Abstract
In the title compound, C9H12N4O, the piperidine ring adopts a chair conformation and makes a dihedral angle of 42.49 (11)° with the approximately planar pyrazole moiety [maximum deviation = 0.038 (2) Å]. In the crystal, N—H⋯O and N—H⋯N hydrogen bonds and a weak C—H⋯O interaction link the molecules into sheets lying parallel to (110).
Related literature
For pharmacological background, see: Patel et al. (1990 ▶); Morimoto et al. (1990 ▶). For related structures see: Zaharan et al. (2001 ▶); Elgemeie et al. (2007 ▶); Gouda et al. (2010 ▶); Shelton et al. (2011 ▶). For standard bond lengths, see: Allen et al. (1987 ▶).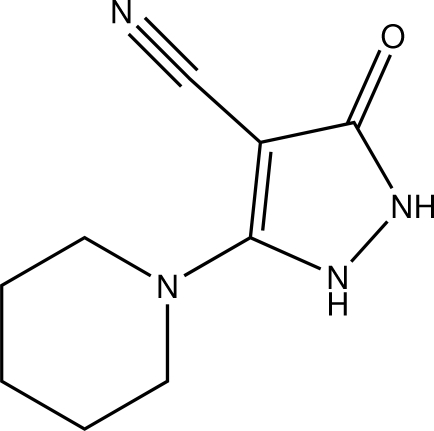
Experimental
Crystal data
C9H12N4O
M r = 192.23
Triclinic,

a = 7.2667 (5) Å
b = 7.9624 (5) Å
c = 8.8306 (8) Å
α = 89.280 (6)°
β = 75.934 (7)°
γ = 71.906 (6)°
V = 470.01 (6) Å3
Z = 2
Cu Kα radiation
μ = 0.77 mm−1
T = 150 K
0.22 × 0.19 × 0.13 mm
Data collection
Oxford Diffraction Gemini diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2006 ▶) T min = 0.849, T max = 0.906
5083 measured reflections
1803 independent reflections
1627 reflections with I > 2σ(I)
R int = 0.013
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.166
S = 1.11
1803 reflections
127 parameters
H-atom parameters constrained
Δρmax = 0.74 e Å−3
Δρmin = −0.61 e Å−3
Data collection: Gemini User Manual (Oxford Diffraction, 2006 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2002 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL, PARST (Nardelli, 1995 ▶), PLATON (Spek, 2009 ▶) and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811047714/hb6450sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811047714/hb6450Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811047714/hb6450Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯N4i | 0.86 | 2.32 | 2.875 (3) | 123 |
| N3—H3⋯O1ii | 0.86 | 2.07 | 2.772 (2) | 138 |
| C4—H4A⋯O1iii | 0.97 | 2.54 | 3.258 (3) | 131 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
The authors thank Universiti Kebangsaan Malaysia for providing facilities and the Ministry of Higher Education Malaysia for the research fund (UKM-GGPM-KPB-098–2010). A scholarship from the Libyan Government to WMA is also greatly appreciated.
supplementary crystallographic information
Comment
In this paper, we report the synthesis and structure of the new derivative of 3-oxo-5-(piperidin-1-yl)-2,3-dihydro-1H- pyrazole-4-carbonitrile. The compound was obtained by cyclization reaction between ethyl 2-cyano-3-(methylthio)-3-(piperidin-1-yl)acrylate and hydrazine.
In the title compound (I), the mean plane of the pyrazole O1/N1/N2/N4/C5/C6/C7/C8/C9 is essentially planar with maximum deviation of -0.038 (2)° for C8 and forms a dihedral angle of 42.49 (11)° with that of the piperidine mean plane N1/C1/C2/C3/C4/C5 (Fig. 1 & Scheme 1). Consequently, a short non-bonding intra D—H..H—X contact forms between the N2—H2 of the pyrazole and the H5B—C5 of the piperidine moeities.
The carbonyl C8=O1 [1.246 (2)] and C6=C7 [1.407 (3) Å] are longer than the average [C=O(1.200 Å)] and C=C [(1.340 Å)] bond lengths, respectively. Whereas the C6—N2 [1.363 (2) Å] and C8—N3 [1.375 (2) Å] bond lengths are shorter than the average C—N [(1.47 Å)] indicative of electron-donating effects of the amino groups. Other bond lengths and angle in the molecules are in the normal ranges (Allen et al.,1987).
