Abstract
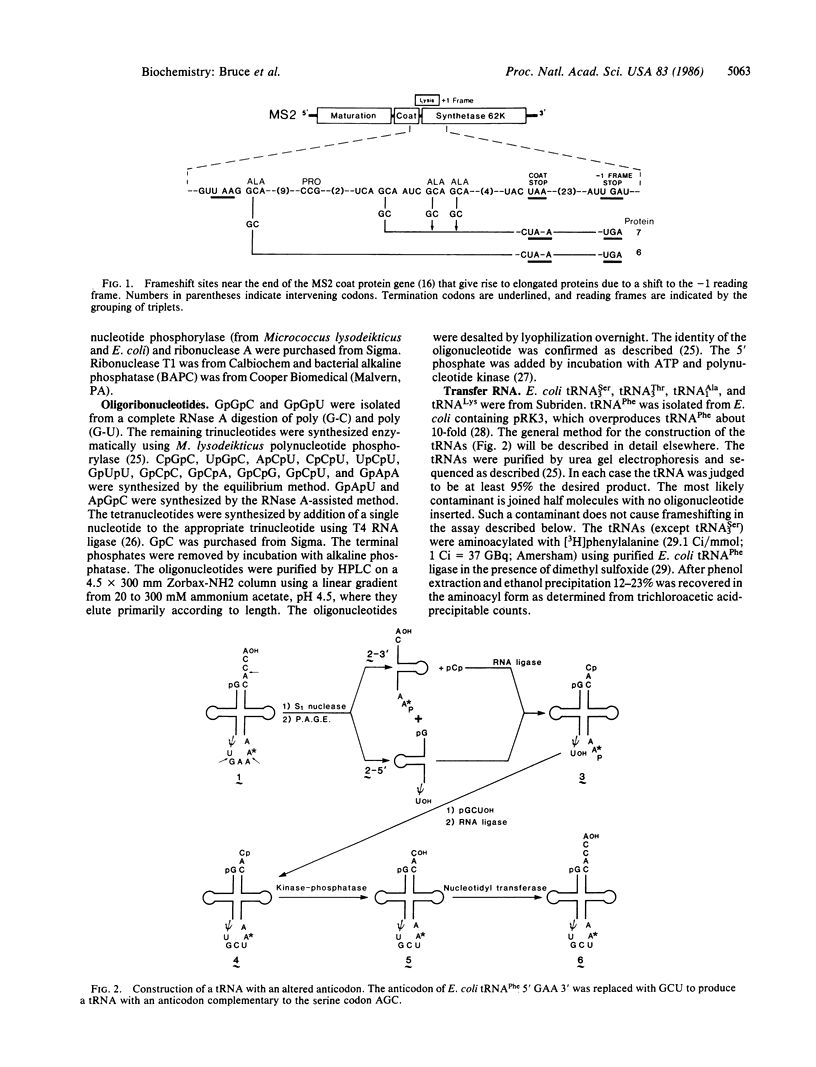
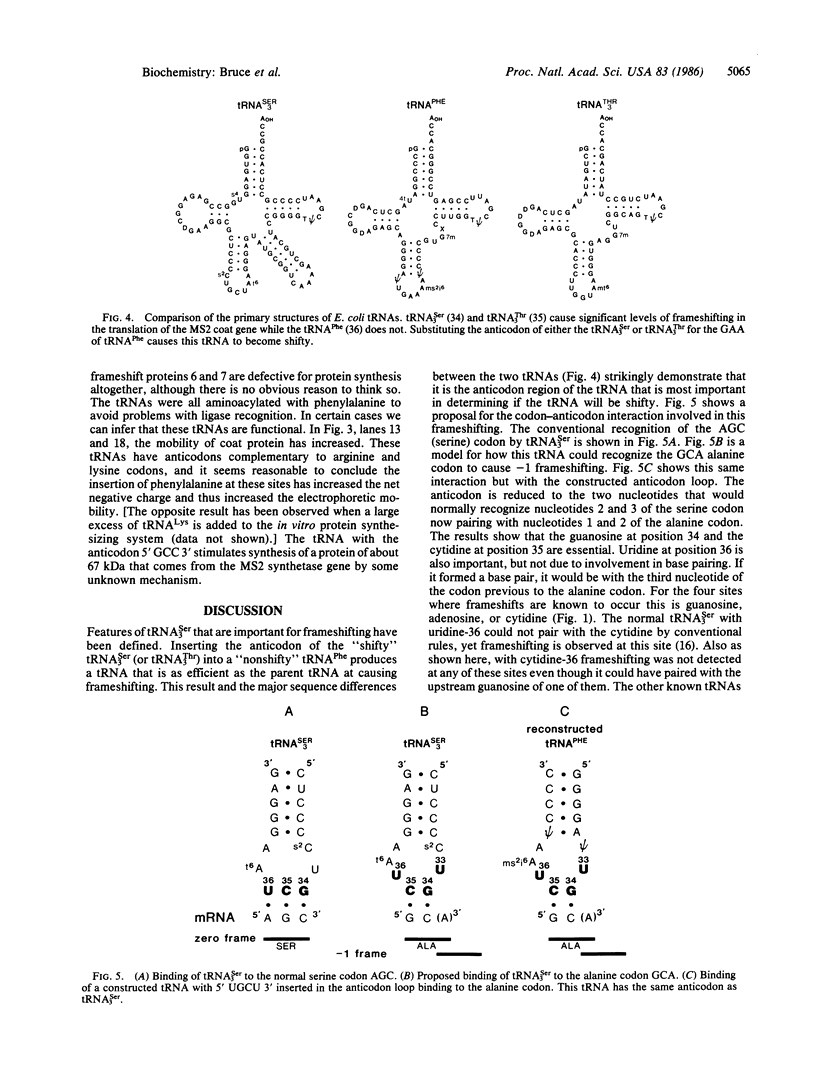
The expression of certain normal genes requires a specific ribosomal frameshift event because the mRNA has the coding information for one protein in two different reading frames. One of several possible mechanisms for this involves recognition of a nontriplet codon by a noncognate tRNA. The AGUC-decoding Escherichia coli tRNASer3 reads a GCA alanine codon to cause a -1 frameshift. Replacement of the anticodon of tRNAPhe with the anticodon of tRNASer3 allows the constructed tRNA to cause this frameshifting. By altering the anticodon loop nucleotides at positions 33-36 in the constructed tRNAPhe molecules, the tRNA was found to recognize a 2-base codon. Instead of the usual anticodon, positions 34-36, the nucleotides in positions 34 and 35 form essential base pairs with the first two positions of the alanine codon. The uridine in position 36 is also required but not for base pairing.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Gesteland R. F., Reid B. R., Anderson C. W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979 Dec;18(4):1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Atkins J. F., Nichols B. P., Thompson S. The nucleotide sequence of the first externally suppressible--1 frameshift mutant, and of some nearby leaky frameshift mutants. EMBO J. 1983;2(8):1345–1350. doi: 10.1002/j.1460-2075.1983.tb01590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer D., Yarus M. The context effect does not require a fourth base pair. Science. 1986 Jan 24;231(4736):393–395. doi: 10.1126/science.3510456. [DOI] [PubMed] [Google Scholar]
- Bare L., Bruce A. G., Gesteland R., Uhlenbeck O. C. Uridine-33 in yeast tRNA not essential for amber suppression. Nature. 1983 Oct 6;305(5934):554–556. doi: 10.1038/305554a0. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Sanger F. The sequence of phenylalanine tRNA from E. coli. FEBS Lett. 1969 Jun;3(4):275–278. doi: 10.1016/0014-5793(69)80157-2. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6105–6109. doi: 10.1073/pnas.81.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Manthey A. E., Gumport R. I. Using T4 RNA ligase with DNA substrates. Methods Enzymol. 1983;100:38–52. doi: 10.1016/0076-6879(83)00044-0. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Atkins J. F., Wills N., Uhlenbeck O., Gesteland R. F. Replacement of anticodon loop nucleotides to produce functional tRNAs: amber suppressors derived from yeast tRNAPhe. