Abstract
Previous studies have shown that pairs of closely-linked Ac/Ds transposable elements can induce various chromosomal rearrangements in plant genomes. To study chromosomal rearrangements in rice, we isolated a line (OsRLG5-161) that contains two inversely-oriented Ds insertions in OsRLG5 (Oryza sativa Receptor like kinase Gene 5). Among approximately 300 plants regenerated from OsRLG5-161 heterozygous seeds, 107 contained rearrangements including deletions, duplications and inversions of various sizes. Most rearrangements were induced by previously identified alternative transposition mechanism. Furthermore, we also detected a new class of rearrangements that contain juxtaposed inversions and deletions on the same chromosome. We propose that these novel alleles were generated by a previously unreported type of alternative transposition reactions involving the 5′ and 3′ termini of two inversely-oriented Ds elements located on the same chromatid. Finally, 11% of rearrangements contained inversions resulting from homologous recombination between the two inverted Ds elements in OsRLG5-161. The high frequency inheritance and great variety of rearrangements obtained suggests that the rice regeneration system results in a burst of transposition activity and a relaxation of the controls which normally limit the transposition competence of individual Ds termini. Together, these results demonstrate a greatly enlarged potential of the Ac/Ds system for plant chromosome engineering.
INTRODUCTION
The maize Ac/Ds transposable element system consists of the autonomous element Activator (Ac) and the non-autonomous element Dissociation (Ds). Pairs of closely linked Ac/Ds elements are known to induce chromosomal breakage and rearrangements such as deletions, duplications, inversions and translocations in maize and Arabidopsis. In maize, two Ds elements in an inverted orientation induce sister chromatid fusion in the presence of Ac (1). In addition, pairs of closely linked Ac and Ds elements have been reported to cause chromosome breakage (2) at frequencies inversely correlated with the inter-transposon distance (2,3). Subsequent reports indicate that, in addition to chromosome breakage, pairs of Ac/Ds elements can generate flanking deletions and inverted duplications via Sister Chromatid Transposition (SCT) (4,5). In SCT, Ac transposase binds to the directly-oriented 5′ and 3′ terminal sequences of transposons on sister chromatids, and these termini are subsequently excised and re-inserted into genomic target sites to generate deletions and corresponding inversions (4,5). Another type of alternative transposition reaction utilizes the reverse-oriented Ac/Ds termini located on the same chromatid; this so-called reversed-ends transposition reaction can generate deletions, inversions and translocations (6,7). The same transposon configuration also generated novel chimeric genes by joining the coding and regulatory sequences of two linked paralogous genes (8). Another recent study (9) identified these and other types of rearrangements including transposition of a macrotransposon (MTn) which extends from the external 5′- and 3′-ends of two separated TEs and includes the segment between them. Transposition of the two external termini leads to MTn excision, with or without subsequent reinsertion in the maize genome. Finally, pairs of Ac and fAc elements at the maize p1 locus have been shown to induce chromosome breakage at frequencies inversely proportional to the inter-transposon distance (10).
Here, we tested the ability of a closely-linked pair of Ds elements to generate chromosomal rearrangements in a transgenic rice system. The results indicate that two Ds elements in an inverted orientation can undergo frequent alternative transposition reactions, as well as homologous recombination (HR). In addition, we identified a new type of single chromatid transposition (SLCT) event involving the directly-oriented 5′ and 3′ termini of different Ds elements on the same chromatid. These results confirm the potential of Ac/Ds elements as agents for genome restructuring, and provide new information on the spectrum of possible rearrangements.
MATERIALS AND METHODS
T-DNA vectors, Agrobacterium transformation and rice lines
Ac and Ds elements were cloned in the T-DNA vector pSB11 (Figure 1) by standard molecular biology methods. Detailed information on the Ac and Ds gene trap cassettes was described in a previous report (11). T-DNA vectors were transformed into LBA4404 cells carrying a pSB11 vector. Co-integration of the DNA was confirmed by SalI digestion. Rice calli were transformed with T-DNA carrying hygromycin phosphotransferase (HPT) according to a previously published method (12) with slight modifications. For the selection of PPT-resistant calli, MS-based media was used (13). Ds lines were developed using Dongjin, an elite japonica variety.
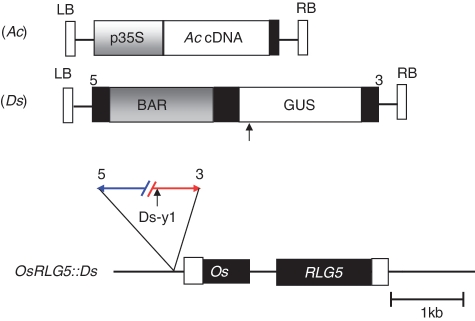
Figure 1.
Structures of the Ac and Ds T-DNA vectors and the OsRLG5::Ds allele. Expression of Ac cDNA is driven by the CaMV 35S promoter in the Ac T-DNA vector. A BAR selection marker and GUS reporter gene were inserted inside the Ds T-DNA vector. The OsRLG5::Ds allele carries a single Ds insertion in the promoter region of the OsRLG5 gene. White and black boxes indicate UTRs and exons of OsRLG5, respectively. Short vertical arrow indicates an EcoRI restriction site inside the GUS coding region. The numbers 5 and 3 above the Ds T-DNA vector or Ds-y1 indicate the 5′ and 3′ Ds-ends, respectively.
Tissue culture for regeneration of plants from seeds
To isolate lines carrying a closely-linked pair of Ds elements, OsRLG5::Ds seeds heterozygous for a single Ds element insertion were utilized to regenerate plants (Figure 2). This method has been described previously (14). Briefly, dry mature seeds were hulled and sterilized with 75% EtOH. To produce plantlets from calli, sequential incubations were performed using four types of tissue culture media: (a) NB medium for callus induction; (b) N6-7-CH medium for Pre-Regeneration; (c) N6S3-CH-I medium for Regeneration I; (d) N6S3-CH-II medium for Regeneration II. Before transfer to a greenhouse, regenerated plants were transplanted into a bottle containing 0.5-stength MS media (15).
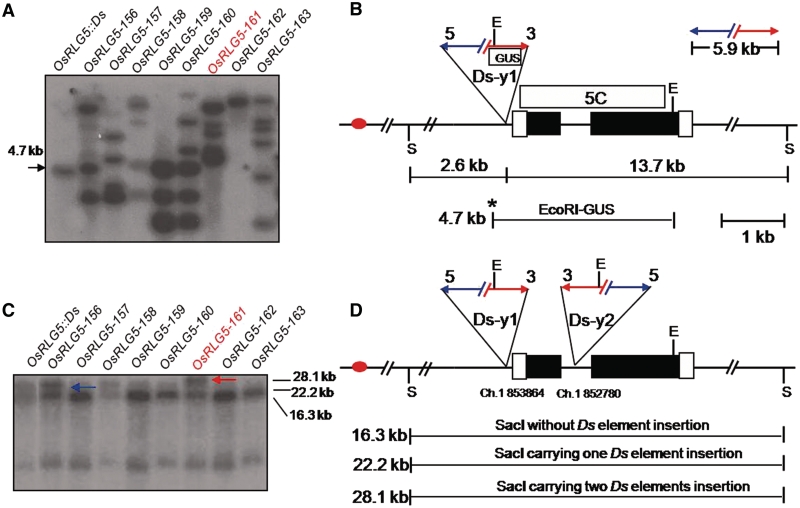
Figure 2.
