Abstract
In the title compound, C11H8ClN3O4, the dihedral angle between benzene and isoxazole rings is 9.92 (1) °. The nitro group is almost coplanar with the benzene ring with an O—N—C—C torsion angle of 8.4 (3)°. The molecular conformation is stabilized by an intramolecular N—H⋯O hydrogen bond, closing a six-membered ring.
Related literature
For applications of leflunomide [systematic name: 5-methyl-N-[4-(trifluoromethyl) phenyl]-isoxazole-4-carboxamide] in the treatment of rheumatoid arthritis, see: Shaw et al. (2011 ▶); Schattenkirchner (2000 ▶). The title compound was synthesized as an immunomodulating leflunomide analog; for another immunomodulating leflunomide analog, see: Huang et al. (2003 ▶).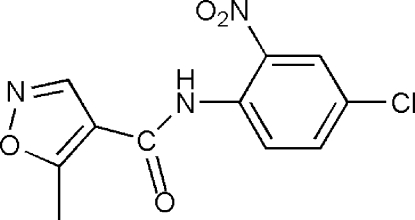
Experimental
Crystal data
C11H8ClN3O4
M r = 281.65
Monoclinic,

a = 5.3870 (11) Å
b = 23.537 (5) Å
c = 9.4600 (19) Å
β = 99.86 (3)°
V = 1181.8 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.34 mm−1
T = 293 K
0.30 × 0.20 × 0.10 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: ψ scan (North et al., 1968 ▶) T min = 0.905, T max = 0.967
4645 measured reflections
2121 independent reflections
1558 reflections with I > 2σ(I)
R int = 0.044
3 standard reflections every 200 reflections intensity decay: 1%
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.143
S = 1.02
2121 reflections
174 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.22 e Å−3
Data collection: CAD-4 EXPRESS (Enraf–Nonius, 1994 ▶); cell refinement: CAD-4 EXPRESS; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811047994/ld2035sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811047994/ld2035Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2B⋯O2 | 0.86 | 1.94 | 2.629 (3) | 136 |
Acknowledgments
The work was supported by the Center of Testing and Analysis, Nanjing University.
supplementary crystallographic information
Comment
Leflunomide is one of the most effective isoxazole-containing disease-modifying drugs for treating rheumatoid arthritis(Shaw et al., 2011; Schattenkirchner, 2000). Many leflunomide analogs have been synthesized and exhibit potent immunomodulating effect (Huang, et al., 2003). The title compound, 5-methyl-N-(4-chloro-2-nitrophenyl)isoxazole-4-carboxamide (I), was synthesized as a novel and potent immunomodulating leflunomide analog. We report herein its crystal structure. The molecular structure of the title compound is shown in Fig. 1. The nitro group is approximately coplanar with the benzene ring, as indicated by the torsion angle O1—N1—C5—C4 of 8.4 (3)°. The amide group is also coplanar with the benzene and isoxazole rings [torsion angles N2—C7—C8—C9 and C7—N2—C6—C1 are -8.9 (4) and -3.1 (4)°], respectively. The molecular conformation is stabilized by an intra-molecular N—H···O hydrogen bond (Table 1). In the crystal, infinite zigzag chain are formed via short inter-molecular Cl···O contacts of 3.089 (3) Å.
Experimental
A solution of 0.05 mol of 5-methylisoxazole-4-carboxylic acid chloride (7.3 g) in 20 ml of acetonitrile was added dropwise, while stirring, to 0.1 mol of 4-chloro-2-nitroaniline (17.2 g), dissolved in 150 ml of acetonitrile, at room temperature. After stirring for 40 more minutes, the precipitated 4-chloro-2-nitroaniline hydrochloride was filtered off and washed with 100 ml portions of acetonitrile, and the combined filtrates were concentrated under reduced pressure. 9.6 g (65% of theory) of yellow crytalline 5-methyl-N-(4-Chloro-2-nitrophenyl)isoxazole-4-carboxamide were thus obtained. Crystals of (I) suitable for X-ray diffraction were obtained by slow evaporation of toluene solution.
Refinement
Carbon- and nitrogen-bound H atoms were placed in calculated positions and were treated as riding on the parent C or N atoms with C—H = 0.96 (methyl), 0.97 (methylene) and N—H = 0.86 Å, Uiso(H) = 1.2 Ueq(C, N). The positions of methyl hydrogens were optimized rotationally with Uiso(H) = 1.5 Ueq(C).
Figures
Fig. 1.
