and the predictors’ ability to improve the starting model (not shown), we can find few convincing correlations of any kind. The only clear trend is that it is easy to improve the MolProbity score when it is initially very bad—a rather trivial conclusion. We note, however, that the inability to predict difficulty may not be true for individual groups. We are examining global results across all groups. It could be that individual groups may be able to better predict how well their strategy will work for a given target. For example, the KNOWMIN group has demonstrated better performance when the GDT-HA of the starting model is higher [2]. With only 14 refinement targets—and many groups not even attempting all targets—it is difficult for us to assess the difficulty for individual groups in a reliable way.
4.2 Group Performance: Model One
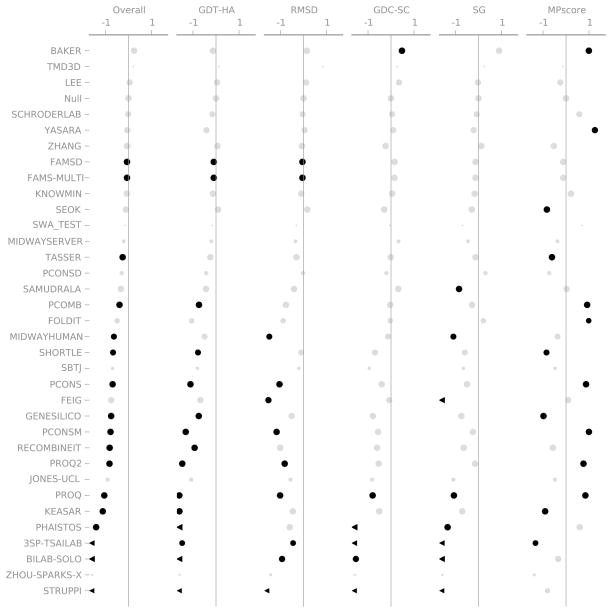
Figure 7 shows the overall results for each group considering only Model 1. The most striking feature is the lack of statistical significance. No group is significantly better than the Null group for Overall Score, GDT-HA, RMSD, or SphereGrinder. The BAKER group is able to consistently improve GDC-SC and many groups are able to improve MPScore. We believe that the MPscore is likely to be the easiest aspect of refinement to improve, although this may not be true for every prediction method. This observation also applies to the standard template-based modeling portion of CASP. If the TBM predictors begin to improve the physicality of their models, the refinement category will become more difficult and relative performance of different groups will change.
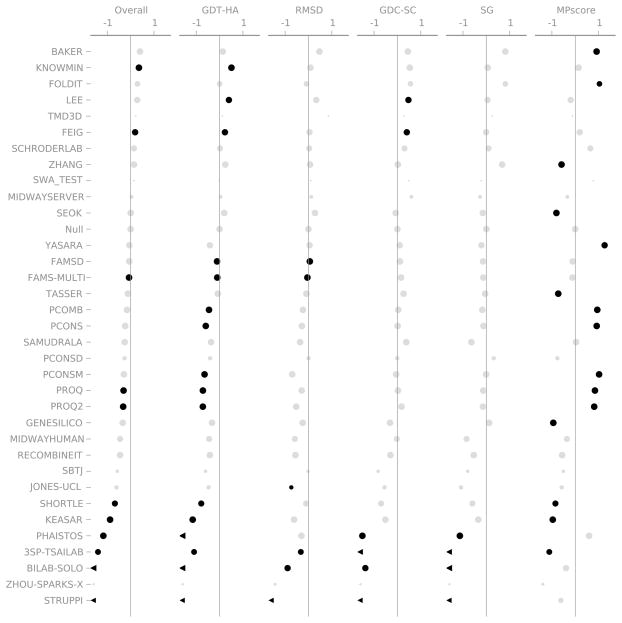
Figure 7.
Summary of the results of the CASP9 Refinement Experiment. Only the models designated as “Model 1” are included. Each column shows one of the metrics we used to evaluate performance. The scales are marked at +/− 1 median absolute deviation (MAD) relative to the Null group. Black points are statistically distinguishable from the Null group; grey points are indistinguishable (Wilcoxon signed-rank test, p = 0.05). The area of each point is proportional to the number of targets that group attempted. A chevron indicates that the corresponding score was off the scale.
The lack of statistical significance is not surprising given the small number of targets. This is obvious if we consider a simplified version of the Wilcoxon test, called the sign test. Here, we only count the number of times that a group improves or worsens the starting model while ignoring the magnitude. The null hypothesis is that a particular group will will improve or worsen each target with equal probability, which leads to a binomial distribution. The null hypothesis for 14 targets is the same as flipping a fair coin 14 times. We achieve a statistically significant p-value of 0.05 only when 11 out of 14 trials come up heads—it’s relatively easy to get 9 or 10 heads. So, in order for the results for a group to be statistically significant, the group would have to improve a given metric 11 or more times out of 14. None of the groups in CAPS9 is that consistent, so almost none of the results are significant. The Wilcoxon test is more sophisticated and takes the magnitudes into account, but the intuition is still the same. There are a small number of targets, so a group must be very consistent to be distinguishable from Null.