Selection of plants containing two Ds elements at the OsRLG5 locus. (A) Southern blot hybridization to identify Ds transpositions in plants regenerated from OsRLG5::Ds seeds. EcoRI-digested genomic DNA samples from the indicated lines were hybridized with a 1.8-kb DNA fragment from the GUS coding region. The progenitor OsRLG5::Ds line exhibits a 4.7-kb band, while most regenerated lines contain multiple novel bands. The OsRLG5-161 line (red) lacks the 4.7-kb band. (B) Structure of the OsRLG5::Ds locus. Red oval indicates centromere, black boxes indicate coding sequences of OsRLG5, and white boxes indicate UTRs. Red and blue arrows indicate the 5′ and 3′ Ds termini. Letters E and S indicate EcoRI and SacI restriction sites, respectively. Boxes labeled GUS and 5C indicate fragments used as probes for Southern analysis. The 2.6 and 13.7 kb indicate distances from the Ds-y1 insertion site to upstream and downstream SacI sites, respectively. Asterisk indicates EcoRI site located inside the Ds-y1 element. The 5.9-kb marked segment above the map indicates the size of Ds. (C) Southern blot hybridization to identify lines containing two copies of Ds inserted in the OsRLG5 locus. SacI-digested DNA samples from the indicated lines were hybridized with probe 5C from the OsRLG5 gene. The 16.3-kb band represents the reference OsRLG5 allele lacking Ds. The original OsRLG5::Ds allele produced a 22.2-kb fragment (blue arrow), while the OsRLG5-161 allele has a 28.1-kb SacI fragment (red arrow). (D) The OsRLG5-161 allele carries two Ds elements in inverted orientation and separated by a distance of 1.1 kb. The numbers below the map indicate the locations of Ds-y1 and Ds-y2 insertion sites on chromosome 1. Horizontal lines indicate the sizes of restriction fragments generated by SacI digestion. Other symbols as in 2B.
Plant growth conditions and propagation of R1 plants
R1 plants were grown under greenhouse conditions at Gyeongsang National University (GNU) and Iowa State University (ISU). The daily high greenhouse temperatures were typically 30°C, while the daily low temperatures were typically 20°C and 23°C at GNU and ISU, respectively. R1 plants regenerated via tissue culture were propagated by selfing. If necessary, R1 plants were ratooned by cutting off the shoots. Re-shooting and maintenance of R1 plants were performed in the greenhouse during winter.
DNA manipulation
Rice genomic DNA was prepared from young leaves using either a urea extraction procedure or a modified CTAB extraction protocol (16). Aliquots of 5 μg of purified DNA were digested with the appropriate restriction endonucleases, size-fractionated on 0.8% agarose gel and transferred onto a nylon membrane. The blots were then hybridized to probes in hybridization buffer containing 6× SSC, 5× Denhardts, 0.5% SDS, 50 nM Tris (pH 8.0), 10 nM EDTA, 0.1 mg/ml salmon sperm DNA (heat denatured) and 5% dextran sulfate. Final washes of the filters were carried out in 0.2× SSC and 0.1% SDS solution for 15 min at 65°C. The membranes were then exposed to X-ray film.
PCR amplifications
PCR amplifications were performed using the oligonucleotide primers listed in Supplementary Table S1. Taq DNA polymerase and dNTPs (SOLGENT: http://www.solgent.co.kr and 5 PRIME: http://www.5prime.com) were used for PCR with 500 ng of genomic DNA as a template. Annealing temperatures in the range of 55–58°C were used, depending on the primer sequences. A typical reaction consisted of an initial denaturation at 95°C for 3 min, followed by 33 cycles of denaturation for 30 s, annealing for 30 s and extension at 72°C or 65°C for 30–90 s, followed by a final extension at 72°C for 7 min.
Identification of rearrangements and cloning of rearrangement breakpoints
Chromosome rearrangements at the OsRLG5-161 locus were identified by PCR using primer pairs 1 + 3 and 2 + 4 (Figure 3). Among 300 R1 plants, 107 plants carried either or both PCR products. To analyze deletions with breakpoints in the proximal and distal regions, a series of primers located in highly conserved regions of the OsRLG gene family were utilized. Inverse-PCR was used to detect breakpoints in some rearrangement events in which Ds-y1 or Ds-y2 had excised. Genomic DNA was digested with the four-base cutter NlaIII, and PCR-amplified using two sets of OsRLG5-specific primers. The locations of PCR primers from the OsRLG5-161 locus are shown in Figure 3, and sequences of all primers used in this study are listed in Supplementary Table S1.
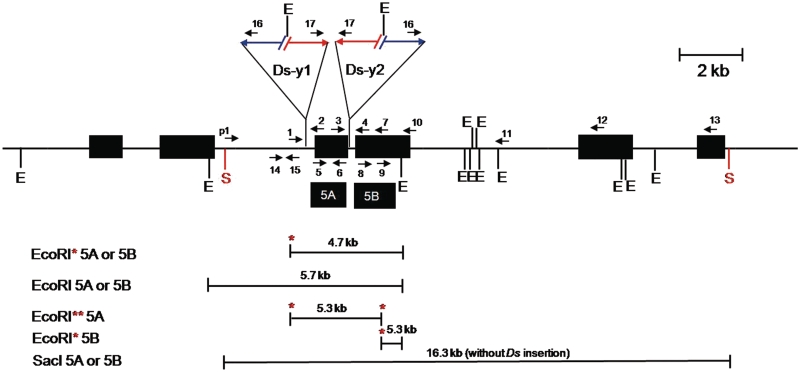
Figure 3.
Genomic structure of OsRLG5-161 and adjacent genes. The OsRLG5-161 allele is flanked on the proximal side by OsRLG20 and the distal side by OsRLG18 (each OsRLG gene consists of two exons). Numbered arrows indicate the orientations and positions of PCR primers. Black boxes marked 5A and 5B indicate fragments used as probes for Southern analysis. Horizontal lines below the map indicate EcoRI or SacI restriction fragments which hybridize with the indicated probes; red asterisks indicate EcoRI sites located inside the Ds-y1 and Ds-y2 elements. Other symbols as in Figure 2.
RESULTS
Generation of rice lines carrying a closely-linked pair of Ds elements at the OsRLG5 locus
We previously established a system for Ac/Ds transposition in japonica rice (15). Ac transposase is provided by a T-DNA construct containing the CaMV 35S promoter driving Ac cDNA, while a second T-DNA construct contains a modified Ds element (Figure 1). Rice plants containing these constructs were screened to identify plants with new Ds insertions. We isolated one line (OsRLG5::Ds) that contained a single copy Ds insertion in the promoter region of the OsRLG5 (Receptor Like kinase Gene 5), which encodes a Serine/Threonine protein kinase closely related to Lrk10 (17). The OsRLG5 gene is located within a cluster of 36 copies of related receptor-like kinase genes on the short arm of rice chromosome 1. Steady state levels of OsRLG5 transcripts are severely diminished by the Ds insertion, but homozygous mutant plants exhibit no obvious mutant phenotype (Xuan, unpublished data).