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7127–7131. doi: 10.1073/pnas.79.23.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Specific interaction of anticodon loop residues with yeast phenylalanyl-tRNA synthetase. Biochemistry. 1982 Aug 17;21(17):3921–3926. doi: 10.1021/bi00260a003. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Elseviers D., Gallagher P., Hoffman A., Weinberg B., Schwartz I. Molecular cloning and regulation of expression of the genes for initiation factor 3 and two aminoacyl-tRNA synthetases. J Bacteriol. 1982 Oct;152(1):357–362. doi: 10.1128/jb.152.1.357-362.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Kern D., Ebel J. P. Incorrect aminoacylations catalysed by E. coli valyl-tRNA synthetase. Biochimie. 1972;54(10):1245–1255. doi: 10.1016/s0300-9084(72)80065-8. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Brakenhoff J., De Vries B. F., Sloof P., Tromp M. C., Van Boom J. H., Benne R. The sequence of the gene for cytochrome c oxidase subunit I, a frameshift containing gene for cytochrome c oxidase subunit II and seven unassigned reading frames in Trypanosoma brucei mitochondrial maxi-circle DNA. Nucleic Acids Res. 1984 Oct 11;12(19):7327–7344. doi: 10.1093/nar/12.19.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U. "Two out of three": an alternative method for codon reading. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1759–1762. doi: 10.1073/pnas.75.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Fulton S. M., Dobson M. J., Wilson W., Kingsman S. M., Kingsman A. J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985 Jan 17;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- Michael N. L., Rothbard J. B., Shiurba R. A., Linke H. K., Schoolnik G. K., Clayton D. A. All eight unassigned reading frames of mouse mitochondrial DNA are expressed. EMBO J. 1984 Dec 20;3(13):3165–3175. doi: 10.1002/j.1460-2075.1984.tb02275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand K. N., Gait M. J. Sequence and cloning of bacteriophage T4 gene 63 encoding RNA ligase and tail fibre attachment activities. EMBO J. 1984 Feb;3(2):397–402. doi: 10.1002/j.1460-2075.1984.tb01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz I., Klotsky R. A., Elseviers D., Gallagher P. J., Krauskopf M., Siddiqui M. A., Wong J. F., Roe B. A. Molecular cloning and sequencing of pheU, a gene for Escherichia coli tRNAPhe. Nucleic Acids Res. 1983 Jul 11;11(13):4379–4389. doi: 10.1093/nar/11.13.