In the crystal, intermolecular hydrogen bonds N2—H2···O4 and N3—H3···O1 and a weak C4—H4···O1 interaction link the molecules forming a two-dimensional polymeric network parallel to (110) (Fig. 2).
Experimental
A mixture of ethyl 2-cyano-3-(methylthio)-3-(piperidin-1-yl)acrylate (4 mmol) and hydrazine hydrate (4 mmol) was heated on a water-bath for 2 h. Then, ethanol (20 ml) was added and the mixture was refluxed for another 2 h. The solvent was evaporated and the product was collected, washed with ethanol, and dried. Colourless blocks of (I) were formed by slow evaporation of the compound from ethanol solution. Yield = 90%.
Refinement
H atoms of both C and N atoms were positioned geometrically and allowed to ride on their parent atoms, with Uiso = 1.2Ueq (C) for CH2 0.97 Å. Hydrogen atoms attached to N were also positioned geometrically and allowed to ride on their parent atoms and with Uiso(H) = 1.2Ueq(N) for N–H 0.86 Å.
Figures
Fig. 1.
The molecular structure of (I), with displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
Crystal packing of (I) viewed down the a axis. Hydrogen bonds [N—H···O (x-1, y, z & -x + 2, -y, -z) N—H···N (x-1, y + 1, z)] are drawn as dashed lines.
Crystal data
| C9H12N4O | Z = 2 |
| Mr = 192.23 | F(000) = 204 |
| Triclinic, P1 | Dx = 1.358 Mg m−3 |
| Hall symbol: -P 1 | Melting point: 527 K |
| a = 7.2667 (5) Å | Cu Kα radiation, λ = 1.54178 Å |
| b = 7.9624 (5) Å | Cell parameters from 3129 reflections |
| c = 8.8306 (8) Å | θ = 5–71° |
| α = 89.280 (6)° | µ = 0.77 mm−1 |
| β = 75.934 (7)° | T = 150 K |
| γ = 71.906 (6)° | Block, colourless |
| V = 470.01 (6) Å3 | 0.22 × 0.19 × 0.13 mm |
Data collection
| Oxford Diffraction Gemini diffractometer | 1803 independent reflections |
| Radiation source: fine-focus sealed tube | 1627 reflections with I > 2σ(I) |
| graphite | Rint = 0.013 |
| ω/2θ scans | θmax = 70.9°, θmin = 5.2° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2006) | h = −8→8 |
| Tmin = 0.849, Tmax = 0.906 | k = −9→9 |
| 5083 measured reflections | l = −10→10 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.166 | H-atom parameters constrained |
| S = 1.11 | w = 1/[σ2(Fo2) + (0.0971P)2 + 0.2641P] where P = (Fo2 + 2Fc2)/3 |
| 1803 reflections | (Δ/σ)max < 0.001 |
| 127 parameters | Δρmax = 0.74 e Å−3 |
| 0 restraints | Δρmin = −0.61 e Å−3 |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems open-flow nitrogen cryostat (Cosier & Glazer, 1986) with a nominal stability of 0.1 K.Cosier, J. & Glazer, A.M., 1986. J. Appl. Cryst. 105 107. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 1.2334 (2) | 0.03312 (18) | 0.39572 (17) | 0.0334 (4) | |
| N1 | 0.8756 (2) | 0.6211 (2) | 0.2901 (2) | 0.0306 (4) | |
| N2 | 0.8011 (2) | 0.3667 (2) | 0.38329 (19) | 0.0273 (4) | |
| H2 | 0.6730 | 0.4130 | 0.4070 | 0.033* | |
| N3 | 0.9078 (2) | 0.1884 (2) | 0.39566 (19) | 0.0285 (4) | |
