Abstract
The title dammarane triterpenoid, C30H50O4, assigned the name chrysura, was isolated from an ethyl acetate extract of Walsura chrysogyne leaves (Meliaceae). It has 20S*,24S* relative stereochemistry and an oxepanone ring with two methyl groups at position 4. The two cyclohexane rings adopt chair conformations. The cyclopentane and tetrahydrofuran rings have envelope conformations; their mean planes make a dihedral angle of 13.1 (3)°, indicating that the rings are only slightly tilted with respect to each other. There is an intramolecular C—H⋯O hydrogen bond in the molecule, which forms S(6) and S(7) ring motifs. In the crystal, molecules are linked via O—H⋯O and C—H⋯O hydrogen bonds, forming chains propagating along [001] which stack along the b-axis direction.
Related literature
For related structures, see: Pan et al. (2010 ▶). For graph-set analysis, see: Bernstein et al. (1995 ▶). For the biological activity of related compounds, see: Burkill (1966 ▶); Hegnauer (1990 ▶); Fujiwara et al. (1982 ▶).
Experimental
Crystal data
C30H50O4
M r = 474.70
Orthorhombic,
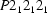
a = 6.9881 (1) Å
b = 11.0108 (2) Å
c = 34.9733 (7) Å
V = 2691.01 (8) Å3
Z = 4
Cu Kα radiation
μ = 0.59 mm−1
T = 100 K
0.40 × 0.08 × 0.07 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.801, T max = 0.960
48305 measured reflections
5058 independent reflections
5040 reflections with I > 2σ(I)
R int = 0.045
Refinement
R[F 2 > 2σ(F 2)] = 0.092
wR(F 2) = 0.233
S = 1.21
5058 reflections
316 parameters
H-atom parameters constrained
Δρmax = 0.47 e Å−3
Δρmin = −0.38 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811047337/su2332sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811047337/su2332Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C19—H19A⋯O32 | 0.96 | 2.44 | 3.082 (6) | 124 |
| O34—H34A⋯O32i | 0.82 | 2.20 | 3.010 (5) | 170 |
| C26—H26A⋯O31i | 0.96 | 2.53 | 3.392 (7) | 150 |
Symmetry code: (i) 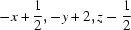 .
.
Acknowledgments
This research work was supported financially by the Research University Grant Scheme (RUGS: 05–01–09–0732RU) of Universiti Putra Malaysia, Malaysia.
supplementary crystallographic information
Comment
Meliaceae or Mahogany is a plant family, in the order of Sapindales, which consists of flowering plants of mostly trees, shrubs and a few herbaceous plants (Burkill, 1966). This family is noted for the wide range of compounds of different classes of which it is compossed, for example, terpenoids (triterpenoids, monoterpenes, sesquiterpenes, limonoids), saponins, alkaloids, polyphenols, quinines, fatty and hydroxyl acids (Hegnauer, 1990). Among these groups of constituents, some are responsible for biological activities such as antiviral, anthelmintic, antitumor, anti-inflammatory and anti-rheumatic, which have been scientifically proven (Fujiwara et al., 1982). Walsura chrysogyne is a Meliaceae species which is among the least explored of higher plants.
The title dammarane triterpenoid, namely chrysura (1), has been isolated for the first time from the ethyl acetate extract of the leaves of Walsura chrysogyne (Meliaceae). Recently, the same compound was reported to have been obtained from Aglaia foveolata, but in resin form (compound 5 in reference Pan et al., 2010). They determined its relative stereochemistry by Nuclear Magnetic Resonance (NMR) spectroscopy. Herein, we describe the crystal structure of the title compound, chrysura (1), whose relative configuration was also obtained by two-dimensional NMR spectroscopy. By a close comparison of the 13C NMR signals at C-20, C-21, C-22, C-23 and C-24 reported for compound 5 (δ 86.5, 27.2, 34.8, 26.3 and 86.4; Pan et al., 2010) and those obtained for the title compound, chrysura (1) (δ 86.5, 27.2, 35.0, 26.4 and 86.5), it was shown that these two compounds are identical. This is substantiated by the 1H NMR signal at H-24 of chrysura (1), which is a doublet of doublet with J values of 10 and 5.5 Hz, comparable to the values observed for compound 5, that is 9.9 and 5.6 Hz. Hence, the relative configuration at C20 and C24 of chrysura (1), was determined by NMR to be the same as that of compound 5 [Pan et al., 2010].
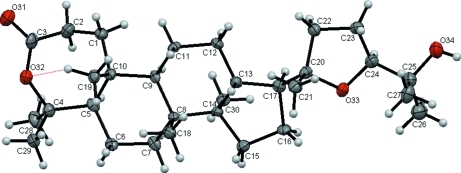
The molecular structure of the title molecule, chrysura (1), is shown in Fig. 1. The two cyclohexane rings, B (C5-C10) and C (C8,C9,C11-C14), adopt chair conformations. The cyclopentane ring D (C13-C17) and the tetrahydrofuran ring E (O33,C20, C22-C24) have envelope conformations, with atoms C14 and C23 at the flap of rings D and E, respectively. The mean planes through rings D and E make a dihedral angle of 13.1 (3)°, indicating that they are only slightly twisted with respect to each other. As shown in Fig. 1, the structure of the molecule is stabilized by an intramolecular C—H···O hydrogen bond (Table 1), which forms S(6) and S(7) ring motifs (Bernstein et al., 1995).