To obtain lines containing two Ds elements in a cis-configuration near the OsRLG5 locus, we screened plants that were regenerated from callus cultures. A previous report described extensive activation of Ds elements during plant regeneration from calli: in one callus-derived regenerated population, >70% of the plants carried independent Ds insertions, and a high proportion (36%) of the transposed Ds elements were inserted into nearby donor sites (11,15). Therefore, we subjected 270 seeds produced by self-pollination of OsRLG5::Ds/OsRLG5 plants to callus induction and plant regeneration. The Ds insertion in the OsRLG5::Ds line was tightly linked to the Ac transgene (data not shown); therefore, half of the seeds from self-pollinated OsRLG5::Ds/OsRLG5 plants were expected to be hemizygous for both Ac and Ds. Regenerated plants were analyzed by Southern blot hybridization to detect transposed Ds elements. Genomic DNA was digested with EcoRI and hybridized with a GUS coding sequence probe. The GUS-hybridizing fragments result from cutting at a site within the modified Ds element, and at EcoRI sites in the flanking genomic DNA. The progenitor OsRLG5::Ds line shows a single 4.7-kb GUS-hybridizing band, while most of the regenerated plants exhibit new bands indicating Ds insertions at new loci (Figure 2A). To identify plants carrying a second Ds insertion in the OsRLG5 gene, genomic DNA was digested with SacI and hybridized with probe 5C as shown in Figure 2B. SacI does not cut within the Ds element, but it cuts in the OsRLG5 locus at 2.6-kb upstream and 13.7-kb downstream of the original Ds insertion site to generate a 22.2-kb SacI fragment in the progenitor OsRLG5::Ds line (Figure 2B). Insertion of a second Ds element within this fragment would generate a 28.1-kb SacI fragment; this was observed in two lines among the 270 lines screened (0.7%). One of these lines (OsRLG5-161) exhibited the 28.1-kb SacI fragment (Figure 2C), but not the 4.7-kb EcoRI-GUS fragment (Figure 2A). These results indicate that OsRLG5-161 may contain a transposed Ds element inserted within the 4.7-kb fragment. Through subsequent PCR analysis (data not shown), the transposed Ds element was found to have inserted 1.1-kb downstream of the original Ds element, in an inverted orientation. For clarity, the original Ds element present in OsRLG5::Ds was designated Ds-y1, while the second transposed Ds element present in OsRLG5-161 was termed Ds-y2 (Figure 2D). Both Ds elements are flanked by characteristic 8-bp target site duplication sequences (TGACTGCA and CCCTGGCT for Ds-y1 and Ds-y2, respectively).
Ds-y1 and Ds-y2 in OsRLG5-161 are competent for SCT
Previous research showed that a 5′ Ac/Ds-end and a 3′ Ac/Ds-end in direct orientation can undergo SCT in maize, tobacco and Arabidopsis (1,4,5,18). The OsRLG5-161 allele contains two pairs of directly-oriented 5′ and 3′ Ds termini: (a) 5′ of Ds-y1 and 3′ of Ds-y2, and (b) 3′ of Ds-y1 and 5′ of Ds-y2. To test whether either of these paired ends can induce flanking inversions as expected from SCT, we performed PCR using primer pairs that could generate products only if the flanking DNA is inverted. As shown in Figure 3, primers 1 + 3 flanking Ds-y1 are both oriented in the distal direction, while primers 2 and 4 flanking Ds-y2 are both oriented in the proximal direction. Thus, PCR using primers 1 + 3 or 2 + 4 should not amplify products from unrearranged OsRLG5-161 genomic DNA. However, a chromosome inversion produced by SCT of the Ds elements could reorient one of the primers and thus produce a positive PCR product (Figure 4A and Supplementary Figure S1). Out of 300 R1 plants screened, 61 generated PCR products with primers 1 + 3, 13 generated PCR products with primers 2 + 4, and 33 R1 plants generated PCR products with both primer pairs (Table 1).
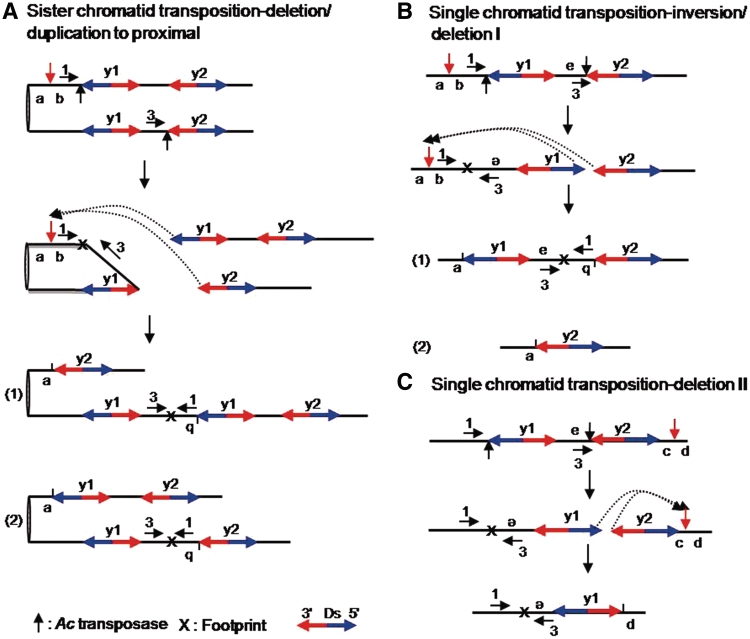
Figure 4.
Models for SCT with a proximal target site and SLCT. (A) SCT is depicted in two steps. In all the diagrams, sister chromatids are attached at the centromere (left). Element y1 and y2 indicate Ds-y1 and Ds-y2, respectively. Transposases cut the 5′-end of y1 on the upper chromatid and the 3′-end of y2 on the lower chromatid, as indicated by the black vertical arrows in the top diagram. The two ends are then reinserted into a new target site between a and b, as shown by the dotted arrows in the second diagram. A red vertical arrow indicates the new target site. Part (1) of the third diagram shows that insertion of the 3′-end of y2 next to a and the 5′-end of y1 next to b generates one chromatid (upper) containing a flanking proximal deletion and a single copy of Ds, and a second chromatid (lower) containing an inverted duplication and three copies of Ds. Part (2) of the fourth diagram shows that insertion of the 5′-end of y1 next to a and the 3′-end of y2 next to b generates one chromatid (upper) containing a flanking proximal deletion and two copies of Ds, and a second chromatid (lower) containing an inverted duplication and two copies of Ds. Note both outcomes result in inversion of sequences flanking Ds. Blue and red arrows indicate the 5′ and 3′ directions of Ds elements, respectively. X indicates a footprint. Primers used to detect rearrangements were shown as horizontal arrows with numbers. (B) The inversion/deletion I process derived from SLCT is depicted in three steps. Transposases cut the 5′-end of y1 and the 3′-end of y2, as indicated by the black vertical arrows in the top diagram. The 5′ and 3′ termini of y1 and y2, respectively, are re-inserted into the proximal region with respect to the original Ds sites, as shown by the dotted arrows and red vertical arrow in the second diagram. Consequently, the fragment from the reinsertion site on the 5′-end of y1 was inverted and was jointed to the 3′-end of y2. Reinsertion of the 5′ and 3′ termini of y1 and y2, respectively, at the target site between a and b leads to two configurations, as shown in parts (1) and (2) in the diagram. Part (1) shows inversions of fragment carrying b located between e and the 3′-end of y2. Part (2) indicates deletions, referred to as deletion I, including fragments carrying b and e, and y1. (C) Deletion II derived from a SLCT is shown in two steps. Transposases cut the 5′-end of y1 and the 3′-end of y2, as indicated by black vertical arrows in the top diagram. The 5′ and 3′ termini of y1 and y2, respectively, are re-inserted into the distal region with respect to the original Ds sites, as shown by the dotted arrows and red vertical arrowhead in the second diagram. Consequently, the 5′-end of y1 reinserted next to d causes inversion of fragments containing e and y1 and deletion of fragment containing c and y2, which is called deletion II.
Table 1.