4.3 Group Performance: Cherry-Picked
Figure 8 shows the results for each group considering the cherry-picked best models for each target. There are now a few statistically significant positive results. BAKER, KNOWMIN, FOLDIT, LEE, and FEIG are significantly better than the Null group for at least one score.
Figure 8.
Summary of the results of the CASP9 Refinement Experiment. For each target and each group, the best overall performing model is selected. Each column shows one of the metrics we used to evaluate performance. The scales are marked at +/− 1 median absolute deviation (MAD) relative to the “Null” group. Black points are statistically distinguishable from the Null group; grey points are indistinguishable (Wilcoxon signed-rank test, p = 0.05). The area of each point is proportional to the number of targets that group attempted. A chevron indicates that the corresponding score was off the scale..
One of the groups to improve more under “cherry-picking” is the KNOWMIN group, due to large (relative to other predictions) improvements in GDT-HA. Virtually every group has better better performance when we cherry-pick models, illustrated by a shift to the right of Figure 8 compared to Figure 7.
One striking observation is that even after cherry-picking the best overall models for each group, only two groups are significantly better than the Null group. The rest are either indistinguishable from (20 groups) or worse than the Null group (10 groups).
4.4 Ranking
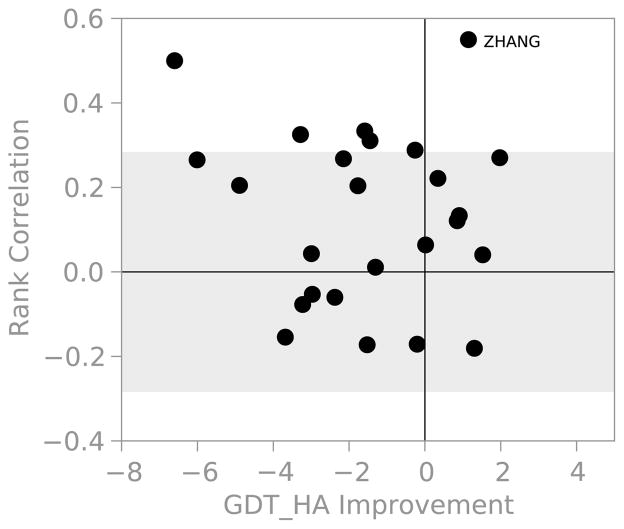
We observed a significant improvement in scores when cherry-picking the best models from each group, which indicates that the predictors have difficulty ranking their models. Figure 9 compares the ability of each group to rank their submissions with their performance on cherry-picked GDT-HA. There are few groups that are able to rank order their structures better than random. However, the few successes don’t necessarily reflect an improvement in the ranking ability. With the exception of ZHANG, all of the groups that excelled at ranking actually made the starting model worse on average. Only the ZHANG group was able to improve GDT-HA and rank order their models. Upon examining the predictions in detail, however, it is clear that some groups used different sampling strategies for their five models. This appears true for the ZHANG group, where their Model 1 follows a conservative strategy and is always less than 1 Å from the starting model. Their Model 2 seems to also correspond to a conservative approach, while the rest of their predictions are more “adventurous”. We feel that the ZHANG group’s ability to rank order their submissions reflects that they know that their Model 1 submission can never be much worse than the starting model, instead of knowing that some prediction is an improvement.
Figure 9.
Groups are unable to correctly rank order their submissions. For each group we plot the average Spearman rank correlation coefficient versus the average change in GDT-HA considering the best model for each target. The shaded region is statistically indistinguishable from randomly ranking five predictions for twelve targets. For groups that did less than twelve targets or less than five predictions for each target, the shaded region would be wider than shown.
In discussions with several groups, we also discovered that they had produced models that were better than those submitted. This is further evidence that the groups have difficulty ranking their predictions. Of course, this information is unavailable to us during the assessment. In the future, it might be helpful if predictors were able to submit a large ensemble of models—in addition to the usual five rank ordered ones. The assessors could use this set of additional models to separate sampling from scoring. That is, to separate the problem into two questions: (1) Can we make better models? (2) Can we tell when we generate them?
The issues between ranking, scoring, and sampling are not clear cut. The fact that groups cannot rank order models does not necessarily mean that there are problems with scoring functions. We don’t expect to see a linear—or even monotonic—relationship between the scoring function and some measure of how geometrically accurate the model is—like GDT-HA or RMSD. In particular, for a “physical” energy function, we don’t expect the energy to go down much until the structure is very close to native. If a structure changes from 5 to 4 Å the energy will not necessarily go down. Further, even if the backbone atoms were all in the correct location, the energy will not go down unless the sidechains and all of the other details of the system are also correct. This constitutes a sampling problem, not a scoring problem. Ultimately, the issues of sampling, scoring, and ranking are intertwined, which makes it difficult to understand what improvements are required for refinement to become more successful.