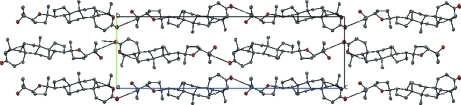
In the crystal of chrysura (1), molecules are linked via intermolecular O—H···O and C—H···O hydrogen bonds (Table 1), forming chains propagating along [001]. These chains stack along the b-axis, as shown in Fig. 2.
Hence, in the title compound, chrysura (1), the relative configurations at C20 and C24 of the epoxy unit (ring E) have been confirmed to be S-methyl configurations.
Experimental
The air-dried ground leaves of Walsura chrysogyne (8.94 kg) collected at Pasir Raja, Terengganu, Malaysia, were macerated in methanol at room temperature (3 × 1000 ml). The crude extract (230 g) was partitioned into hexane (12.2 g), ethyl acetate (EtOAc; 16.6 g), and water (16.8 g). A portion (9.0 g) of the EtOAc extract was further fractionated by using vacuum column chromatography on silica gel normal phase (7.5 × 20 cm) eluted with CHCl3, and CHCl3—MeOH in 10% increasing amounts of MeOH. Fraction MeOH-CHCl3 [9:1] (2.0 g) was subjected to another column chromatography on Sephadex LH-20 (2 × 30 cm) with CHCl3–MeOH (9:1) to yield four fractions. The fraction obtained by hexane-EtOAc [7:3] (85.3 mg) was further purified on silica gel normal phase (1 × 20 cm) eluted with hexane-acetone (9:1) to afford the title compound (134.8 mg, 0.059%). Colourless needle-shaped crystals of the title compound, suitable for X-ray diffraction analysis, were recrystallized from ethyl aceate-acetone. The 1H- and 13C-NMR spectral data were consistent with those reported by (Pan et al., 2010).
Refinement
All the H atoms were positioned geometrically and refined using a riding model: O—H = 0.82 Å and C—H = 0.93 – 0.98 Å with Uiso~(H) = 1.5Ueq(O, Cmethyl), and = 1.2Ueq(C) for all other C-bound H atoms. A rotating-group model was applied for the methyl groups. The anomalous dispersion effects of the atoms in the molecule are not sufficient to determine the absolute structure of the molecule in the crystal [Flack parameter = 0.1 (5)].
Figures
Fig. 1.
The molecular structure of the title molecule, chrysura (1), showing 50% probability displacement ellipsoids and the atom-numbering scheme. The intramolecular C-H···O hydrogen bond is shown as a dashed red line.
Fig. 2.
The crystal packing of the title compound, chrysura (1), viewed along the a axis, showing the formation of the hydrogen bonded chains (see Table 1 for details). H atoms not involved in the hydrogen bonds (dashed lines) have been omitted for clarity.
Crystal data
| C30H50O4 | F(000) = 1048 |
| Mr = 474.70 | Dx = 1.172 Mg m−3 |
| Orthorhombic, P212121 | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 9811 reflections |
| a = 6.9881 (1) Å | θ = 3–69° |
| b = 11.0108 (2) Å | µ = 0.59 mm−1 |
| c = 34.9733 (7) Å | T = 100 K |
| V = 2691.01 (8) Å3 | Needle, colourless |
| Z = 4 | 0.40 × 0.08 × 0.07 mm |
Data collection
| Bruker APEXII CCD diffractometer | 5058 independent reflections |
| Radiation source: fine-focus sealed tube | 5040 reflections with I > 2σ(I) |
| graphite | Rint = 0.045 |
| φ and ω scans | θmax = 69.9°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −7→8 |