| H3 | 0.8563 | 0.1043 | 0.4146 | 0.034* | |
| N4 | 1.4774 (3) | 0.3703 (3) | 0.2487 (2) | 0.0397 (5) | |
| C1 | 1.0124 (3) | 0.7167 (3) | 0.2139 (3) | 0.0349 (5) | |
| H1A | 1.1496 | 0.6418 | 0.2014 | 0.042* | |
| H1B | 0.9907 | 0.8218 | 0.2789 | 0.042* | |
| C2 | 0.9771 (4) | 0.7692 (3) | 0.0547 (3) | 0.0394 (5) | |
| H2A | 1.0165 | 0.6635 | −0.0144 | 0.047* | |
| H2B | 1.0596 | 0.8413 | 0.0094 | 0.047* | |
| C3 | 0.7585 (4) | 0.8722 (3) | 0.0673 (3) | 0.0424 (6) | |
| H3A | 0.7234 | 0.9851 | 0.1253 | 0.051* | |
| H3B | 0.7386 | 0.8954 | −0.0366 | 0.051* | |
| C4 | 0.6237 (3) | 0.7685 (3) | 0.1497 (3) | 0.0380 (5) | |
| H4A | 0.4847 | 0.8390 | 0.1628 | 0.046* | |
| H4B | 0.6491 | 0.6610 | 0.0865 | 0.046* | |
| C5 | 0.6632 (3) | 0.7218 (3) | 0.3084 (2) | 0.0333 (5) | |
| H5A | 0.6282 | 0.8293 | 0.3742 | 0.040* | |
| H5B | 0.5808 | 0.6517 | 0.3588 | 0.040* | |
| C6 | 0.9374 (3) | 0.4540 (2) | 0.3267 (2) | 0.0249 (4) | |
| C7 | 1.1293 (3) | 0.3363 (2) | 0.3210 (2) | 0.0252 (4) | |
| C8 | 1.1054 (3) | 0.1702 (2) | 0.3729 (2) | 0.0262 (4) | |
| C9 | 1.3185 (3) | 0.3601 (3) | 0.2816 (2) | 0.0289 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0258 (7) | 0.0273 (7) | 0.0420 (8) | −0.0030 (6) | −0.0067 (6) | 0.0098 (6) |
| N1 | 0.0276 (9) | 0.0285 (9) | 0.0356 (9) | −0.0062 (7) | −0.0116 (7) | 0.0092 (7) |
| N2 | 0.0200 (8) | 0.0254 (8) | 0.0343 (9) | −0.0032 (6) | −0.0082 (6) | 0.0071 (6) |
| N3 | 0.0254 (8) | 0.0232 (8) | 0.0371 (9) | −0.0070 (7) | −0.0092 (7) | 0.0072 (6) |
| N4 | 0.0283 (10) | 0.0412 (11) | 0.0545 (11) | −0.0131 (8) | −0.0169 (8) | 0.0118 (8) |
| C1 | 0.0353 (11) | 0.0276 (10) | 0.0462 (12) | −0.0126 (9) | −0.0152 (9) | 0.0095 (9) |
| C2 | 0.0445 (13) | 0.0297 (11) | 0.0387 (12) | −0.0095 (9) | −0.0038 (9) | 0.0084 (9) |
| C3 | 0.0532 (14) | 0.0321 (11) | 0.0331 (11) | −0.0007 (10) | −0.0116 (10) | 0.0076 (9) |
| C4 | 0.0364 (11) | 0.0332 (11) | 0.0388 (11) | 0.0013 (9) | −0.0154 (9) | 0.0018 (9) |
| C5 | 0.0295 (11) | 0.0274 (10) | 0.0382 (11) | −0.0016 (8) | −0.0093 (8) | 0.0061 (8) |
| C6 | 0.0271 (10) | 0.0274 (10) | 0.0215 (8) | −0.0078 (8) | −0.0095 (7) | 0.0035 (7) |
| C7 | 0.0244 (9) | 0.0259 (10) | 0.0257 (9) | −0.0066 (7) | −0.0090 (7) | 0.0029 (7) |
| C8 | 0.0255 (9) | 0.0261 (9) | 0.0252 (9) | −0.0050 (7) | −0.0071 (7) | 0.0016 (7) |
| C9 | 0.0298 (11) | 0.0269 (10) | 0.0318 (10) | −0.0070 (8) | −0.0141 (8) | 0.0065 (8) |
Geometric parameters (Å, °)
| O1—C8 | 1.246 (2) | C2—H2A | 0.9700 |
| N1—C6 | 1.329 (3) | C2—H2B | 0.9700 |
| N1—C1 | 1.467 (3) | C3—C4 | 1.520 (3) |
| N1—C5 | 1.471 (3) | C3—H3A | 0.9700 |
| N2—C6 | 1.376 (2) | C3—H3B | 0.9700 |
| N2—N3 | 1.408 (2) | C4—C5 | 1.517 (3) |
| N2—H2 | 0.8600 | C4—H4A | 0.9700 |
| N3—C8 | 1.362 (3) | C4—H4B | 0.9700 |
| N3—H3 | 0.8600 | C5—H5A | 0.9700 |
| N4—C9 | 1.148 (3) | C5—H5B | 0.9700 |