Classification of chromosomal rearrangements based on PCR patternsa
| No of lines | Primer 1 and 3a | Primer 2 and 4a | Expected rearrangement models | Proportion (%) |
|---|---|---|---|---|
| 61 | + | − | A(1), B(1), E | 20.3 |
| 13 | − | + | A(2), B (2), F | 4.3 |
| 33 | + | + | D | 11 |
| Total 300 | 35.6 |
aThe locations of the primer sets are shown in Figure 3.
+, PCR product; −, no PCR product.
Models A(1) and A(2) are shown in Figure 6A and Figure 4B(1) and models B(1) and B(2) are shown in Figure 4B(2), 4C and Supplementary Figure S3A. Models E and F are shown in Figure 4A and Supplementary Figure S1, respectively, and model D is shown in Supplementary Figure S4A.
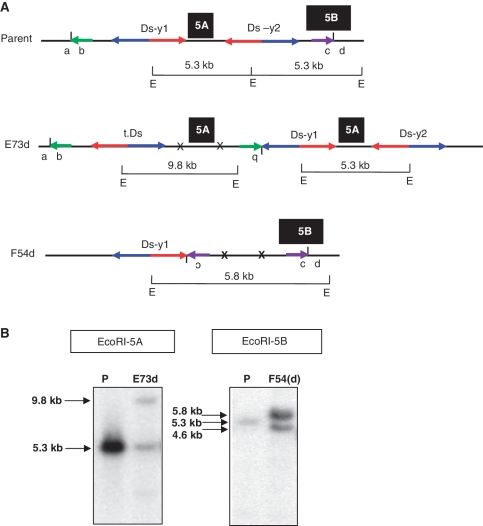
R1 Plants which produced PCR products using a single pair of primers (either 1 + 3 or 2 + 4) will be discussed first. Among the 74 (61 + 13) lines that were positive for either primer pairs 1 + 3 or 2 + 4, some lines also carried deletions in the same R1 plants. The deletion break points were cloned and sequenced (see ‘Materials and Methods’ section), and a series of proximal and distal primers were designed to identify the duplication breakpoints. In this way we cloned the breakpoints of two inverted duplication candidates (E73d and F54d). For line E73d, we used proximal primer 73 paired with either Ds 5′ primer 16 or OsRLG5-specific primer 2. For line F54d, we used distal primer 8 paired with Ds 3′-end primer 17 or OsRLG5-specific primer 1 (Supplementary Figure S2A). PCR products of ∼1.1 and 0.8 kb were obtained from lines E73d and F54d, respectively (Supplementary Figure S2B). Direct sequencing of these PCR products indicate that E73d and F54d contain inverted duplications of ∼520 and ∼1.2 kb, respectively. PCR results showed that one transposed Ds element was identified, which was located between original Ds-y1 insertion and the EcoRI recognition site in the proximal region (Supplementary Figure S2B). The presence of these duplications was confirmed by the results of Southern blot hybridizations using genomic fragments 5A and 5B flanking the Ds element as probes (Figure 5A). As shown in Figure 5B, E73d contains two EcoRI fragments that hybridize with probe 5A: one band of 5.3 kb which matches the size of the progenitor allele, and a second novel band of 9.8 kb. These results are consistent with the presence of a proximal inverted duplication in the E73d line. For F54d, Southern blot analysis shows that probe 5B hybridizes with two EcoRI bands: one of 5.8 kb (putative duplication chromosome) and one of 4.6 kb (putative deletion chromosome). These results suggest that the F54d duplication chromosome was recovered in the heterozygous state with a corresponding deletion chromosome (deletion events will be described in the following section). Together, the PCR, sequencing and Southern blot data indicate that E73d carries a proximal inverted duplication of 520 kb, while F54d carries a distal inverted duplication of 1.2 kb, plus a deletion. Moreover, sequencing of the PCR products generated by primers 1 + 3 (E73d) and 2 + 4 (F54d) confirm the expected rearrangement junctions and transposition footprints. We conclude that E73d and F54d both contain inverted duplications generated by SCT.
Figure 5.
Genomic structure and Southern blot hybridization of SCT-induced duplications. (A) Genomic structures of parent and SCT-induced duplications (E73d and F54d) are shown. Short vertical lines between a and b and between c and d indicate new junctions of E73d and F54d, respectively. X indicates Ds excision footprints and t.Ds means translocated Ds element. Green and purple short horizontal arrows indicate the orientations of the duplicated regions. Inversion of duplicated regions (b of E73d and c of F54d) is depicted as upside-down characters. Black boxes 5A and 5B indicate probe locations as described in Figure 3. The sizes of EcoRI fragments that span the probes are shown above horizontal bars marked with E, which stands for the EcoRI recognition site. The 5′ and 3′-ends of both Ds elements are indicated by blue and red arrows, respectively. (B) Duplication line E73d contains two bands: one band of 5.3 kb (same as parental, P), and a second band measuring 9.8 kb. These bands correspond to the fragments shown in (A). Line F54d contains a 5.8-kb band produced from the duplication chromosome whose structure is shown in Figure 5A, and a 4.6-kb band derived from a chromosome deletion. The parental (P) line produces a band of 5.3 kb. Breakpoints of the deletion (F54) and duplication F54d are shown in Table 2.
Directly-oriented Ds-y1 and Ds-y2 termini in the same chromatid can undergo alternative transposition
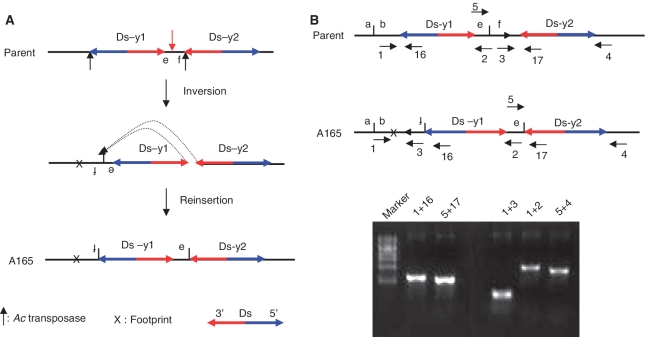
We identified several cases containing a segmental inversion and deletion on the same chromosome. We hypothesized that these may have been generated by alternative transposition reactions involving the directly-oriented 5′- and 3′-ends of different Ds elements on the same chromatid (Figures 4B, C and 6A). In one case (line A165), we performed PCR on genomic DNA with primer pair 1 + 16; surprisingly, the size of the PCR product is larger than expected. Sequencing of the PCR product revealed that fragment contain f between Ds-y1 and Ds-y2 was inverted and inserted between b and Ds-y1 (Figure 6B); an apparent Ac/Ds footprint was identified at the junction of b and f. A second PCR was performed using the primer pair 5 + 17. Sequencing this PCR product revealed that fragment contain e joined to Ds-y2, and the 8-bp sequences flanking the 5′-end of Ds-y1 and the 8-bp sequence flanking the 3′-end of Ds-y2 are the same. These results strongly suggest that A165 originated via an alternative transposition mechanism in which the 5′-end of Ds-y1 and the 3′-end of Ds-y2 on the same chromatid are recognized and cut by Ac transposase. The sequences flanking the excised Ds termini were ligated together, followed by insertion of the Ds termini back into the same chromatid as described in Figure 6. Because the Ds termini involved in the transposition reaction are from the same chromatid, we designate this type of transposition reaction as SLCT.
Figure 6.