| Tmin = 0.801, Tmax = 0.960 | k = −13→13 |
| 48305 measured reflections | l = −41→42 |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.092 | w = 1/[σ2(Fo2) + (0.0677P)2 + 8.7996P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.233 | (Δ/σ)max < 0.001 |
| S = 1.21 | Δρmax = 0.47 e Å−3 |
| 5058 reflections | Δρmin = −0.38 e Å−3 |
| 316 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0021 (7) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack, H. D. (1983). Acta Cryst. A39, 876–881 |
| Secondary atom site location: difference Fourier map | Flack parameter: 0.1 (5) |
Special details
| Experimental. The needle-shape crystal was placed in the cold stream of an Oxford Cyrosystems Cobra open-flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100.0 (1) K. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O31 | 0.0510 (6) | 0.8536 (4) | 1.00267 (11) | 0.0404 (10) | |
| O32 | 0.2497 (5) | 0.9976 (3) | 0.98533 (9) | 0.0275 (8) | |
| O33 | 0.3411 (5) | 1.0371 (3) | 0.66313 (9) | 0.0277 (8) | |
| O34 | 0.2551 (6) | 0.9264 (3) | 0.56801 (10) | 0.0345 (9) | |
| H34A | 0.2593 | 0.9556 | 0.5464 | 0.052* | |
| C1 | 0.1697 (7) | 0.8667 (4) | 0.90988 (13) | 0.0237 (10) | |
| H1A | 0.0311 | 0.8708 | 0.9109 | 0.028* | |
| H1B | 0.2030 | 0.8116 | 0.8893 | 0.028* | |
| C2 | 0.2412 (9) | 0.8102 (5) | 0.94789 (14) | 0.0306 (12) | |
| H2A | 0.1914 | 0.7284 | 0.9505 | 0.037* | |
| H2B | 0.3799 | 0.8056 | 0.9477 | 0.037* | |
| C3 | 0.1774 (8) | 0.8847 (5) | 0.98059 (13) | 0.0287 (12) | |
| C4 | 0.4229 (7) | 1.0448 (5) | 0.96570 (13) | 0.0236 (10) | |
| C5 | 0.4357 (7) | 1.0211 (4) | 0.92154 (13) | 0.0209 (10) | |
| H5A | 0.5138 | 0.9478 | 0.9185 | 0.025* | |
| C6 | 0.5520 (6) | 1.1256 (4) | 0.90319 (13) | 0.0203 (10) | |
| H6A | 0.4782 | 1.2001 | 0.9046 | 0.024* | |
| H6B | 0.6691 | 1.1376 | 0.9176 | 0.024* | |
| C7 | 0.6018 (7) | 1.0995 (4) | 0.86170 (13) | 0.0204 (9) | |
| H7A | 0.6752 | 1.0249 | 0.8603 | 0.024* | |
| H7B | 0.6808 | 1.1648 | 0.8518 | 0.024* | |
| C8 | 0.4216 (6) | 1.0873 (4) | 0.83692 (12) | 0.0183 (9) | |
| C9 | 0.2945 (6) | 0.9860 (4) | 0.85516 (12) | 0.0165 (9) | |
| H9A | 0.3707 | 0.9117 | 0.8529 | 0.020* | |
| C10 | 0.2470 (7) | 0.9967 (4) | 0.89916 (12) | 0.0195 (9) | |
| C11 | 0.1148 (6) | 0.9619 (4) | 0.83075 (13) | 0.0190 (9) | |
| H11A | 0.0369 | 1.0348 | 0.8302 | 0.023* | |
| H11B | 0.0400 | 0.8981 | 0.8426 | 0.023* | |
| C12 | 0.1632 (7) | 0.9243 (4) | 0.78936 (13) | 0.0208 (10) | |
| H12A | 0.2262 | 0.8458 | 0.7894 | 0.025* | |
| H12B | 0.0463 | 0.9173 | 0.7746 | 0.025* | |
| C13 | 0.2941 (7) | 1.0189 (4) | 0.77122 (12) | 0.0203 (10) | |
| H13A | 0.2229 | 1.0956 | 0.7717 | 0.024* | |
| C14 | 0.4758 (6) | 1.0405 (4) | 0.79509 (13) | 0.0167 (9) | |
| C15 | 0.5831 (7) | 1.1312 (4) | 0.76941 (13) | 0.0235 (10) | |
| H15A | 0.7189 | 1.1311 | 0.7751 | 0.028* | |
| H15B | 0.5337 | 1.2128 | 0.7728 | 0.028* | |
| C16 | 0.5460 (7) | 1.0854 (5) | 0.72793 (13) | 0.0217 (10) | |
| H16A | 0.6548 | 1.0393 | 0.7187 | 0.026* | |
| H16B | 0.5246 | 1.1535 | 0.7108 | 0.026* | |
| C17 | 0.3625 (6) | 1.0024 (4) | 0.72988 (13) | 0.0191 (10) | |