The structure of the title compound, showing the atomic numbering scheme. Non-H atoms are shown with 30% probability displacement ellipsoids.
Crystal data
| C11H8ClN3O4 | F(000) = 576 |
| Mr = 281.65 | Dx = 1.583 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 5.3870 (11) Å | θ = 9–13° |
| b = 23.537 (5) Å | µ = 0.34 mm−1 |
| c = 9.4600 (19) Å | T = 293 K |
| β = 99.86 (3)° | Block, white |
| V = 1181.8 (4) Å3 | 0.30 × 0.20 × 0.10 mm |
| Z = 4 |
Data collection
| Enraf–Nonius CAD-4 diffractometer | 1558 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.044 |
| graphite | θmax = 25.2°, θmin = 1.7° |
| ω/2θ scans | h = 0→6 |
| Absorption correction: ψ scan (North et al., 1968) | k = −28→28 |
| Tmin = 0.905, Tmax = 0.967 | l = −11→11 |
| 4645 measured reflections | 3 standard reflections every 200 reflections |
| 2121 independent reflections | intensity decay: 1% |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H-atom parameters constrained |
| wR(F2) = 0.143 | w = 1/[σ2(Fo2) + (0.090P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 2121 reflections | Δρmax = 0.18 e Å−3 |
| 174 parameters | Δρmin = −0.22 e Å−3 |
| 0 restraints | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.010 (3) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl | 1.14810 (15) | 0.27507 (3) | 0.51209 (9) | 0.0712 (3) | |
| N1 | 0.5249 (4) | 0.33771 (10) | 0.8313 (2) | 0.0534 (6) | |
| C1 | 0.8613 (5) | 0.42551 (11) | 0.5906 (3) | 0.0476 (6) | |
| H1A | 0.8768 | 0.4628 | 0.5606 | 0.057* | |
| O1 | 0.5463 (5) | 0.28905 (9) | 0.8742 (3) | 0.0868 (8) | |
| O2 | 0.3751 (4) | 0.37087 (9) | 0.8716 (2) | 0.0611 (5) | |
| C2 | 0.9988 (5) | 0.38350 (11) | 0.5390 (3) | 0.0507 (6) | |
| H2A | 1.1070 | 0.3927 | 0.4757 | 0.061* | |
| N2 | 0.5564 (4) | 0.45566 (9) | 0.7398 (2) | 0.0469 (5) | |
| H2B | 0.4559 | 0.4440 | 0.7951 | 0.056* | |
| N3 | 0.0876 (5) | 0.56302 (12) | 0.9317 (3) | 0.0656 (7) | |
| O3 | 0.6784 (4) | 0.53680 (8) | 0.6379 (2) | 0.0586 (5) | |
| C3 | 0.9774 (5) | 0.32777 (11) | 0.5806 (3) | 0.0475 (6) | |
| O4 | 0.1878 (4) | 0.61421 (9) | 0.8857 (2) | 0.0658 (6) | |
| C4 | 0.8225 (5) | 0.31427 (11) | 0.6765 (3) | 0.0501 (6) | |
| H4A | 0.8099 | 0.2768 | 0.7060 | 0.060* | |
| C5 | 0.6850 (5) | 0.35645 (10) | 0.7294 (2) | 0.0434 (6) | |
| C6 | 0.6978 (4) | 0.41348 (10) | 0.6875 (2) | 0.0406 (5) | |
| C7 | 0.5541 (5) | 0.51330 (10) | 0.7158 (2) | 0.0433 (6) | |
| C8 | 0.3842 (4) | 0.54381 (11) | 0.7965 (2) | 0.0444 (6) | |
| C9 | 0.2056 (5) | 0.52340 (12) | 0.8774 (3) | 0.0555 (7) | |
| H9A | 0.1763 | 0.4850 | 0.8901 | 0.067* | |
| C10 | 0.3644 (5) | 0.60153 (11) | 0.8058 (3) | 0.0522 (7) | |
| C11 | 0.4990 (7) | 0.65000 (12) | 0.7545 (4) | 0.0731 (9) | |