Inversion in line A165 induced by SLCT. (A) SLCT model for formation of A165 is depicted in two steps. The parent chromosome is cut by Ac transposase at the 5′-end of Ds-y1 and at the 3′-end of Ds-y2, as shown by the short black arrows in the top diagram. Subsequently, the sequences flanking the 5′ and 3′ Ds termini are ligated, which results in the formation of a footprint (X) and inversion of a segment including Ds-y1 and fragments containing e and f, as shown in the second diagram. The third diagram shows that the excised Ds termini are reinserted into a site between e and f, as shown by the red vertical arrow and dotted arrows in the second diagram. Consequently, inverted fragments containing f and e are joined to the 5′-end of Ds-y1 and to the 3′-end of Ds-y2, respectively. (B) Genomic structures of OsRLG5 locus in parent and A165 lines. Short horizontal arrows (numbered) indicate the orientations and approximate positions of the primers at the OsRLG5 locus. Short vertical line between two Ds elements indicates the e-f target site. The orientation of ITS in the parent is indicated by the arrow. Gel photo shows the products of five PCR reactions using genomic A165 DNA template and the primer pairs indicated above each lane.
Both SCT and SLCT can produce nested interstitial deletions
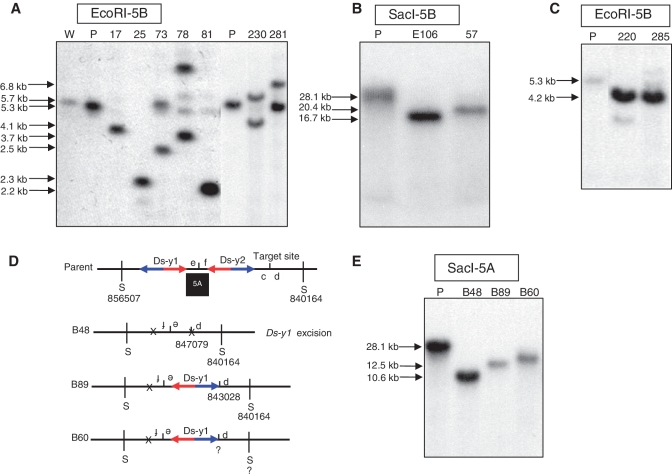
As predicted in Figure 4 and Supplementary Figure S1, SCT and SLCT can generate segmental deletions. We isolated 28 deletions from OsRLG5-161, ranging in size from 184 bp to 520 kb; all the deletions originate at the OsRLG5 locus, and extend to either proximal or distal sites as listed in Table 2. In E106, the Ds-y1 5′-end joined to a site 84.5-kb proximal; we propose that this deletion was generated via SCT (Figure 4A). Southern analysis with SacI-5B detected a 16.7-kb band in homozygous deletion line E106 which is shorter than parental chromosome (Figure 7B). Lines E59 and E69 contain two somatic SCT-induced deletions which joined the Ds-y1 5′-end to sites 3.5- and 6.0-kb proximal, respectively. Sequences of the junctions of E59 and E69 are shown in Supplementary Table S2. In line F54, the Ds-y2 5′-end is joined to a site 750 bp distal; we propose that this deletion was generated via SCT (Supplementary Figure S1). As shown in Figure 5B, line F54(d) contained a 4.6-kb deletion and a 5.8-kb duplication.
Table 2.
Lines of each model of chromosomal rearrangement
| Model | Line number | Nature of rearrangement | Location of new junction sitea | Size of the rearrangement (bp) |
|---|---|---|---|---|
| A | A165 | Inversion(Germinal) | Ch.1 853640 | 224 |
| B | B89 | Deletion (Germinal) | Ch.1 843028 | 9752 |
| B48 | Deletion (Germinal) | Ch.1 847079 | 5701 | |
| B60 | Deletion (Germinal) | |||
| B3 | Deletion (Somatic) | Ch.1 854719 | 855 | |
| B38 | Deletion (Somatic) | Ch.1 854646 | 782 | |
| B54 | Deletion (Somatic) | Ch.1 854822 | 958 | |
| D | D4 | Inversion (Germinal) | 1100 | |
| D35 | Inversion (Germinal) | 1100 | ||
| D7 | Inversion (Germinal) | 1100 | ||
| D64 | Inversion (Germinal) | 1100 | ||
| D5 | Inversion (Germinal) | 1100 | ||
| E | E106 | Deletion (Germinal) | Ch.1938355 | 84 491 |
| E59 | Deletion (Somatic) | Ch.1 857384 | 3520 | |
| E69 | Deletion (Somatic) | Ch.1 859908 | 6044 | |
| E73d | Duplication (Germinal) | Ch.1 1372985 | 526 104 | |
| B or E | 17 | Deletion (Germinal) | Ch.1 860732 | 7952 |
| 25 | Deletion (Germinal) | Ch.1 976290 | 123 510 | |
| 57 | Deletion (Germinal) | Ch.1 854594 | 1814 | |
| 73 | Deletion (Germinal) | Ch.1 1372994 | 520 214 | |
| 78 | Deletion (Germinal) | Ch.1 937320 | 84 540 | |
| 81 | Deletion (Germinal) | Ch.1 856237 | 3257 | |
| 230 | Deletion (Germinal) | Ch.1 860745 | 7965 | |
| 281 | Deletion (Germinal) | Ch.1 857481 | 4701 | |
| 44 | Deletion (Somatic) | Ch.1 853780 | 184 | |
| 71 | Deletion (Somatic) | Ch.1 860946 | 8166 | |
| 83 | Deletion (Somatic) | Ch.1 912296 | 59 516 | |
| 89 | Deletion (Somatic) | Ch.1 919442 | 66 662 | |
| 242 | Deletion (Somatic) | Ch.1 857590 | 4810 | |
| 269 | Deletion (Somatic) | Ch.1 857655 | 4875 | |
| F | F54 | Deletion (Germinal) | Ch.1 852030 | 750 |
| F54d | Duplication (Germinal) | Ch.1 851564 | 1216 | |
| B or F | 220 | Deletion (Germinal) | Ch.1 853380 | 600 |
| 285 | Deletion (Germinal) | Ch.1 852347 | 1517 | |
| 33 | Deletion (Somatic) | Ch.1 846052 | 7812 |
aNumbers indicate the position on chromosome 1.
Figure 7.
Transposition-induced deletions in the OsRLG5 locus. (A) Southern blot analysis of lines E106 and 57. Genomic DNA was cut with SacI and hybridized with genomic probe 5B (Figure 3). Chromosomal rearrangement in line E106 was induced by Ds-y1 reinsertion in the proximal region. Ds-y2 was not excised from these two lines. Shorter DNA fragments are detected in these two lines compared with the parental line. (B) Southern blot analysis of deletions in the proximal region. Genomic DNA from the indicated lines was cut with EcoRI and hybridized with probe 5B. Sizes of hybridizing bands are shown at the left of the Southern blot. (C) Southern blot analysis of lines 220 and 285 which contain distal deletions. Genomic DNA was cut with EcoRI and hybridized with probe 5B. Deletion lines produce 4.2-kb fragments which are <5.3-kb band in parental lane. (D) Inferred structures of parental chromosome and distal deletions. Short vertical line between c and d indicates insertion target site. S indicates SacI digestion sites, whose positions are indicated. Ds-y1 was retained in line B89, but excised in line B48. The breakpoint of line B60 was not obtained. (E) SLCT-induced deletions distal to the original Ds position were examined by Southern blot. SacI cut genomic DNA was hybridized with probe 5A (Figure 3). Deletion chromosomes produced shorter bands than the parental line. W, wild-type; P, parental line.