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Squires C., Konrad B., Kirschbaum J., Carbon J. Three adjacent transfer RNA genes in Escherichia coli. Proc Natl Acad Sci U S A. 1973 Feb;70(2):438–441. doi: 10.1073/pnas.70.2.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternbach H., von der Haar F., Schlimme E., Gaertner E., Cramer F. Isolation and properties of tRNA nucleotidyl transferase from yeast. Eur J Biochem. 1971 Sep 24;22(2):166–172. doi: 10.1111/j.1432-1033.1971.tb01528.x. [DOI] [PubMed] [Google Scholar]
- Stulberg M. P. The isolation and properties of phenylalanyl ribonucleic acid synthetase from Escherichia coli B. J Biol Chem. 1967 Mar 10;242(5):1060–1064. [PubMed] [Google Scholar]
- Sugiura M. Purification of the T4 DNA ligase by Blue Sepharose chromatography. Anal Biochem. 1980 Nov 1;108(2):227–229. doi: 10.1016/0003-2697(80)90573-4. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Cameron V. Equimolar addition of oligoribonucleotides with T4 RNA ligase. Nucleic Acids Res. 1977 Jan;4(1):85–98. doi: 10.1093/nar/4.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Brummer B., Hüttenhofer A., Kaudewitz F. Leakiness of termination codons in mitochondrial mutants of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1984;198(2):62–68. doi: 10.1007/BF00328702. [DOI] [PubMed] [Google Scholar]
- Weiss R. B. Molecular model of ribosome frameshifting. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5797–5801. doi: 10.1073/pnas.81.18.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R., Gallant J. Mechanism of ribosome frameshifting during translation of the genetic code. 1983 Mar 31-Apr 6Nature. 302(5907):389–393. doi: 10.1038/302389a0. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Ishikura H. Nucleotide sequence of tRNA(Ser)(3) from Escherichia coli. FEBS Lett. 1973 Feb 1;29(3):231–234. doi: 10.1016/0014-5793(73)80026-2. [DOI] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T. D., Smythers G. W., Oroszlan S. Bovine leukemia virus protease: purification, chemical analysis, and in vitro processing of gag precursor polyproteins. J Virol. 1986 Mar;57(3):826–832. doi: 10.1128/jvi.57.3.826-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz V. F., Neckelmann N., Simpson L. Sequences of six genes and several open reading frames in the kinetoplast maxicircle DNA of Leishmania tarentolae. J Biol Chem. 1984 Dec 25;259(24):15136–15147. [PubMed] [Google Scholar]