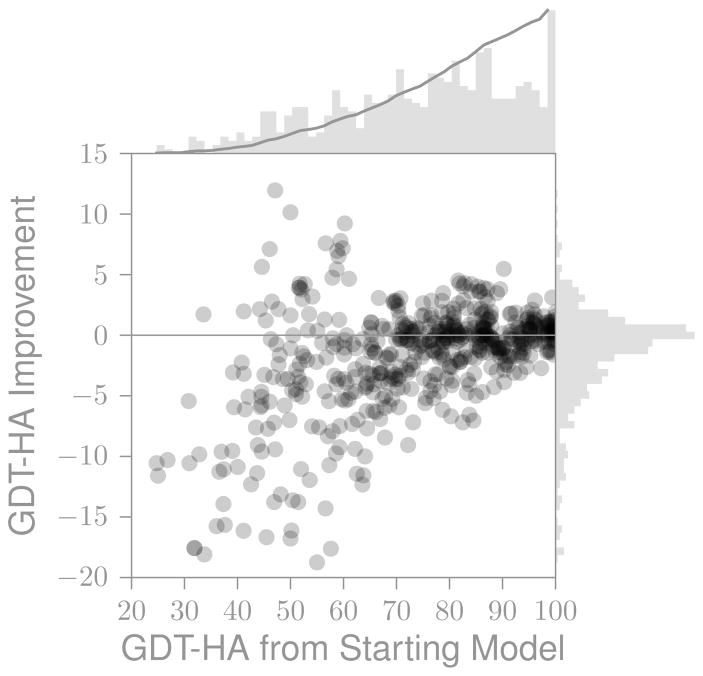
4.5 Breadth of sampling
Figure 10 shows the relationship between the breadth of sampling and improvement in GDT-HA for all predictions from the top ten groups. Even considering only the best groups, the average prediction is worse than the starting model (the GDT-HA Improvement is skewed to negative values). Most of the predictions are conservative. Approximately 50 percent of the predictions have a GDT-HA to template of 75 or higher. In contrast, 6 of 14 targets have a starting model that has a GDT-HA of less than 50. Half of the targets require changes more than 50 GDT-HA to be correct, but only 12 percent of the submitted structures have a GDT-HA to template of below 50.
Figure 10.
Relationship between breadth of sampling and improvement for the top groups (all models, top ten groups by cherry-picked overall score). The histograms indicate the marginal distribution along that axis. The solid line in the upper histogram shows the cumulative distribution
From the predictions of the top groups we can distinguish several kinds of strategies (not shown). Some predictors (e.g. KNOWMIN) are conservative. Their predictions are close to each other and are relatively close to the starting model. Other predictors (e.g. BAKER) are more adventurous. Their predictions still generally cluster together, but they are farther from the starting model. A number of groups appear to use a mixture of these two strategies (e.g. ZHANG). Here the predictions are often quite different from one another. Some of the predictions remain close to the starting model, while others are much farther away.
Depending on the target each strategy has its own strengths and weaknesses. For example, conservative strategies almost never make the starting model much worse. However, they will never produce a dramatic improvement in some structures because they do not sample broadly enough. More adventurous search strategies have the potential of finding large improvements, but also have challenges because the size of the search space increases rapidly as the breadth of sampling is increased. There are many more structures that must be sampled and more structures that must be correctly rank ordered—which, as we have seen, is a challenge. This is illustrated by comparing the results for BAKER and KNOWMIN in Figure 5. The distributions for KNOWMIN are narrower and the results more consistent, while the best improvements for BAKER are larger, but the overall results are less consistent.
We chose three structures of increasing difficulty (TR569, TR557, and TR624) to serve as case studies to demonstrate the successes and pitfalls of different sampling strategies.
Target TR569 (Figure 11) is the easiest of the three targets. The starting model is largely correct, but a loop is slightly misplaced and two short β-strands do not have the correct geometry. Even though TR569 is the target for which the most groups improved GDT-HA, less than half of the predictions were an improvement over the starting model (Figure 6). We expect that increasing the secondary structure around the loop region, should greatly improve the stability of the resulting structure. The amount of sampling required is not as large as for other targets since no changes in topology are required. The success in this target might also be due to scoring functions becoming more useful very close to the native structure. Multiple kinds of search strategies perform well here.
Figure 11.

Example of an easily refinable target: TR569..