In some deletions, the Ds-y1 3′-end joined to a distal site, or the Ds-y2 3′-end joined to a proximal site; these could be generated via either SCT or SLCT. Putative germinal deletions in the proximal region were confirmed by Southern blot hybridization. Southern analysis with SacI-5B showed that line 57 contained a 20.4-kb deletion (Figure 7B). In line 57, the Ds-y2 element remained at the reinsertion site, while it was excised from the breakpoints in line 17, 25, 73, 78, 81, 230 and 281. Southern analysis using EcoRI digestion and hybridization with probe 5B indicated that deletions in lines 17, 25 and 81 were maintained in homozygous condition, while deletions in lines 73, 78, 230 and 281 were maintained as heterozygotes with parental or duplication chromosomes. Deletions in lines 17, 25, 73, 78, 81, 230 and 281 were 4.1, 2.3, 2.5, 3.7, 2.2, 4.1 and 6.8 kb in size, respectively (Figure 7A). Finally, deletions in the distal region were also identified; these include lines 220 and 285 which contained deletions of ∼4.2 kb in homozygous condition (Figure 7C).
In lines B48 and B89, fragments carrying e and f, and Ds-y1 were inverted, and the Ds-y1 5′-end is joined to sites 5.7 and 9.7 kb distal, respectively. In B48, Ds-y1 was excised after rearrangement. Sequencing of the junction between d and e identified the expected Ds excision footprint, indicating that it originated via SLCT as shown in Figure 4C. Line B60 contained a deletion and an inversion that were suspected to have resulted from SLCT, but we failed to clone the breakpoint. Genomic structures of three deletions are shown in Figure 7D. Southern blot analysis using SacI and probe 5A was used to detect deletion homozygotes from three deletion lines. The results showed that deletion homozygotes B48 and B89 contained SacI fragments of 10.6 and 12.5 kb in size, respectively. Deletion homozygote B60 exhibited a band <28.1-kb parental line (Figure 7E), although we did not clone the breakpoint. Deletions in the proximal region of OsRLG5 as shown in Supplementary Figure S3A were analyzed and three somatic SLCT deletions were identified. In F3, F38 and F54, fragments carrying e and f, and Ds-y2 were inverted, and the Ds-y2 5′-end joined to sites 855, 782 and 958-bp proximal, respectively (Supplementary Figure S3). Sequences of the junctions between 5′ of Ds-y2 and f are shown in Supplementary Table S2.
HR of Ds elements can invert the inter-transposon segments
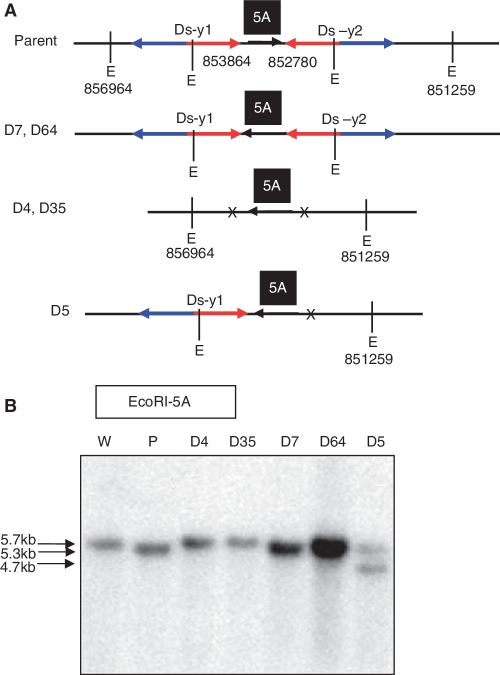
As shown in Supplementary Figure S4, HR between two Ds elements in inverted orientation could invert the inter-transposon segment (ITS). Putative recombination events could be detected by PCR using two sets of primer pairs flanking the Ds-y1 and Ds-y2 elements (1 + 3 and 2 + 4; Figure 3). These primer pairs would produce products only upon inversion of the ITS DNA. Among 300 R1 plants tested, 33 exhibited evidence of ITS inversion (Table 1). Because the inversions detected in the R1 plants could be either somatic or germinal events, we further tested R2 progeny plants derived from each of the 33 candidate inversion lines. For each candidate line, genomic DNA was isolated from ten R2 plants and analyzed by PCR. ITS inversions were detected among the progeny of the 33 candidate lines (data not shown). Five inversion-positive lines were further examined by analysis of individual R2 plants. The genomic structures of these five inversions and the primer locations are shown in Supplementary Figure S5B. Two PCR primer pairs (1 + 17 and 4 + 17; Figure 3) were used to determine whether Ds-y1 and Ds-y2 remained at the original insertion sites. PCR results showed that both Ds-y1 and Ds-y2 elements were excised in lines D4 and D35, whereas only the Ds-y2 element was excised in line D5 (Supplementary Figure S5A). These candidate inversions were further tested by Southern blot hybridizations with genomic fragment 5A as probe. As shown in Figure 3, EcoRI restriction sites located either within Ds or 1.5 kb distal to the Ds-y2 insertion site in OsRLG5-161 can be utilized to detect the presence of Ds elements in the five ITS events. The Southern blot results indicate that two plants homozygous for ITS inversions were identified among ten progeny of self-pollinated plant D5 (Supplementary Figure S5C). These two plants produced a single 4.7-kb band (EcoRI*5A shown in Figure 3) that was smaller than the band in the parent D5 line (Supplementary Figure S5C). In addition, both Ds-y1 and Ds-y2 were present in line D7 and D64. The chromosome configuration of line D7 is shown in Figure 8A and Supplementary Figure S5B. PCR results were verified by Southern blot hybridization using 5A as a probe. In lines D4 and D35, EcoRI-5A probed Southern blot showed bands of the same size as those from wild-type (5.7-kb-EcoRI 5A shown in Figure 3) because both Ds were excised. Southern blot analysis of lines D7 and D64, which carried both Ds elements, revealed bands of the same size as the parental line (5.3-kb-EcoRI**5A shown in Figure 3). Line D5 contained, in addition to the 5.3-kb parental band, a smaller band of 4.7 kb produced by excision of Ds-y2 (Figure 8B). For lines D4, D35 and D5, the new junctions were cloned and their sequences are presented in Supplementary Table S2.
Figure 8.
Inversions induced by HR between Ds elements. (A) Genomic structures of parent and inversion lines are shown. Numbers indicate positions of Ds-y1, Ds-y2 and EcoRI sites in chromosome 1. Other symbols as in Figure 3. (B) Southern blot analysis of putative HR-induced inversion lines. Genomic DNA from the indicated lines was digested with EcoRI and hybridized with probe 5A. Lines D4 and D35 showed bands of the same size as the wild type allele (5.7 kb), consistent with excision of both Ds-y1 and Ds-y2. Lines D7 and D64 showed bands of the same size (5.3 kb) as parental line (OsRLG5-161), consistent with retention of both Ds-y1 and Ds-y2. Line D5 contained one parental band (5.3 kb) and one shorter band (4.7 kb), suggesting heterozygosity for a parental allele (both Ds elements retained) and the D5 allele in which Ds-y2 had excised. W, wild-type; P, parental line.