| C1—C2 | 1.520 (3) | C6—C7 | 1.407 (3) |
| C1—H1A | 0.9700 | C7—C9 | 1.406 (3) |
| C1—H1B | 0.9700 | C7—C8 | 1.442 (3) |
| C2—C3 | 1.521 (3) | ||
| C6—N1—C1 | 123.24 (17) | C2—C3—H3B | 109.5 |
| C6—N1—C5 | 122.91 (17) | H3A—C3—H3B | 108.1 |
| C1—N1—C5 | 113.60 (16) | C5—C4—C3 | 109.84 (19) |
| C6—N2—N3 | 108.06 (14) | C5—C4—H4A | 109.7 |
| C6—N2—H2 | 126.0 | C3—C4—H4A | 109.7 |
| N3—N2—H2 | 126.0 | C5—C4—H4B | 109.7 |
| C8—N3—N2 | 109.28 (15) | C3—C4—H4B | 109.7 |
| C8—N3—H3 | 125.4 | H4A—C4—H4B | 108.2 |
| N2—N3—H3 | 125.4 | N1—C5—C4 | 110.09 (17) |
| N1—C1—C2 | 109.99 (17) | N1—C5—H5A | 109.6 |
| N1—C1—H1A | 109.7 | C4—C5—H5A | 109.6 |
| C2—C1—H1A | 109.7 | N1—C5—H5B | 109.6 |
| N1—C1—H1B | 109.7 | C4—C5—H5B | 109.6 |
| C2—C1—H1B | 109.7 | H5A—C5—H5B | 108.2 |
| H1A—C1—H1B | 108.2 | N1—C6—N2 | 120.17 (17) |
| C1—C2—C3 | 111.40 (19) | N1—C6—C7 | 132.01 (18) |
| C1—C2—H2A | 109.3 | N2—C6—C7 | 107.82 (16) |
| C3—C2—H2A | 109.3 | C9—C7—C6 | 131.41 (17) |
| C1—C2—H2B | 109.3 | C9—C7—C8 | 121.20 (17) |
| C3—C2—H2B | 109.3 | C6—C7—C8 | 107.35 (16) |
| H2A—C2—H2B | 108.0 | O1—C8—N3 | 124.15 (18) |
| C4—C3—C2 | 110.68 (18) | O1—C8—C7 | 129.26 (18) |
| C4—C3—H3A | 109.5 | N3—C8—C7 | 106.58 (16) |
| C2—C3—H3A | 109.5 | N4—C9—C7 | 176.5 (2) |
| C4—C3—H3B | 109.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···N4i | 0.86 | 2.32 | 2.875 (3) | 123 |
| N3—H3···O1ii | 0.86 | 2.07 | 2.772 (2) | 138 |
| C4—H4A···O1iii | 0.97 | 2.54 | 3.258 (3) | 131 |
Symmetry codes: (i) x−1, y, z; (ii) −x+2, −y, −z+1; (iii) x−1, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6450).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Elgemeie, G. H., Elghandour, A. H. & Abd Elaziz, G. W. (2007). Synth. Commun. 37, 2827–2834.
- Gouda, M. A., Berghot, M. A., Abd El-Ghani, G. E. & Khalil, A. M. (2010). Eur. J. Med. Chem. 45, 1338–1345. [DOI] [PubMed]
- Morimoto, K., Makino, K., Yamamoto, S. & Sakata, G. (1990). J. Heterocycl. Chem. 27, 807–810.
- Nardelli, M. (1995). J. Appl. Cryst. 28, 659.
- Oxford Diffraction (2002). CrysAlis RED Oxford Diffraction Ltd, Abingdon, England.
- Oxford Diffraction (2006). Gemini User Manual Oxford Diffraction Ltd, Abingdon, England.
- Patel, H. V., Fernandes, P. S. & Vyas, K. A. (1990). Indian J. Chem. Sect. B, 29, 135–141.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shelton, A. H., Stephenson, A., Ward, M. D. & Kassim, M. B. (2011). Acta Cryst. E67, o2445. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zaharan, M. A., El-Sharief, A. M. S., El-Gaby, M. S. A., Ammar, Y. A. & El-Said, U. H. (2001). Farmaco, 56, 277–283. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811047714/hb6450sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811047714/hb6450Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811047714/hb6450Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