In contrast, target TR557 poses a more difficult challenge because it requires rearrangement of two β-sheets into an alpha helix (Figure 12). Few groups were able to capture this and the ones that did had the helix in a completely wrong orientation. This target is significantly challenging to local sampling strategies. Few groups were able to capture the change in secondary structure (BILAB-SOLO, RECOMBINEIT, ZHOU-SPARKS-X), although the resulting orientation of the helix was wrong. We suspect that these algorithms found additional reference templates and were able to bypass the problems of having to disrupt the β-sheet—which would be very unfavorable—to then reform an alpha helix.
Figure 12.
Example of a hard refinable target: TR557. Several difficulties arise, especially near one of the termini where changes in secondary structure are needed..
Target TR624 was especially challenging because large movements of two β-hairpins were required. One of the hairpins must tilt “up” whereas the other must tilt “down”, thus interchanging the position of the refined areas. This is an example where local sampling cannot accomplish much because independently each of the hairpins is in its correct secondary structure. This conformational change was only solved by a few groups (Figure 13). Sampling such a large conformational change requires an adventurous search strategy. However, often such strategies make unrelated parts of the model worse (e.g BILAB-solo, marked B in Figure 13), thus masking the positive aspects of their sampling strategy. Only the FOLDIT and BAKER groups managed this interchange resulting in large GDT-HA improvements.
Figure 13.
Refinement of TR624 was a particularly hard challenge. Local environments near the refinement areas are correct, but a topology change is needed: the β-sheets leading to the red and blue loops have to tilt in opposite ways. Only three groups captured this.
The issues of sampling and scoring are intertwined. It is possible that the predictors sampled a wider range of structures, but did not score them in the top 5. Again, it would be beneficial if predictors could submit a much larger ensemble of structures to help the assessors separate the issues of sampling and scoring.
4.6 Improvement since CASP8
As with the template-based modeling portion of CASP[27], it is very difficult to judge improvement in refinement compared to previous CASPs. It does not appear that there has been a significant step forward in refinement since CASP8. However, it is hard to assess if there has been any small improvement because it would be masked by the fact that targets change from year to year and we have no way of assessing target difficulty. Also, we judge the improvement from models that have already been refined during the TBM category. As the TBM methods improve, there should be less room for improvement, or at least, new strategies for refinement should be used. Finally, the small number of targets means that it would be virtually impossible to establish statistically significant improvement since CASP8.
If we focus on the top group (BAKER, at least by our overall score) in both CASP8 and CASP9 and examine their average cherry-picked GDT-HA values in both competitions, we find that the BAKER group actually did worse in CASP9. Did they change strategies or were the targets more difficult? Are the original templates better refined during TBM during CASP9? Or, is this just statistical noise? There are few targets and the variances of the scores are large. Since there’s no single strategy that works on all targets, the problems to be solved during refinement on such a small set of targets might favor one strategy over another.
Overall, if there is any improvement since CASP8, it is hard to detect. Since potential improvements to prediction algorithms are obscured by changes in evaluation methodologies and targets, the field would benefit from the registration and use of old versions of the refinement codes. This will make it easier to measure progress. We also suggest that the Prediction Center and the CASP organizers work with the predictors to select one or more reference algorithms from the top performing CASP groups that can be frozen in their current state and used as a benchmark to gauge improvement. It is also clear that the refinement category needs more targets if statistically meaningful conclusions are to be drawn.
5 Comparison with experimental data
Proteins are flexible molecules. Flexibility is often crucial for a protein’s biological role [28, 29, 30]. Currently, this information is not taken into account during CASP assessment. Any flexible regions are not included in the assessment and proteins are parsed into domains. We believe a potentially better way to deal with flexibility is to quantify how well the predicted models reproduce the experimental data. That is to say, are groups submitting predictions compatible with the experimental information we have available? We have followed a similar procedure as in the last CASP [1] by comparing to either the NMR information coming from nuclear Overhauser effect data [31, 32] or by doing molecular replacement likelihood analysis [24, 25] on X-ray structures. We have found only partial correlation between experimental results and GDT-HA, further showing the difficulty in choosing one metric alone as the gold standard to evaluate model similarity. Finally, we show that refinement is useful for producing structures that are used as input for molecular replacement.
5.1 Comparison with NMR data
Direct comparison with NMR data is valuable for two reasons. First, NMR is an ensemble experiment and and the experimental data capture the flexibility of the entire ensemble. Second, NMR experiments are performed near biologically relevant conditions. NMR experiments should be free of artifacts due to crystal packing and are performed close to physiological temperatures.
In well structured large molecules like proteins, inter-proton distances are directly related to nuclear Overhauser effect (NOE) intensities [33]. These intensities give insight into inter-proton distances that can be used to derive structures. In the previous CASP8 [1] experiment, the NOE upper-bound distance violation (UBV) was used as an alternative way to evaluate the success of refinement. It helped identify refined models that have better agreement with NMR data but were regarded as worse models by the more traditional metric GDT-TS. Normally, a hydrogen pair (i,j) is considered violating the NOE upper bound distance when is positive [31], otherwise the violation vi,j is considered zero. Since groups are not told to submit ensembles but individual structures the above expression reduces to: . The average UBV was then calculated as where N is the number of experimentally determined NOE distances (for more details see Ref [1]). The upper bound distances (UBD) are derived from experimental NOE data and inherently allows for structural flexibility (all distances smaller than the UBD are equally “good”). Thus, several models might be in accordance with this metric.