| H11A | 0.4181 | 0.6848 | 0.7737 | 0.110* | |
| H11B | 0.4958 | 0.6464 | 0.6532 | 0.110* | |
| H11C | 0.6705 | 0.6503 | 0.8035 | 0.110* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl | 0.0730 (5) | 0.0583 (5) | 0.0884 (6) | 0.0099 (4) | 0.0308 (4) | −0.0103 (4) |
| N1 | 0.0613 (14) | 0.0467 (13) | 0.0561 (12) | −0.0111 (11) | 0.0211 (11) | 0.0031 (10) |
| C1 | 0.0504 (14) | 0.0448 (14) | 0.0502 (13) | −0.0018 (12) | 0.0158 (11) | 0.0077 (11) |
| O1 | 0.121 (2) | 0.0491 (13) | 0.1062 (18) | −0.0060 (12) | 0.0648 (16) | 0.0174 (11) |
| O2 | 0.0612 (12) | 0.0607 (12) | 0.0687 (12) | −0.0006 (10) | 0.0320 (10) | 0.0094 (9) |
| C2 | 0.0515 (15) | 0.0515 (16) | 0.0530 (14) | −0.0043 (12) | 0.0199 (12) | 0.0013 (11) |
| N2 | 0.0524 (12) | 0.0432 (12) | 0.0493 (11) | −0.0006 (9) | 0.0210 (10) | 0.0049 (9) |
| N3 | 0.0568 (14) | 0.0717 (18) | 0.0722 (15) | 0.0055 (12) | 0.0221 (12) | −0.0037 (12) |
| O3 | 0.0713 (13) | 0.0431 (11) | 0.0691 (11) | −0.0025 (9) | 0.0335 (10) | 0.0048 (8) |
| C3 | 0.0449 (13) | 0.0467 (15) | 0.0520 (14) | 0.0003 (11) | 0.0112 (11) | −0.0044 (11) |
| O4 | 0.0620 (12) | 0.0666 (14) | 0.0703 (12) | 0.0135 (10) | 0.0152 (10) | −0.0107 (10) |
| C4 | 0.0541 (15) | 0.0379 (14) | 0.0587 (15) | −0.0039 (11) | 0.0107 (13) | 0.0027 (11) |
| C5 | 0.0434 (13) | 0.0429 (14) | 0.0454 (13) | −0.0068 (11) | 0.0123 (11) | 0.0015 (10) |
| C6 | 0.0430 (13) | 0.0411 (13) | 0.0377 (11) | −0.0023 (10) | 0.0069 (10) | 0.0009 (9) |
| C7 | 0.0459 (13) | 0.0408 (13) | 0.0431 (13) | −0.0014 (11) | 0.0069 (11) | 0.0017 (10) |
| C8 | 0.0433 (13) | 0.0464 (15) | 0.0428 (12) | 0.0017 (11) | 0.0053 (10) | −0.0003 (10) |
| C9 | 0.0513 (15) | 0.0590 (18) | 0.0577 (15) | −0.0006 (13) | 0.0140 (13) | −0.0033 (13) |
| C10 | 0.0521 (15) | 0.0536 (16) | 0.0506 (14) | 0.0088 (13) | 0.0080 (12) | −0.0030 (12) |
| C11 | 0.089 (2) | 0.0471 (17) | 0.086 (2) | 0.0019 (16) | 0.0222 (18) | −0.0016 (15) |
Geometric parameters (Å, °)
| Cl—C3 | 1.734 (3) | O3—C7 | 1.210 (3) |
| N1—O1 | 1.214 (3) | C3—C4 | 1.371 (4) |
| N1—O2 | 1.230 (3) | O4—C10 | 1.346 (3) |
| N1—C5 | 1.467 (3) | C4—C5 | 1.382 (3) |
| C1—C2 | 1.374 (4) | C4—H4A | 0.9300 |
| C1—C6 | 1.405 (3) | C5—C6 | 1.405 (3) |
| C1—H1A | 0.9300 | C7—C8 | 1.475 (3) |
| C2—C3 | 1.380 (4) | C8—C10 | 1.367 (4) |
| C2—H2A | 0.9300 | C8—C9 | 1.412 (4) |
| N2—C7 | 1.375 (3) | C9—H9A | 0.9300 |
| N2—C6 | 1.393 (3) | C10—C11 | 1.477 (4) |
| N2—H2B | 0.8600 | C11—H11A | 0.9600 |
| N3—C9 | 1.285 (3) | C11—H11B | 0.9600 |
| N3—O4 | 1.419 (3) | C11—H11C | 0.9600 |
| O1—N1—O2 | 121.7 (2) | C6—C5—N1 | 122.4 (2) |
| O1—N1—C5 | 118.0 (2) | N2—C6—C1 | 122.0 (2) |
| O2—N1—C5 | 120.3 (2) | N2—C6—C5 | 121.6 (2) |
| C2—C1—C6 | 121.5 (2) | C1—C6—C5 | 116.3 (2) |