DISCUSSION
Research on transposon-mediated chromosomal rearrangements and their heritability is important not only for understanding the relationship between transposon activity and genome evolution, but also in order to enable chromosomal engineering for crop improvement. Rice is rich in genetic materials and has a completely sequenced genome, which is a great advantage in the study of genomic DNA and its rearrangement (19,20). Tissue culture systems are well established and large numbers of transgenic plants can be obtained in a relatively short time. Furthermore, rice plants can be maintained by ratoon culture for prolonged periods (15). These factors confer great advantages to rice over other grain crops such as maize and wheat for studying chromosomal rearrangements and their heritability. Even though transposon-mediated gene tagging systems have been successfully applied in rice, there are very few reports that describe the extent and scope of transposon-induced chromosomal rearrangements in rice genomes. Previous studies have shown that pairs of closely-linked Ac/Ds elements can induce chromosome rearrangements in other plant species (1,7,9,18). Therefore we aimed to identify and analyze a rice stock containing two Ds insertions in a single locus. In a previous transposon-tagging project, we obtained a line (OsRLG5::Ds) containing a single Ds element inserted in the OsRLG5 locus, which encodes a receptor-like kinase gene of unknown function. From OsRLG5::Ds we regenerated and analyzed 270 progeny plants; among these we identified two events where an additional copy of intact Ds was inserted within a 22.2-kb SacI fragment including OsRLG5. If this frequency (0.7%) can be extended to other loci, then screening approximately 200 progeny of plants containing a single Ds insertion may be sufficient to yield stocks containing two linked Ds elements.
An exceptionally high frequency of chromosomal rearrangements was obtained in regenerated plants
Line OsRLG5-161 contains two identical Ds elements inserted at the OsRLG5 locus in inverted orientation and separated by 1.1 kb. Among 300 R1 progeny of OsRLG5-161 produced by tissue-culture regeneration, 107 (36%) contained chromosome rearrangements including deletions, duplications and inversions. A subset of events was tested by R2 progeny analysis to distinguish between somatic and germinal rearrangements. Among 34 rearrangements tested, 22 events (65%) were transmitted to the R2 generation and thus were considered to be germinal events. Inheritance of rearrangements was examined in the R2 generation by PCR using the same primers as those used for R1 plants. Supplementary Figure S6 shows that deletions were maintained in R2 plants. Also, Southern blot analysis confirmed PCR data (Figures 5, 7 and 8). The data suggested that rearrangement in the derivatives (22 germinal events) were quite stable in the following generations regardless of the presence of Ac transposase. Failure to detect the remaining 12 events (35%) in the R2 progeny could be attributed to two major reasons: First, some rearrangements may have been restricted to the R1 vegetative cells and were not included in the cells that give rise to gametophytes. Second, some rearrangements may cause gametophyte or seedling lethality and thus would not be transmitted. Even so, the frequency of heritable rearrangements obtained in this study is much higher than that reported for pairs of linked Ac/Ds elements in maize and Arabidopsis. The high frequency observed in our study most likely reflects a high rate of Ds transposition induced by tissue-culture regeneration. In a previous study, >70% of Ds elements were mobilized in independent rice calli cell lines before the initiation of plantlets (15). A number of reports have shown that tissue culture conditions can reactivate silent transposable elements in maize (21) and rice (22–24); moreover, reactivation is associated with alteration of methylation patterns in the Ds termini (24). Together, these observations suggest that the high frequencies of Ds transposition and chromosome rearrangements observed are due in part to tissue-culture induced alleviation of epigenetic silencing of transposon activity. Moreover, chromosomal rearrangements that would be expected to cause gamete lethality could be detected by re-growing shoots via ratoon culture, a technique that allowed us to identify additional aberrant chromosomes.
Identification of SLCT in association with SCT, standard transposition and HR
Genetic and molecular analyses have shown that Ac/Ds elements often transpose during or shortly after DNA replication. Following replication of a single Ac/Ds element, only one of the two daughter elements is competent for transposition (25–28). Wang and Kunze (29) proposed that competence for transposition is determined by strand-specific methylation patterns: immediately following replication, the two daughter Ds elements will be hemi-methylated on opposite DNA strands; one element will have coding-strand methylation, while the other has non-coding-strand methylation. In vitro binding assays show that Ac transposase binds more strongly to Ds ends with coding-strand methylation than to Ds ends with noncoding-strand methylation (30). For a standard Ac/Ds element, the transposition-competent 5′ and 3′ termini are located in one chromatid. However, configurations of Ac/Ds elements containing 5′ and 3′ termini in direct orientation can undergo SCT because the transposition-competent termini are located in different sister chromatids (4,5). In this study, we identified a number of novel deletions and inversions which can be explained best by SLCT of a pair of directly-oriented 5′ and 3′ Ds-ends (e.g. the 5′-end of Ds-y1 and the 3′-end of Ds-y2). SLCT has not been previously reported for Ac/Ds elements, although we have observed a low frequency of SLCT of modified Ac/Ds elements in transgenic maize (Yu et al., unpublished data). The high frequency of SLCT we observed at OsRLG5-161 provides even greater potential for generation of genome diversity by the Ac/Ds system.
Interestingly, we observed high frequencies of both alternative transposition reactions (SLCT and SCT) together with standard transposition (Ds excision) at the OsRLG5-161 locus. Among 34 chromosome rearrangement events induced by alternative transposition, 24 were accompanied by excision of one or both Ds elements: 5 lines had excision of Ds-y1, 16 lines had excision of Ds-y2 and 3 lines had excision of both Ds elements. These observations do not necessarily contradict the methylation model; for example, tissue culture and/or regeneration steps may result in substantial demethylation of Ds termini (24). Ac transposase can bind unmethylated Ds ends fairly well (30), and thus it is possible that demethylated Ds elements at OsRLG5-161 can undergo all possible transposition reactions. Alternatively, Ds methylation may vary in different cells and/or developmental stages, such that SLCT, SCT and standard transposition each occur at particular times or stages of plant regeneration.
In addition to rearrangements induced by Ac/Ds transposition, we observed a high frequency of cases (11%; 33 of 300 R1 plants tested) that contained an inversion of the 1.1-kb ITS in OsRLG5-161. The simplest explanation for these ITS inversions is that they were generated by HR between the inversely oriented flanking Ds-y1 and Ds-y2 elements. Possibly, the short (1.1 kb) inter-transposon distance in the OsRLG5-161 line may facilitate high levels of HR. Previous studies in maize and Arabidopsis have shown that transposition of Ac/Ds from sites located between two direct repeats can stimulate HR (31,32).
Significance of highly diverse and frequent genomic rearrangements
McClintock proposed that genomes respond to stresses by undergoing structural and functional modifications, which could contribute to the diversification of species or subspecies (33). Although McClintock did not describe a specific mechanism for genome structural modifications, various researchers have proposed that alternative transposition reactions involving closely linked transposable elements could play a role in driving genomic rearrangements during species diversification (8,9,34–36). Chromosomal rearrangements including deletions and inversions have been implicated in the creation of new functional genes and in the prevention of genome enlargement (8,37), both of which might keep genome smaller and complete. Extensive comparative genomic maps have revealed a large number of genomic structural differences between modern varieties and their ancestry (38). Many of these differences might be attributed to alternative transpositions.
The results of our study provide further support for alternative transposition as a direct cause of genome rearrangements in response to stress. The rearrangements identified here were generated via both standard and alternative transposition pathways, as well as HR of the inversely-oriented Ds elements. These results are apparently due to hyper-activation of transposable elements as a consequence of tissue-culture conditions that impose significant genome stress as described by McClintock. The outcome of our system could be extrapolated as a model for the evolutionary impact of stress-induced TE activation.