| H17A | 0.4059 | 0.9182 | 0.7274 | 0.023* | |
| C30 | 0.5988 (7) | 0.9245 (4) | 0.79638 (13) | 0.0220 (10) | |
| H30A | 0.6375 | 0.9032 | 0.7709 | 0.033* | |
| H30B | 0.5254 | 0.8593 | 0.8072 | 0.033* | |
| H30C | 0.7101 | 0.9388 | 0.8118 | 0.033* | |
| C19 | 0.0918 (7) | 1.0887 (4) | 0.90822 (13) | 0.0226 (10) | |
| H19A | 0.0491 | 1.0775 | 0.9341 | 0.034* | |
| H19B | −0.0139 | 1.0775 | 0.8910 | 0.034* | |
| H19C | 0.1421 | 1.1693 | 0.9053 | 0.034* | |
| C20 | 0.2217 (7) | 1.0257 (4) | 0.69762 (13) | 0.0202 (10) | |
| C21 | 0.1120 (7) | 1.1431 (5) | 0.70141 (14) | 0.0261 (11) | |
| H21A | 0.2003 | 1.2093 | 0.7042 | 0.039* | |
| H21B | 0.0308 | 1.1393 | 0.7235 | 0.039* | |
| H21C | 0.0353 | 1.1554 | 0.6790 | 0.039* | |
| C22 | 0.0884 (7) | 0.9175 (5) | 0.68812 (14) | 0.0245 (10) | |
| H22A | 0.1426 | 0.8414 | 0.6970 | 0.029* | |
| H22B | −0.0370 | 0.9283 | 0.6995 | 0.029* | |
| C23 | 0.0774 (8) | 0.9211 (4) | 0.64427 (14) | 0.0255 (10) | |
| H23A | 0.0413 | 0.8428 | 0.6338 | 0.031* | |
| H23B | −0.0120 | 0.9824 | 0.6355 | 0.031* | |
| C24 | 0.2804 (7) | 0.9535 (4) | 0.63416 (14) | 0.0257 (11) | |
| H24A | 0.3588 | 0.8800 | 0.6362 | 0.031* | |
| C25 | 0.3219 (8) | 1.0139 (5) | 0.59506 (15) | 0.0294 (11) | |
| C26 | 0.5357 (9) | 1.0307 (6) | 0.59086 (17) | 0.0382 (14) | |
| H26A | 0.5632 | 1.0659 | 0.5664 | 0.057* | |
| H26B | 0.5981 | 0.9533 | 0.5929 | 0.057* | |
| H26C | 0.5815 | 1.0835 | 0.6107 | 0.057* | |
| C27 | 0.2159 (9) | 1.1313 (5) | 0.58997 (15) | 0.0351 (13) | |
| H27A | 0.2411 | 1.1632 | 0.5649 | 0.053* | |
| H27B | 0.2579 | 1.1886 | 0.6089 | 0.053* | |
| H27C | 0.0811 | 1.1173 | 0.5928 | 0.053* | |
| C28 | 0.5949 (8) | 0.9895 (6) | 0.98605 (15) | 0.0344 (12) | |
| H28A | 0.5901 | 1.0098 | 1.0127 | 0.052* | |
| H28C | 0.7106 | 1.0210 | 0.9751 | 0.052* | |
| H28D | 0.5923 | 0.9028 | 0.9832 | 0.052* | |
| C29 | 0.4088 (9) | 1.1798 (5) | 0.97708 (14) | 0.0302 (12) | |
| H29C | 0.3941 | 1.1862 | 1.0043 | 0.045* | |
| H29D | 0.3003 | 1.2159 | 0.9647 | 0.045* | |
| H29A | 0.5232 | 1.2213 | 0.9694 | 0.045* | |
| C18 | 0.3211 (7) | 1.2099 (4) | 0.83435 (14) | 0.0237 (10) | |
| H18A | 0.2779 | 1.2337 | 0.8593 | 0.036* | |
| H18B | 0.2133 | 1.2036 | 0.8174 | 0.036* | |
| H18C | 0.4087 | 1.2696 | 0.8247 | 0.036* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O31 | 0.043 (2) | 0.049 (2) | 0.029 (2) | −0.013 (2) | 0.0095 (18) | 0.0026 (18) |
| O32 | 0.0313 (18) | 0.0309 (19) | 0.0202 (15) | −0.0033 (17) | 0.0022 (14) | 0.0007 (14) |
| O33 | 0.0307 (18) | 0.0328 (19) | 0.0195 (16) | −0.0063 (15) | 0.0047 (14) | 0.0008 (15) |
| O34 | 0.046 (2) | 0.035 (2) | 0.0225 (17) | −0.0042 (19) | −0.0020 (17) | 0.0001 (15) |
| C1 | 0.021 (2) | 0.028 (3) | 0.022 (2) | −0.006 (2) | −0.0023 (19) | 0.001 (2) |
| C2 | 0.036 (3) | 0.031 (3) | 0.025 (2) | −0.006 (2) | 0.002 (2) | 0.006 (2) |
| C3 | 0.034 (3) | 0.036 (3) | 0.017 (2) | −0.002 (2) | −0.005 (2) | 0.010 (2) |
| C4 | 0.025 (2) | 0.027 (2) | 0.020 (2) | −0.001 (2) | −0.003 (2) | 0.0022 (19) |
| C5 | 0.019 (2) | 0.022 (2) | 0.021 (2) | 0.0041 (19) | −0.0062 (19) | −0.0019 (19) |