| C2—C1—H1A | 119.2 | O3—C7—N2 | 124.1 (2) |
| C6—C1—H1A | 119.2 | O3—C7—C8 | 123.3 (2) |
| C1—C2—C3 | 120.4 (2) | N2—C7—C8 | 112.5 (2) |
| C1—C2—H2A | 119.8 | C10—C8—C9 | 103.6 (2) |
| C3—C2—H2A | 119.8 | C10—C8—C7 | 125.4 (2) |
| C7—N2—C6 | 129.4 (2) | C9—C8—C7 | 131.0 (2) |
| C7—N2—H2B | 115.3 | N3—C9—C8 | 113.6 (3) |
| C6—N2—H2B | 115.3 | N3—C9—H9A | 123.2 |
| C9—N3—O4 | 104.7 (2) | C8—C9—H9A | 123.2 |
| C4—C3—C2 | 119.9 (2) | O4—C10—C8 | 109.1 (2) |
| C4—C3—Cl | 120.2 (2) | O4—C10—C11 | 116.5 (3) |
| C2—C3—Cl | 119.8 (2) | C8—C10—C11 | 134.3 (3) |
| C10—O4—N3 | 109.1 (2) | C10—C11—H11A | 109.5 |
| C3—C4—C5 | 119.8 (2) | C10—C11—H11B | 109.5 |
| C3—C4—H4A | 120.1 | H11A—C11—H11B | 109.5 |
| C5—C4—H4A | 120.1 | C10—C11—H11C | 109.5 |
| C4—C5—C6 | 121.9 (2) | H11A—C11—H11C | 109.5 |
| C4—C5—N1 | 115.7 (2) | H11B—C11—H11C | 109.5 |
| C6—C1—C2—C3 | 0.7 (4) | C4—C5—C6—C1 | −1.0 (3) |
| C1—C2—C3—C4 | −1.5 (4) | N1—C5—C6—C1 | 179.2 (2) |
| C1—C2—C3—Cl | 178.87 (19) | C6—N2—C7—O3 | 2.0 (4) |
| C9—N3—O4—C10 | −0.4 (3) | C6—N2—C7—C8 | −177.5 (2) |
| C2—C3—C4—C5 | 1.1 (4) | O3—C7—C8—C10 | −7.0 (4) |
| Cl—C3—C4—C5 | −179.31 (19) | N2—C7—C8—C10 | 172.5 (2) |
| C3—C4—C5—C6 | 0.2 (4) | O3—C7—C8—C9 | 171.5 (2) |
| C3—C4—C5—N1 | 180.0 (2) | N2—C7—C8—C9 | −8.9 (4) |
| O1—N1—C5—C4 | 8.4 (3) | O4—N3—C9—C8 | 0.4 (3) |
| O2—N1—C5—C4 | −171.8 (2) | C10—C8—C9—N3 | −0.2 (3) |
| O1—N1—C5—C6 | −171.8 (2) | C7—C8—C9—N3 | −179.0 (2) |
| O2—N1—C5—C6 | 7.9 (4) | N3—O4—C10—C8 | 0.3 (3) |
| C7—N2—C6—C1 | −3.1 (4) | N3—O4—C10—C11 | −177.0 (2) |
| C7—N2—C6—C5 | 176.7 (2) | C9—C8—C10—O4 | −0.1 (3) |
| C2—C1—C6—N2 | −179.6 (2) | C7—C8—C10—O4 | 178.8 (2) |
| C2—C1—C6—C5 | 0.6 (3) | C9—C8—C10—C11 | 176.5 (3) |
| C4—C5—C6—N2 | 179.2 (2) | C7—C8—C10—C11 | −4.6 (5) |
| N1—C5—C6—N2 | −0.6 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2B···O2 | 0.86 | 1.94 | 2.629 (3) | 136. |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LD2035).
References
- Enraf–Nonius (1994). CAD-4 EXPRESS Enraf–Nonius, Delft, The Netherlands.
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- Huang, W. H., Yang, C. L., LEE, A. R. & Chiu, H. F. (2003). Chem. Pharm. Bull. 51, 313–314. [DOI] [PubMed]
- North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968). Acta Cryst. A24, 351–359.
- Schattenkirchner, M. (2000). Immunopharmacology, 47, 291–298. [DOI] [PubMed]
- Shaw, J. J., Chen, B., Wooley, P., Palfey, B., Lee, A. R., Huang, W. H. & Zeng, D. (2011). Am. J. Biomed. Sci. 3, 218–227. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811047994/ld2035sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811047994/ld2035Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