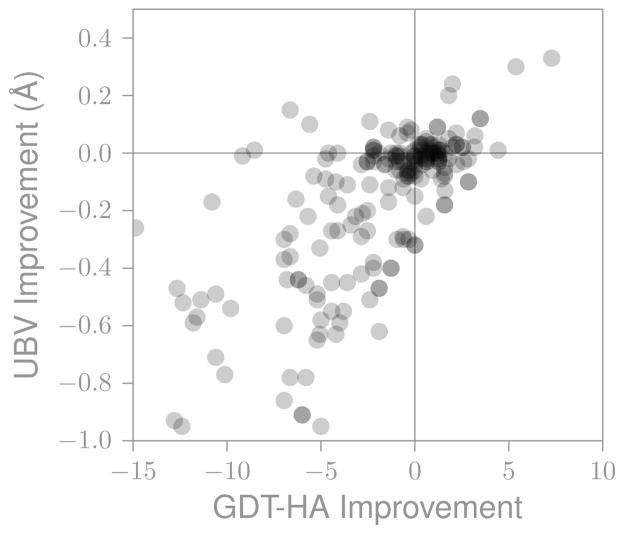
Figure 14 shows results for the present CASP. Overall, GDT-HA and NMR data are correlated, but there are cases where the model has improved agreement with experiment and worse GDT-HA (upper-left quadrant) and vice versa (lower-right quadrant). These results follow the trend found in CASP8. Bear in mind that NOE violations are measured from an ensemble of structures, meaning that at any point it is possible for a structure to not obey some of the NOE restraints. Although this is a fairly simple implementation of experimental information on the predicted models, it is still quite informative beyond the information obtained by comparing with a single structure. In the future, deeper insights into the qualties of structures may be obtainable from NMR ensembles.
Figure 14.
Changes in GDT-HA and agreement with NOE data are partially correlated (all models, all groups)
5.2 Comparison with X-ray data
Structure prediction methods are already benefiting biomolecular X-ray crystallography by providing input structures to molecular replacement (MR) likelihood algorithms [24, 25] as implemented in the Phaser software package [34]. The purpose of molecular replacement is to use predicted homologous structures to solve the phase problem [35, 36, 37, 38]. We estimate the quality of the predictions for this purpose by calculating the z-score of the best orientation of the model in the unit cell of the crystal with respect to placing it in a set of random orientations (for more details see supplementary information and ref. [39]). We performed this calculation for all predictions on each of the eleven targets where the structure was determined experimentally by X-ray crystallography. Intuitively, if the prediction matches the data poorly, then the orientation does not matter very much. However, if the prediction is a good fit to the data, then the optimal alignment is much better than random and the resulting Z-scores are much higher. Empirically, Z-scores below 6 usually do not provide accurate enough initial phases for crystallographic refinement to be successful, while scores above 8 usually result in success.
Table 2 shows the best molecular replacement results for each target. Out of the eleven X-ray derived targets with available diffraction data, five starting models were already reliable MR solutions. However, there were six starting models with Z-scores below 4.5 that are not of sufficient quality to solve the phase problem. In these latter cases, the best predictions provide useful or even dramatic improvements as MR inputs as demonstrated by a increase of the Z-scores in all cases (with two dramatic improvements to > 27). Overall, the average Z-score of the best predictions is 8.5 standard deviations better than the average of the starting model. However, the Z-scores are much higher when the structure from the PDB is used as input (Control), indicating that there is significant room for further improvements to structure prediction algorithms. For example, the best predictions only slightly reduced the number of clashes (PACK) between non-crystallographic symmetry (NCS) copies and/or symmetry mates, whereas the PDB models have no C-alpha atoms within 3 Å of each other. Accordingly, it might be helpful in future CASP refinement events to give information regarding the crystal symmetry, since this missing information will have some influence on the final refined structures. A recent experiment demonstrated that structure prediction is more accurate when unit cell and NCS information is available [40].
Table 2.