| C6 | 0.012 (2) | 0.028 (2) | 0.021 (2) | 0.0015 (19) | −0.0016 (17) | −0.0021 (19) |
| C7 | 0.016 (2) | 0.023 (2) | 0.022 (2) | 0.0068 (19) | −0.0033 (18) | −0.0054 (18) |
| C8 | 0.016 (2) | 0.022 (2) | 0.017 (2) | −0.0077 (19) | 0.0012 (18) | −0.0014 (18) |
| C9 | 0.011 (2) | 0.023 (2) | 0.0159 (19) | 0.0030 (18) | 0.0004 (16) | −0.0053 (18) |
| C10 | 0.016 (2) | 0.023 (2) | 0.019 (2) | −0.0015 (19) | 0.0020 (17) | −0.0020 (18) |
| C11 | 0.013 (2) | 0.021 (2) | 0.022 (2) | −0.0026 (17) | −0.0007 (17) | 0.0058 (18) |
| C12 | 0.021 (2) | 0.023 (2) | 0.018 (2) | −0.0068 (19) | −0.0034 (18) | −0.0029 (18) |
| C13 | 0.024 (2) | 0.018 (2) | 0.019 (2) | −0.0029 (19) | −0.0040 (19) | 0.0013 (17) |
| C14 | 0.016 (2) | 0.017 (2) | 0.017 (2) | −0.0048 (17) | −0.0023 (17) | −0.0003 (17) |
| C15 | 0.022 (2) | 0.026 (2) | 0.023 (2) | −0.004 (2) | 0.002 (2) | 0.0021 (19) |
| C16 | 0.013 (2) | 0.028 (2) | 0.024 (2) | −0.0043 (19) | 0.0007 (18) | 0.000 (2) |
| C17 | 0.019 (2) | 0.015 (2) | 0.023 (2) | 0.0042 (18) | −0.0006 (18) | −0.0004 (18) |
| C30 | 0.016 (2) | 0.026 (2) | 0.024 (2) | 0.0000 (19) | −0.0011 (19) | −0.0006 (19) |
| C19 | 0.019 (2) | 0.029 (2) | 0.020 (2) | 0.000 (2) | 0.0014 (18) | −0.0009 (19) |
| C20 | 0.019 (2) | 0.021 (2) | 0.021 (2) | 0.0000 (19) | −0.0021 (18) | 0.0050 (18) |
| C21 | 0.022 (2) | 0.031 (3) | 0.025 (2) | −0.003 (2) | −0.005 (2) | 0.004 (2) |
| C22 | 0.023 (2) | 0.027 (2) | 0.023 (2) | −0.001 (2) | 0.000 (2) | −0.0035 (19) |
| C23 | 0.028 (3) | 0.023 (2) | 0.026 (2) | −0.007 (2) | 0.000 (2) | 0.002 (2) |
| C24 | 0.030 (3) | 0.024 (2) | 0.023 (2) | −0.004 (2) | 0.000 (2) | 0.0011 (19) |
| C25 | 0.032 (3) | 0.031 (3) | 0.025 (2) | −0.002 (2) | 0.000 (2) | 0.000 (2) |
| C26 | 0.040 (3) | 0.043 (3) | 0.032 (3) | −0.006 (3) | −0.007 (2) | 0.007 (3) |
| C27 | 0.040 (3) | 0.040 (3) | 0.025 (2) | 0.003 (3) | 0.002 (2) | 0.007 (2) |
| C28 | 0.033 (3) | 0.045 (3) | 0.025 (2) | 0.000 (3) | −0.013 (2) | 0.000 (2) |
| C29 | 0.034 (3) | 0.035 (3) | 0.021 (2) | −0.010 (2) | 0.002 (2) | −0.006 (2) |
| C18 | 0.023 (2) | 0.023 (2) | 0.025 (2) | −0.002 (2) | 0.000 (2) | −0.004 (2) |
Geometric parameters (Å, °)
| O31—C3 | 1.223 (7) | C15—C16 | 1.558 (6) |
| O32—C3 | 1.352 (7) | C15—H15A | 0.9700 |
| O32—C4 | 1.485 (6) | C15—H15B | 0.9700 |
| O33—C24 | 1.433 (6) | C16—C17 | 1.576 (6) |
| O33—C20 | 1.472 (5) | C16—H16A | 0.9700 |
| O34—C25 | 1.428 (6) | C16—H16B | 0.9700 |
| O34—H34A | 0.8200 | C17—C20 | 1.519 (6) |
| C1—C2 | 1.551 (7) | C17—H17A | 0.9800 |
| C1—C10 | 1.576 (7) | C30—H30A | 0.9600 |
| C1—H1A | 0.9700 | C30—H30B | 0.9600 |
| C1—H1B | 0.9700 | C30—H30C | 0.9600 |
| C2—C3 | 1.477 (8) | C19—H19A | 0.9600 |
| C2—H2A | 0.9700 | C19—H19B | 0.9600 |
| C2—H2B | 0.9700 | C19—H19C | 0.9600 |
| C4—C28 | 1.523 (7) | C20—C21 | 1.509 (7) |
| C4—C29 | 1.542 (7) | C20—C22 | 1.548 (7) |
| C4—C5 | 1.569 (6) | C21—H21A | 0.9600 |
| C5—C6 | 1.548 (7) | C21—H21B | 0.9600 |
| C5—C10 | 1.557 (6) | C21—H21C | 0.9600 |
| C5—H5A | 0.9800 | C22—C23 | 1.536 (6) |
| C6—C7 | 1.520 (6) | C22—H22A | 0.9700 |
| C6—H6A | 0.9700 | C22—H22B | 0.9700 |
| C6—H6B | 0.9700 | C23—C24 | 1.505 (7) |