Compared with previous reports, the tissue-culture and plant regeneration method described here yields much higher frequencies and greater varieties of TE-induced genomic rearrangements. Indeed, the transposition and recombination frequency in regenerated plants is so high that rearrangement events were easily identified using standard PCR techniques. In contrast, previous studies that did not involve tissue culture and regeneration identified candidate Ac/Ds-induced rearrangements by the loss of marker genes located near the transposon termini. In maize, chromosomal aberrations induced by the Ac/Ds system were initially identified by loss of kernel pericarp or aleurone pigmentation specified by the p1 and bz1 genes, respectively (5,7,9). In Arabidopsis and rice, candidate rearrangement events were detected through loss of selectable marker genes included in the Ds-containing transgene construct (39; C. Yu et al. 2011, submitted for publication). The ability to identify rearrangements directly by PCR analysis, together with the great variety of possible rearrangements, makes the rice tissue-culture system a powerful model for analysis of Ac/Ds transposition, as well as for the generation of chromosome aberrations for rice functional genomics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Next-Generation BioGreen 21 Program, Rural Development Administration, Republic of Korea (grant PJ008215 and PJ008168); National Science Foundation, USA (grant 0450243 to T.P. and J.Z.); BK21 program scholarship (to Y.H.X.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Chuanhe Yu and Xianyan Kuang for advice on experiments and comments on the manuscript.
REFERENCES
- 1.Weil CF, Wessler SR. Molecular evidence that chromosome breakage by Ds element is caused by aberrant transposition. Plant Cell. 1993;5:515–522. doi: 10.1105/tpc.5.5.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dooner HK, Belachew A. Chromosome breakage by pairs of closely linked transposable elements of the Ac-Ds family in maize. Genetics. 1991;129:855–862. doi: 10.1093/genetics/129.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ralston EJ, English J, Dooner HK. Chromosome-breaking structure in maize involving a fractured Ac element. Proc. Natl Acad. Sci. USA. 1989;86:9451–9455. doi: 10.1073/pnas.86.23.9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Peterson T. Genome rearrangements by nonlinear transposons in maize. Genetics. 1999;153:1403–1410. doi: 10.1093/genetics/153.3.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Peterson T. A segmental deletion series generated by sister-chromatid transposition of Ac transposable elements in maize. Genetics. 2005;171:333–344. doi: 10.1534/genetics.104.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Peterson T. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics. 2004;167:1929–1937. doi: 10.1534/genetics.103.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Yu C, Pulletikuri V, Lamb J, Danilova T, Weber DF, Birchler J, Peterson T. Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Genes Dev. 2009;23:755–765. doi: 10.1101/gad.1776909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Zhang F, Peterson T. Transposition of reversed Ac element ends generates novel chimeric genes in maize. PLoS Genet. 2006;6:e164. doi: 10.1371/journal.pgen.0020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang JT, Dooner HK. Macrotrasposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell. 2008;20:2019–2032. doi: 10.1105/tpc.108.060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C, Zhang J, Pulletikuri V, Weber DF, Peterson T. Spatial configuration of transposable element Ac termini affects their ability to induce chromosomal breakage in maize. Plant Cell. 2010;22:744–754. doi: 10.1105/tpc.109.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin HG, Choe MS, Lee SH, Park SH, Park SH, Koo JC, Kim NY, Lee JJ, Oh BG, Yi GH, et al. Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 1999;19:615–624. doi: 10.1046/j.1365-313x.1999.00561.x. [DOI] [PubMed] [Google Scholar]
- 12.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 13.Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. High efficiency transformation of maize (Zea mays L.) mediated by agrobacterium tumefaciens. Nat. Biotechnol. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- 14.Park SJ, Piao HL, Xuan YH, Park SH, Je BI, Kim CM, Lee EJ, Park SH, Ryu BC, Lee KH, et al. Analysis of intragenic Ds transpositions and excision events generating novel allelic variation in rice. Mol. Cell. 2006;21:284–293. [PubMed] [Google Scholar]
- 15.Kim CM, Piao HL, Park SJ, Chon NS, Je BI, Sun BY, Park SH, Park JY, Lee EJ, Kim MJ, et al. Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J. 2004;39:252–263. doi: 10.1111/j.1365-313X.2004.02116.x. [DOI] [PubMed] [Google Scholar]
- 16.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl Acad. Sci. USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catherine F, Beat K. High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl Acad. Sci. USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.English JJ, Harrison K, Jones J. Aberrant transpositions of maize DoubleDs-like elements usually involve Ds ends on sister chromatids. Plant Cell. 1995;7:1235–1247. doi: 10.1105/tpc.7.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagamura Y, Antonio BA, Sasaki T. Rice molecular genetic map using RFLPs and its applications. Plant Mol. Biol. 1997;35:79–87. [PubMed] [Google Scholar]
- 20.Krishnan A, Guiderdoni E, An GH, Hsing YC, Han CD, Lee MC, Yu SM, Upadhyaya N, Ramachandran S, Zhang QF, et al. Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 2009;149:165–170. doi: 10.1104/pp.108.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peschke VM, Phillips RL, Gengenbach BG. Discovery of transposable element activity among progeny of tissue culture-derived maize plants. Science. 1987;238:804–807. doi: 10.1126/science.238.4828.804. [DOI] [PubMed] [Google Scholar]
- 22.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl Acad. Sci. USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirochika H. Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol. Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- 24.Kim CM, Je BI, Piao HL, Park SJ, Kim MJ, Park SH, Park JY, Park SH, Lee EK, Chon NS, et al. Reprogramming of the activity of the Activator/Dissociation transposon family during plant regeneration in rice. Mol. Cell. 2002;14:231–237. [PubMed] [Google Scholar]
- 25.Greenblatt IM, Brink RA. Twin mutations in medium variegated pericarp maize. Genetics. 1962;47:489–501. doi: 10.1093/genetics/47.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenblatt IM. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, modulator, in maize. Genetics. 1984;108:471–485. doi: 10.1093/genetics/108.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Greenblatt IM, Dellaporta SL. Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics. 1987;117:109–116. doi: 10.1093/genetics/117.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Greenblatt IM, Dellaporta SL. Molecular analysis of Ac transposition and DNA replication. Genetics. 1992;130:665–676. doi: 10.1093/genetics/130.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Heinlein M, Kunze R. Methylation pattern of activator transposase binding sites in maize endosperm. Plant Cell. 1996;8:747–758. doi: 10.1105/tpc.8.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ros F, Kunze R. Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics. 2001;157:1723–1733. doi: 10.1093/genetics/157.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Y, Li X, Peterson T. Ac insertion site affects the frequency of transposon-induced homologous recombination at the maize p1 Locus. Genetics. 2000;156:2007–2017. doi: 10.1093/genetics/156.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Y, Thomas P. Intrachromosomal homologous recombination in Arabidopsis by a maize transposon. Mol. Gen. Genet. 2000;263:22–29. doi: 10.1007/pl00008672. [DOI] [PubMed] [Google Scholar]
- 33.McClintock B. The significance of responses to the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 34.Gray YH, Tanaka MM, Sved JA. P-element induced recombination in Drosophila melanogaster: Hybrid element insertion. Genetics. 1996;144:1601–1610. doi: 10.1093/genetics/144.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua-Van A, Langin T, Daboussi MJ. Aberranttransposition of a Tc1-mariner element, impala, in the fungus Fusarium oxysporum. Mol. Genet. Genomics. 2002;267:79–87. doi: 10.1007/s00438-002-0638-9. [DOI] [PubMed] [Google Scholar]
- 36.Geurts AM, Collier LS, Geurts JL, Oseth LL, Bell ML, Mu D, Lucito R, Godbout SA, Green LE, Lowe SW, et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2:e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennetzen JL, Kellogg EA. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devos KM. Updating the ‘crop circle’. Curr. Opin. Plant Biol. 2005;8:155–162. doi: 10.1016/j.pbi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Krishnaswamy L, Zhang J, Peterson T. Reversed end Ds element: a novel tool for chromosome engineering in Arabidopsis. Plant Mol. Biol. 2008;68:399–411. doi: 10.1007/s11103-008-9377-6. [DOI] [PubMed] [Google Scholar]