Abstract
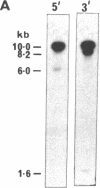
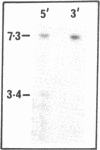
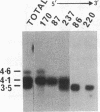
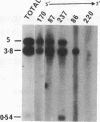
We report here that component members of the keratin intermediate filament (IF) type I and type II gene families of sheep are closely linked but apparently the two families are not. Nine genes, accounting for up to half of the keratin IF gene repertoire, were mapped in four cosmid clones and the linkage between the genes ranged from several kilobases to 20 kilobases. In one cosmid, three tandem type I genes had the same transcriptional arrangement and were regularly spaced. In another cosmid, tandem genes encoding type II keratins were identified and, surprisingly, a solitary exon was discovered in the intergene region between the two type II genes. In a normal gene this exon encodes one of the most conserved amino acid regions of IF proteins, the C-terminal end of the alpha-helical core. Homologous C-terminal protein subdomains were encoded by two wool keratin type II genes and we suggest that this arrangement may also exist in the other wool keratin type II genes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Eichner R., Bonitz P., Sun T. T. Classification of epidermal keratins according to their immunoreactivity, isoelectric point, and mode of expression. J Cell Biol. 1984 Apr;98(4):1388–1396. doi: 10.1083/jcb.98.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. V., Coppock S. M., Green H., Cleveland D. W. Two distinct classes of keratin genes and their evolutionary significance. Cell. 1981 Nov;27(1 Pt 2):75–84. doi: 10.1016/0092-8674(81)90362-7. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Marchuk D. Type I and type II keratins have evolved from lower eukaryotes to form the epidermal intermediate filaments in mammalian skin. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5857–5861. doi: 10.1073/pnas.80.19.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N., Kaufmann E., Fischer S., Plessmann U., Weber K. Neurofilament architecture combines structural principles of intermediate filaments with carboxy-terminal extensions increasing in size between triplet proteins. EMBO J. 1983;2(8):1295–1302. doi: 10.1002/j.1460-2075.1983.tb01584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Hanukoglu I., Fuchs E. The cDNA sequence of a Type II cytoskeletal keratin reveals constant and variable structural domains among keratins. Cell. 1983 Jul;33(3):915–924. doi: 10.1016/0092-8674(83)90034-x. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Johnson L. D., Idler W. W., Zhou X. M., Roop D. R., Steinert P. M. Structure of a gene for the human epidermal 67-kDa keratin. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1896–1900. doi: 10.1073/pnas.82.7.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorcano J. L., Franz J. K., Franke W. W. Amino acid sequence diversity between bovine epidermal cytokeratin polypeptides of the basic (type II) subfamily as determined from cDNA clones. Differentiation. 1984;28(2):155–163. doi: 10.1111/j.1432-0436.1984.tb00278.x. [DOI] [PubMed] [Google Scholar]
- Krieg T. M., Schafer M. P., Cheng C. K., Filpula D., Flaherty P., Steinert P. M., Roop D. R. Organization of a type I keratin gene. Evidence for evolution of intermediate filaments from a common ancestral gene. J Biol Chem. 1985 May 25;260(10):5867–5870. [PubMed] [Google Scholar]
- Lazarides E. Intermediate filaments: a chemically heterogeneous, developmentally regulated class of proteins. Annu Rev Biochem. 1982;51:219–250. doi: 10.1146/annurev.bi.51.070182.001251. [DOI] [PubMed] [Google Scholar]
- Lehnert M. E., Jorcano J. L., Zentgraf H., Blessing M., Franz J. K., Franke W. W. Characterization of bovine keratin genes: similarities of exon patterns in genes coding for different keratins. EMBO J. 1984 Dec 20;3(13):3279–3287. doi: 10.1002/j.1460-2075.1984.tb02290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk D., McCrohon S., Fuchs E. Complete sequence of a gene encoding a human type I keratin: sequences homologous to enhancer elements in the regulatory region of the gene. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1609–1613. doi: 10.1073/pnas.82.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Moll R., Franke W. W., Schiller D. L., Geiger B., Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982 Nov;31(1):11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Parry D. A., Steven A. C., Steinert P. M. The coiled-coil molecules of intermediate filaments consist of two parallel chains in exact axial register. Biochem Biophys Res Commun. 1985 Mar 29;127(3):1012–1018. doi: 10.1016/s0006-291x(85)80045-0. [DOI] [PubMed] [Google Scholar]
- Quax W., Egberts W. V., Hendriks W., Quax-Jeuken Y., Bloemendal H. The structure of the vimentin gene. Cell. 1983 Nov;35(1):215–223. doi: 10.1016/0092-8674(83)90224-6. [DOI] [PubMed] [Google Scholar]
- Roop D. R., Tsai M. J., O'Malley B. W. Definition of the 5' and 3' ends of transcripts of the ovalbumin gene. Cell. 1980 Jan;19(1):63–68. doi: 10.1016/0092-8674(80)90388-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B., Parker R. C., Davidson N. Representation of DNA sequences in recombinant DNA libraries prepared by restriction enzyme partial digestion. Gene. 1982 Sep;19(2):201–209. doi: 10.1016/0378-1119(82)90007-5. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Steinert P. M., Parry D. A., Idler W. W., Johnson L. D., Steven A. C., Roop D. R. Amino acid sequences of mouse and human epidermal type II keratins of Mr 67,000 provide a systematic basis for the structural and functional diversity of the end domains of keratin intermediate filament subunits. J Biol Chem. 1985 Jun 10;260(11):7142–7149. [PubMed] [Google Scholar]
- Steinert P. M., Parry D. A., Racoosin E. L., Idler W. W., Steven A. C., Trus B. L., Roop D. R. The complete cDNA and deduced amino acid sequence of a type II mouse epidermal keratin of 60,000 Da: analysis of sequence differences between type I and type II keratins. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5709–5713. doi: 10.1073/pnas.81.18.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert P. M., Steven A. C., Roop D. R. The molecular biology of intermediate filaments. Cell. 1985 Sep;42(2):411–420. doi: 10.1016/0092-8674(85)90098-4. [DOI] [PubMed] [Google Scholar]
- Tyner A. L., Eichman M. J., Fuchs E. The sequence of a type II keratin gene expressed in human skin: conservation of structure among all intermediate filament genes. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4683–4687. doi: 10.1073/pnas.82.14.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]