| C7—C8 | 1.535 (6) | C23—H23A | 0.9700 |
| C7—H7A | 0.9700 | C23—H23B | 0.9700 |
| C7—H7B | 0.9700 | C24—C25 | 1.548 (7) |
| C8—C18 | 1.524 (6) | C24—H24A | 0.9800 |
| C8—C9 | 1.562 (6) | C25—C27 | 1.500 (8) |
| C8—C14 | 1.597 (6) | C25—C26 | 1.513 (8) |
| C9—C11 | 1.541 (6) | C26—H26A | 0.9600 |
| C9—C10 | 1.579 (6) | C26—H26B | 0.9600 |
| C9—H9A | 0.9800 | C26—H26C | 0.9600 |
| C10—C19 | 1.518 (6) | C27—H27A | 0.9600 |
| C11—C12 | 1.543 (6) | C27—H27B | 0.9600 |
| C11—H11A | 0.9700 | C27—H27C | 0.9600 |
| C11—H11B | 0.9700 | C28—H28A | 0.9600 |
| C12—C13 | 1.525 (6) | C28—H28C | 0.9600 |
| C12—H12A | 0.9700 | C28—H28D | 0.9600 |
| C12—H12B | 0.9700 | C29—H29C | 0.9600 |
| C13—C17 | 1.533 (6) | C29—H29D | 0.9600 |
| C13—C14 | 1.538 (6) | C29—H29A | 0.9600 |
| C13—H13A | 0.9800 | C18—H18A | 0.9600 |
| C14—C15 | 1.538 (6) | C18—H18B | 0.9600 |
| C14—C30 | 1.540 (6) | C18—H18C | 0.9600 |
| C3—O32—C4 | 124.7 (4) | C15—C16—C17 | 106.4 (4) |
| C24—O33—C20 | 110.9 (4) | C15—C16—H16A | 110.4 |
| C25—O34—H34A | 109.5 | C17—C16—H16A | 110.4 |
| C2—C1—C10 | 117.2 (4) | C15—C16—H16B | 110.4 |
| C2—C1—H1A | 108.0 | C17—C16—H16B | 110.4 |
| C10—C1—H1A | 108.0 | H16A—C16—H16B | 108.6 |
| C2—C1—H1B | 108.0 | C20—C17—C13 | 118.6 (4) |
| C10—C1—H1B | 108.0 | C20—C17—C16 | 113.4 (4) |
| H1A—C1—H1B | 107.2 | C13—C17—C16 | 103.0 (4) |
| C3—C2—C1 | 110.1 (4) | C20—C17—H17A | 107.0 |
| C3—C2—H2A | 109.6 | C13—C17—H17A | 107.0 |
| C1—C2—H2A | 109.6 | C16—C17—H17A | 107.0 |
| C3—C2—H2B | 109.6 | C14—C30—H30A | 109.5 |
| C1—C2—H2B | 109.6 | C14—C30—H30B | 109.5 |
| H2A—C2—H2B | 108.2 | H30A—C30—H30B | 109.5 |
| O31—C3—O32 | 116.8 (5) | C14—C30—H30C | 109.5 |
| O31—C3—C2 | 123.5 (5) | H30A—C30—H30C | 109.5 |
| O32—C3—C2 | 119.6 (4) | H30B—C30—H30C | 109.5 |
| O32—C4—C28 | 106.7 (4) | C10—C19—H19A | 109.5 |
| O32—C4—C29 | 99.5 (4) | C10—C19—H19B | 109.5 |
| C28—C4—C29 | 108.4 (4) | H19A—C19—H19B | 109.5 |
| O32—C4—C5 | 116.3 (4) | C10—C19—H19C | 109.5 |
| C28—C4—C5 | 110.4 (4) | H19A—C19—H19C | 109.5 |
| C29—C4—C5 | 114.7 (4) | H19B—C19—H19C | 109.5 |
| C6—C5—C10 | 111.4 (4) | O33—C20—C21 | 106.7 (4) |
| C6—C5—C4 | 108.3 (4) | O33—C20—C17 | 104.8 (4) |
| C10—C5—C4 | 118.4 (4) | C21—C20—C17 | 114.1 (4) |
| C6—C5—H5A | 106.0 | O33—C20—C22 | 103.3 (4) |
| C10—C5—H5A | 106.0 | C21—C20—C22 | 111.9 (4) |
| C4—C5—H5A | 106.0 | C17—C20—C22 | 114.8 (4) |
| C7—C6—C5 | 112.1 (4) | C20—C21—H21A | 109.5 |
| C7—C6—H6A | 109.2 | C20—C21—H21B | 109.5 |
| C5—C6—H6A | 109.2 | H21A—C21—H21B | 109.5 |
| C7—C6—H6B | 109.2 | C20—C21—H21C | 109.5 |
| C5—C6—H6B | 109.2 | H21A—C21—H21C | 109.5 |
| H6A—C6—H6B | 107.9 | H21B—C21—H21C | 109.5 |
| C6—C7—C8 | 111.6 (4) | C23—C22—C20 | 103.0 (4) |
| C6—C7—H7A | 109.3 | C23—C22—H22A | 111.2 |
| C8—C7—H7A | 109.3 | C20—C22—H22A | 111.2 |
| C6—C7—H7B | 109.3 | C23—C22—H22B | 111.2 |
| C8—C7—H7B | 109.3 | C20—C22—H22B | 111.2 |
| H7A—C7—H7B | 108.0 | H22A—C22—H22B | 109.1 |
| C18—C8—C7 | 109.5 (4) | C24—C23—C22 | 101.1 (4) |
| C18—C8—C9 | 113.2 (4) | C24—C23—H23A | 111.5 |
| C7—C8—C9 | 107.4 (4) | C22—C23—H23A | 111.5 |
| C18—C8—C14 | 109.9 (4) | C24—C23—H23B | 111.5 |