Best MR scores for each target. Z-scores are computed from applying Equation 1 in SI for the optimal solution and comparing it to the mean and standard deviation for 500 random solutions. Z-scores above 6 may be good enough to solve the phase problem, but generally numbers higher than 8 are preferred. PACK refers to the number of packing violations.
| Target | PDB ID | Template | Best Prediction | Control | ||||
|---|---|---|---|---|---|---|---|---|
| Z | PACK | ID | Z | PACK | Z | PACK | ||
| 517 | 3pnx | 4.4 | 5 | PCONSD | 27.4 | 0 | 54.7 | 0 |
| 530 | 3npp | 9.5 | 5 | SBTJ | 17.0 | 5 | 33.0 | 0 |
| 567 | 3n70 | 4.8 | 4 | FEIG | 27.1 | 1 | 50.8 | 0 |
| 568 | 3n6y | 4.4 | 3 | BILAB-SOLO | 6.3 | 5 | 27.6 | 0 |
| 574 | 3nrf | 4.4 | 3 | PCONSM, RECOMBINEIT | 6.6 | 5 | 31.6 | 0 |
| 576 | 3na2 | 14.5 | 0 | LEE | 18.2 | 2 | 51.9 | 0 |
| 592 | 3nhv | 11.5 | 2 | FEIG | 32.5 | 0 | 60.4 | 0 |
| 594 | 3ni8 | 9.1 | 0 | MIDWAYHUMAN | 11.8 | 2 | 25.4 | 0 |
| 606 | 3noh | 3.6 | 5 | GENESILICO | 6.8 | 0 | 17.3 | 0 |
| 622 | 3nkl | 12.9 | 0 | LEE | 17.6 | 0 | 38.8 | 0 |
| 624 | 3nrl | 4.4 | 1 | FOLDIT | 5.9 | 5 | 28.3 | 0 |
|
| ||||||||
| Mean | 7.6 | 2.5 | 16.1 | 2.3 | 38.2 | 0 | ||
Despite the apparent difficulty of the protein structure refinement problem, these results clearly indicate that refinement is of practical utility for molecular replacement and can make crystallographic refinement more successful. Successful MR only requires that one model results in a viable solution. The inability to correctly rank order predictions does not preclude refinement techniques from being successful at molecular replacement, as the MR-Z score will indicate when a good model has been produced. It is possible to scan through many models until a good solution is found—limited by available computer power and time.
It is difficult to turn the MR Z-score into a single average number per group for two reasons. First, the sensitivity of different targets to the correct orientation is different. In particular, the presence of NCS dramatically increases the sensitivity to correct orientation. Second, the MR Z-score is asymmetric. A worse MR solution only lowers the Z-score slightly, while a better MR solution can lead to a large increase in Z-score. Instead, Table 3 reports how often each group was able to improve Z-scores from the likely unrefinable (< 7) to likely refinable (> 7). Overall, 20 groups were able to improve at least one model to the point where MR was likely to succeed.
Table 3.
Number of targets where molecular replacement results went from unlikely-to-succeed (TFZ < 7) to likely-to-succeed (TFZ > 7). Groups that did not improve any targets are not included.
| Group | Improved | Attempted |
|---|---|---|
| MIDWAYSERVER | 2 | 3 |
| ZHANG | 2 | 6 |
| LEE | 2 | 6 |
| BAKER | 2 | 6 |
| FEIG | 2 | 6 |
| SEOK | 2 | 6 |
| PCONS | 2 | 6 |
| PROQ2 | 2 | 6 |
| GENESILICO | 1 | 6 |
| KEASAR | 1 | 6 |
| KNOWMIN | 1 | 6 |
| SBTJ | 1 | 1 |
| MIDWAYHUMAN | 1 | 5 |
| SCHRODERLAB | 1 | 6 |
| SAMUDRALA | 1 | 6 |
| YASARA | 1 | 6 |
| PCONSM | 1 | 6 |
| PCOMB | 1 | 6 |
| PROQ | 1 | 6 |
| PCONSD | 1 | 3 |
| Null | 0 | 6 |
Figure 15 shows the correlation between GDT-HA and MR Z-score. Models below 30 GDT-HA are not useful for phasing. Above 30 GDT-HA, there is some correlation, but a high GDT-HA does not necessarily imply that a model will be useful for phasing. We have also examined (not shown) the correlations between MR Z-score and our other metrics, as well as weighted combinations of two metrics, and we do not find any clear correlations with MR Z-score. This illustrates, once again, that there is no “right” score for evaluating models. The relative performance of different groups ultimately depends on the end use of the refined model.
Figure 15.
GDT-HA and MR Z-score are only partially correlated (all models, all groups). Structures with low GDT-HA perform poorly in molecular replacement. Models with higher GDT-HA perform better. A high GDT-HA, however, does not guarantee good performance in molecular replacement..
6 Ideas for future refinement
Based on our experience as CASP assessors, we suggest several improvements to the structure of the refinement experiment:
It is critical that refinement have more targets. We realize there are logistical challenges to this, but it is very difficult to reach statistically significant conclusions with so few targets.
If a group wishes to test multiple prediction algorithms or scoring functions, they should register multiple groups. Otherwise any information regarding ranking is lost and it becomes difficult for the assessors to say anything meaningful.
If groups could submit a larger unranked ensemble—in addition to the usual five ranked models—it would help to separate the refinement problem into two questions: (1) Can we generate better models? and (2) Can we pick them out?