| C7—C8—C14 | 110.5 (4) | C22—C23—H23B | 111.5 |
| C9—C8—C14 | 106.2 (3) | H23A—C23—H23B | 109.4 |
| C11—C9—C8 | 111.1 (4) | O33—C24—C23 | 105.4 (4) |
| C11—C9—C10 | 112.4 (3) | O33—C24—C25 | 107.1 (4) |
| C8—C9—C10 | 117.7 (4) | C23—C24—C25 | 119.1 (4) |
| C11—C9—H9A | 104.7 | O33—C24—H24A | 108.3 |
| C8—C9—H9A | 104.7 | C23—C24—H24A | 108.3 |
| C10—C9—H9A | 104.7 | C25—C24—H24A | 108.3 |
| C19—C10—C5 | 112.6 (4) | O34—C25—C27 | 110.0 (4) |
| C19—C10—C1 | 108.2 (4) | O34—C25—C26 | 109.9 (5) |
| C5—C10—C1 | 109.1 (4) | C27—C25—C26 | 111.7 (5) |
| C19—C10—C9 | 113.8 (4) | O34—C25—C24 | 103.6 (4) |
| C5—C10—C9 | 109.0 (4) | C27—C25—C24 | 112.5 (4) |
| C1—C10—C9 | 103.7 (4) | C26—C25—C24 | 108.8 (4) |
| C9—C11—C12 | 112.8 (4) | C25—C26—H26A | 109.5 |
| C9—C11—H11A | 109.0 | C25—C26—H26B | 109.5 |
| C12—C11—H11A | 109.0 | H26A—C26—H26B | 109.5 |
| C9—C11—H11B | 109.0 | C25—C26—H26C | 109.5 |
| C12—C11—H11B | 109.0 | H26A—C26—H26C | 109.5 |
| H11A—C11—H11B | 107.8 | H26B—C26—H26C | 109.5 |
| C13—C12—C11 | 109.8 (4) | C25—C27—H27A | 109.5 |
| C13—C12—H12A | 109.7 | C25—C27—H27B | 109.5 |
| C11—C12—H12A | 109.7 | H27A—C27—H27B | 109.5 |
| C13—C12—H12B | 109.7 | C25—C27—H27C | 109.5 |
| C11—C12—H12B | 109.7 | H27A—C27—H27C | 109.5 |
| H12A—C12—H12B | 108.2 | H27B—C27—H27C | 109.5 |
| C12—C13—C17 | 119.9 (4) | C4—C28—H28A | 109.5 |
| C12—C13—C14 | 112.1 (4) | C4—C28—H28C | 109.5 |
| C17—C13—C14 | 105.8 (4) | H28A—C28—H28C | 109.5 |
| C12—C13—H13A | 106.0 | C4—C28—H28D | 109.5 |
| C17—C13—H13A | 106.0 | H28A—C28—H28D | 109.5 |
| C14—C13—H13A | 106.0 | H28C—C28—H28D | 109.5 |
| C13—C14—C15 | 100.7 (4) | C4—C29—H29C | 109.5 |
| C13—C14—C30 | 110.4 (4) | C4—C29—H29D | 109.5 |
| C15—C14—C30 | 106.5 (4) | H29C—C29—H29D | 109.5 |
| C13—C14—C8 | 110.6 (4) | C4—C29—H29A | 109.5 |
| C15—C14—C8 | 116.2 (4) | H29C—C29—H29A | 109.5 |
| C30—C14—C8 | 111.9 (4) | H29D—C29—H29A | 109.5 |
| C14—C15—C16 | 104.6 (4) | C8—C18—H18A | 109.5 |
| C14—C15—H15A | 110.8 | C8—C18—H18B | 109.5 |
| C16—C15—H15A | 110.8 | H18A—C18—H18B | 109.5 |
| C14—C15—H15B | 110.8 | C8—C18—H18C | 109.5 |
| C16—C15—H15B | 110.8 | H18A—C18—H18C | 109.5 |
| H15A—C15—H15B | 108.9 | H18B—C18—H18C | 109.5 |
| C10—C1—C2—C3 | 63.3 (6) | C17—C13—C14—C15 | 44.3 (4) |
| C4—O32—C3—O31 | 169.9 (4) | C12—C13—C14—C30 | 64.5 (5) |
| C4—O32—C3—C2 | −15.1 (7) | C17—C13—C14—C30 | −67.9 (5) |
| C1—C2—C3—O31 | 107.2 (6) | C12—C13—C14—C8 | −59.9 (5) |
| C1—C2—C3—O32 | −67.5 (6) | C17—C13—C14—C8 | 167.7 (4) |
| C3—O32—C4—C28 | −76.9 (5) | C18—C8—C14—C13 | −63.3 (5) |
| C3—O32—C4—C29 | 170.5 (4) | C7—C8—C14—C13 | 175.7 (4) |
| C3—O32—C4—C5 | 46.8 (6) | C9—C8—C14—C13 | 59.5 (5) |
| O32—C4—C5—C6 | 150.4 (4) | C18—C8—C14—C15 | 50.6 (5) |
| C28—C4—C5—C6 | −87.9 (5) | C7—C8—C14—C15 | −70.4 (5) |
| C29—C4—C5—C6 | 34.9 (6) | C9—C8—C14—C15 | 173.5 (4) |
| O32—C4—C5—C10 | 22.4 (6) | C18—C8—C14—C30 | 173.2 (4) |
| C28—C4—C5—C10 | 144.1 (4) | C7—C8—C14—C30 | 52.2 (5) |
| C29—C4—C5—C10 | −93.1 (5) | C9—C8—C14—C30 | −64.0 (4) |
| C10—C5—C6—C7 | −58.1 (5) | C13—C14—C15—C16 | −38.9 (4) |
| C4—C5—C6—C7 | 170.0 (4) | C30—C14—C15—C16 | 76.3 (4) |