We also suggest that the organizers work with predictors to maintain a reference refinement algorithm that can be frozen at its current state. This would make it easier to assess changes in target difficulty and if there has been any progress in refinement.
It has become clear that not knowing the crystal environment poses a challenge for high accuracy predictions in both TBM and refinement. It could be interesting to give the unit cell and non-crystallographic symmetry information to the predictors. This might allow for more successful predictions of targets like TR576, although we realize that this would go beyond the usual CASP test where only the sequence is given.
7 Summary
With CASP10 on the horizon, there appears to be significant room for improvement in the refinement category. Nearly all of the predictors were indistinguishable from or worse than the Null group. Of the groups where there is improvement, most do so at the local level by refining the general physicality and sidechain positions. Few groups can consistently improve backbone positioning (as measured by GDT-HA) and even in successful cases the improvements are typically modest. If the TBM predictors improve the physicality and sidechain modeling of their prediction pipelines, we expect refinement performance to suffer in CASP10 unless more focus is given to broader sampling. Sampling locally around the original model means that most groups will not sample very bad structures but it also limits the ability to produce significant improvements. The inability to rank order models also poses a significant challenge.
Despite current limitations, from the experimental point of view, value was added to the original models that make them more useful for techniques such as molecular replacement. This happened for all targets in this competition, with some models having a very large improvement. Even with all of the current challenges, refinement appears to be useful for molecular replacement. This also illustrates the difficulty faced by CASP assessors: the utility of a particular prediction varies with the intended use and is not easy to capture in a standardized way.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the organization and support of the CASP organizers and the Structure Prediction Center. The authors are also greatful for the participation of the refinement predictors. The authors would also like to thank Vijay Pande, Timothy D. Fenn, and Christopher J. Fennell for helpful discussions. Computational support was provided in part by Pengyu Ren. This work was supported by NIH grants GM090205, GM34993, and GM081210. AP acknowledges support from EMBO long term fellowship (ALTF 1107-2009).
References
- 1.MacCallum Justin L, Hua L, Schnieders MJ, Pande VS, Jacobson MP, Dill KA. Assessment of the protein structure refinement category in CASP8. Proteins: Structure, Function, and Bioinformatics. 2009;77(S9):66–80. doi: 10.1002/prot.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chopra Gaurav, Summa Christopher M, Levitt Michael. Solvent dramatically affects protein structure refinement. P Natl Acad Sci USA. 2008;105:20239–20244. doi: 10.1073/pnas.0810818105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Hao, Mark Alan E. Refinement of homology-based protein structures by molecular dynamics simulation techniques. Protein Sci. 2004;13:211–220. doi: 10.1110/ps.03381404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishitani Ryuichiro, Terada Tohru, Shimizu Kentaro. Refinement of comparative models of protein structure by using multicanonical molecular dynamics simulations. Mol Simulation. 2008;34:327–336. [Google Scholar]
- 5.Jagielska Anna, Wroblewska Liliana, Skolnick Jeffrey. Protein model refinement using an optimized physics-based all-atom force field. P Natl Acad Sci USA. 2008;105:8268–73. doi: 10.1073/pnas.0800054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MR, Tsai J, Baker D, Kollman PA. Molecular dynamics in the endgame of protein structure prediction. J Mol Biol. 2001;313:417–30. doi: 10.1006/jmbi.2001.5032. [DOI] [PubMed] [Google Scholar]
- 7.Lu H, Skolnick Jeffrey. Application of statistical potentials to protein structure refinement from low resolution ab initio models. Biopolymers. 2003;70:575–584. doi: 10.1002/bip.10537. [DOI] [PubMed] [Google Scholar]
- 8.Misura Kira MS, Baker David. Progress and challenges in high-resolution refinement of protein structure models. Proteins. 2005 Apr;59(1):15–29. doi: 10.1002/prot.20376. [DOI] [PubMed] [Google Scholar]
- 9.Misura Kira MS, Chivian Dylan, Rohl Carol A, Kim David E, Baker David. Physically realistic homology models built with ROSETTA can be more accurate than their templates. P Natl Acad Sci USA. 2006;103:5361–6. doi: 10.1073/pnas.0509355103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sellers Benjamin D, Zhu Kai, Zhao Suwen, Friesner Richard A, Jacobson Matthew P. Toward better refinement of comparative models: Predicting loops in inexact environments. Proteins. 2008;72:959–971. doi: 10.1002/prot.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wroblewska Liliana, Skolnick Jeffrey. Can a physics-based, all-atom potential find a protein’s native structure among misfolded structures? i. large scale amber benchmarking. J Comput Chem. 2007;28:2059–2066. doi: 10.1002/jcc.20720. [DOI] [PubMed] [Google Scholar]
- 12.Wroblewska L, Jagielska A, Skolnick J. Development of a physics-based force field for the scoring and refinement of protein models. Biophys J. 2008;94:3227–3240. doi: 10.1529/biophysj.107.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Jiang, Fan Hao, Periole Xavier, Honig Barry, Mark Alan E. Refining homology models by combining replica-exchange molecular dynamics and statistical potentials. Proteins. 2008;72:1171–1188. doi: 10.1002/prot.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Jianhan, Brooks Charles L., III Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins. 2007;67:922–930. doi: 10.1002/prot.21345. [DOI] [PubMed] [Google Scholar]
- 15.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian Bin, Ortiz Angel R, Baker David. Improvement of comparative model accuracy by free-energy optimization along principal components of natural structural variation. P Natl Acad Sci USA. 2004;101:15346–15351. doi: 10.1073/pnas.0404703101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kryshtafovych Andriy, Prlic Andreas, Dmytriv Zinoviy, Daniluk Pawel, Maciej Milostan, Eyrich Volker, Hubbard Tim, Fidelis Krzysztof. New tools and expanded data analysis capabilities at the protein structure prediction center. Proteins. 2007;69(Suppl 8):19–26. doi: 10.1002/prot.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–25. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 20.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. Amber. 2008;10 [Google Scholar]
- 21.Zemla Adam. LGA: A method for finding 3D similarities in protein structures. Nucleic acids research. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keedy Daniel A, Williams Christopher J, Headd Jeffrey J, Bryan Arendall W, Chen Vincent B, Kapral Gary J, Gillespie Robert A, Block Jeremy N, Zemla Adam, Richardson David C, Richardson Jane S. The other 90% of the protein: assessment beyond the Calphas for CASP8 template-based and high-accuracy models. Proteins: Structure, Function, and Bioinformatics. 2009;77(Suppl 9):29–49. doi: 10.1002/prot.22551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Vincent B, Bryan Arendall W, Headd Jeffrey J, Keedy Daniel A, Immormino Robert M, Kapral Gary J, Murray Laura W, Richardson Jane S, Richardson David C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010 jan;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallographica Section D-Biological Crystallography. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 25.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallographica Section D-Biological Crystallography. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 26.Mobley DL, Chodera JD, Dill KA. On the use of orientational restraints and symmetry corrections in alchemical free energy calculations. J Chem Phys. 2006;125(8):084902. doi: 10.1063/1.2221683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryshtafovych A, Venclovas C, Fidelis K, Moult J. Progress over the first decade of CASP experiments. Proteins. 2005;61(Suppl 7):225–36. doi: 10.1002/prot.20740. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen BF, Stock AM, Ringe D, Petsko GA. Crystalline ribonuclease a loses function below the dynamical transition at 220k. Nature. 1992;357:423–424. doi: 10.1038/357423a0. [DOI] [PubMed] [Google Scholar]
- 29.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 30.Benkovic SJ, Hammes-Schiffer SA. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 31.Zagrovic B, Gunsteren WF. Comparing atomistic simulation data with the NMR experiment: How much can noes actually tell us. Proteins. 2006;63:210–218. doi: 10.1002/prot.20872. [DOI] [PubMed] [Google Scholar]
- 32.Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 33.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 34.McCoy Airlie J, Grosse-Kunstleve Ralf W, Adams Paul D, Winn Martyn D, Storoni Laurent C, Read Randy J. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40(4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giorgetti Alejandro, Raimondo Domenico, Miele Adriana Erica, Tramontano Anna. Evaluating the usefulness of protein structure models for molecular replacement. Bioinformatics. 2005;21(Suppl 2):72–6. doi: 10.1093/bioinformatics/bti1112. [DOI] [PubMed] [Google Scholar]
- 36.Raimondo D, Giorgetti A, Bosi S, Tramontano A. Automatic procedure for using models of proteins in molecular replacement. Proteins-Structure Function and Bioinformatics. 2007;66(3):689–696. doi: 10.1002/prot.21225. [DOI] [PubMed] [Google Scholar]
- 37.Das R, Baker D. Prospects for de novo phasing with de novo protein models. Acta Crystallographica Section D-Biological Crystallography. 2009;65:169–175. doi: 10.1107/S0907444908020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian Bin, Raman Srivatsan, Das Rhiju, Bradley Philip, McCoy Airlie J, Read Randy J, Baker David. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450(7167):259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy Airlie. Liking likelihood. Acta Crystallographica Section D. 2004;60(12 Part 1):2169–2183. doi: 10.1107/S0907444904016038. [DOI] [PubMed] [Google Scholar]
- 40.Tyka Michael D, Keedy Daniel A, Andr Ingemar, DiMaio Frank, Song Yifan, Richardson David C, Richardson Jane S, Baker David. Alternate states of proteins revealed by detailed energy landscape mapping. Journal of Molecular Biology. 2011;405(2):607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.