| C5—C6—C7—C8 | 62.3 (5) | C8—C14—C15—C16 | −158.3 (4) |
| C6—C7—C8—C18 | 67.2 (5) | C14—C15—C16—C17 | 20.4 (5) |
| C6—C7—C8—C9 | −56.1 (5) | C12—C13—C17—C20 | 74.5 (6) |
| C6—C7—C8—C14 | −171.5 (4) | C14—C13—C17—C20 | −157.7 (4) |
| C18—C8—C9—C11 | 62.4 (5) | C12—C13—C17—C16 | −159.3 (4) |
| C7—C8—C9—C11 | −176.6 (3) | C14—C13—C17—C16 | −31.5 (4) |
| C14—C8—C9—C11 | −58.3 (4) | C15—C16—C17—C20 | 136.0 (4) |
| C18—C8—C9—C10 | −69.2 (5) | C15—C16—C17—C13 | 6.5 (5) |
| C7—C8—C9—C10 | 51.7 (5) | C24—O33—C20—C21 | −113.7 (4) |
| C14—C8—C9—C10 | 170.0 (4) | C24—O33—C20—C17 | 125.0 (4) |
| C6—C5—C10—C19 | −78.2 (5) | C24—O33—C20—C22 | 4.4 (5) |
| C4—C5—C10—C19 | 48.3 (6) | C13—C17—C20—O33 | 164.2 (4) |
| C6—C5—C10—C1 | 161.6 (4) | C16—C17—C20—O33 | 43.1 (5) |
| C4—C5—C10—C1 | −71.9 (5) | C13—C17—C20—C21 | 47.9 (6) |
| C6—C5—C10—C9 | 49.0 (5) | C16—C17—C20—C21 | −73.2 (5) |
| C4—C5—C10—C9 | 175.6 (4) | C13—C17—C20—C22 | −83.2 (5) |
| C2—C1—C10—C19 | −102.1 (5) | C16—C17—C20—C22 | 155.8 (4) |
| C2—C1—C10—C5 | 20.8 (6) | O33—C20—C22—C23 | −27.2 (5) |
| C2—C1—C10—C9 | 136.8 (4) | C21—C20—C22—C23 | 87.2 (5) |
| C11—C9—C10—C19 | −53.3 (5) | C17—C20—C22—C23 | −140.7 (4) |
| C8—C9—C10—C19 | 77.7 (5) | C20—C22—C23—C24 | 39.0 (5) |
| C11—C9—C10—C5 | −179.9 (4) | C20—O33—C24—C23 | 20.8 (5) |
| C8—C9—C10—C5 | −48.9 (5) | C20—O33—C24—C25 | 148.5 (4) |
| C11—C9—C10—C1 | 64.0 (5) | C22—C23—C24—O33 | −37.0 (5) |
| C8—C9—C10—C1 | −165.0 (4) | C22—C23—C24—C25 | −157.1 (4) |
| C8—C9—C11—C12 | 58.0 (5) | O33—C24—C25—O34 | −179.1 (4) |
| C10—C9—C11—C12 | −167.7 (4) | C23—C24—C25—O34 | −59.9 (6) |
| C9—C11—C12—C13 | −54.2 (5) | O33—C24—C25—C27 | −60.4 (6) |
| C11—C12—C13—C17 | −179.8 (4) | C23—C24—C25—C27 | 58.8 (6) |
| C11—C12—C13—C14 | 55.3 (5) | O33—C24—C25—C26 | 64.0 (6) |
| C12—C13—C14—C15 | 176.7 (4) | C23—C24—C25—C26 | −176.8 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C19—H19A···O32 | 0.96 | 2.44 | 3.082 (6) | 124 |
| O34—H34A···O32i | 0.82 | 2.20 | 3.010 (5) | 170 |
| C26—H26A···O31i | 0.96 | 2.53 | 3.392 (7) | 150 |
Symmetry codes: (i) −x+1/2, −y+2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2332).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555-1573.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Burkill, L. H. (1966). A Dictionary of Economics Products of The Malay Peninsula, Vols. I and II. Kuala Lumpur: Ministry of Agriculture and Cooperatives.
- Fujiwara, T., Takeda, T., Ogihara, Y., Shimizu, M., Nomura, T. & Tomita, Y. (1982). Chem. Pharm. Bull. 30, 4025–4030. [DOI] [PubMed]
- Hegnauer, R. (1990). Editor. Chemotaxonomie der Planzen, Vol. IX. Basel and Stuttgart: Birkhauser Verlag.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Pan, L., Kardono, L. B. S., Riswan, S., Chai, H., Carcache de Blanco, E. J., Pannell, C. M., Soejart, D. D., McCloud, T. G., Newman, D. J. & Kinghorn, A. D. (2010). J. Nat. Prod. 73, 1873–1878. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811047337/su2332sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811047337/su2332Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report