Abstract
Growth factor withdrawal inhibits cell cycle progression by stimulating expression of growth-arresting genes through the activation of Forkhead box O transcription factors such as FOXO3a, which binds to the FHRE-responsive elements of a number of target genes such as PUMA and GADD45a. Following exposure of cells to growth factors FOXO3a-mediated transcription is rapidly repressed. We determined that repression correlates with activation of PI3K/AKT pathway leading to FOXO3a phosphorylation and release of FOXO3a protein from PUMA and GADD45a chromatin. We show here that Myc significantly and selectively contributes to repression of FOXO-mediated expression of PUMA and GADD45a. We found that in Myc deprived cells inhibition of PUMA and GADD45a following serum stimulation is impaired and that Myc does not interfere with p53 induction of PUMA transcription. We observed that following activation, Myc is rapidly recruited to PUMA and GADD45a chromatin, with a concomitant switch in promoter occupancy from FOXO3a to Myc. Myc recruitment stimulates deacetylation of Histone H3 and H4 and methylation of lysine 9 in H3 (H3K9me2) on both PUMA and GADD45 chromatin. These data highlight a Myc role on cell growth by selectively inhibiting FOXO3a induced transcription of PUMA and GADD45.
INTRODUCTION
FOXO transcription factors, through direct binding to the cognate FHRE sites, direct expression of gene targets governing cell cycle arrest, apoptosis, metabolism, differentiation and oxidative defense. FOXO factors are regulated through the phosphatidylinositol 3-kinase (PI3-K) pathway. In the absence of growth factors, FOXOs are localized to the nucleous and transcriptionally active (1–3). Activation of AKT in response to growth factor stimulation induces phosphorylation of FOXOs at three highly conserved serine and threonine residues (2). Because the PI3K-AKT axis is activated in virtually all human cancers, FOXO proteins result frequently inactivated in cancer cells (4) and murine genetic studies provided formal proof of the role of FOXOs in tumor suppression (5,6). Constitutive FOXO3a activation results in the repression of Myc target genes (6–8), suppression of Myc-driven lymphomagenesis and activation of cell cycle arrest and apopotsis in human renal cell carcinoma (6,9). FOXO-mediated inhibition of Myc-target genes appears to operate through induction of Mxi1-SRa and mir-145 (7,8). It has been suggested that FOXO3a and Myc might co-regulate a common set of targets through their recruitment to the respective cognate binding sites present on the same sequences. Accordingly, there is a significant overlap between FOXO3a and Myc targets including growth-promoting factors CyclinD2, CDK4, CyclinE2 (7) and growth-arrest factors such as PUMA (10), p27kip1 (11–14) and GADD45a (15,16) suggesting that FOXO3a and Myc might reciprocally regulate a common set of genes.
Here, we evaluate if Myc can interfere with FOXO3a-dependent transcription of two targets, the proapoptotic BBC3/PUMA (10) and the DNA damage-responsive GADD45a (15). Using a conditional Myc expression system, we determined that in response to serum withdrawal, PUMA and GADD45 expression is upregulated by FOXO3a-dependent activation. Following cell growth stimulation and Myc activation, we found that Myc is rapidly recruited to PUMA and GADD45a chromatin, with a concomitant switch in promoter occupancy from FOXO3a to Myc. Myc recruitment significantly and selectively contributes to repression of FOXO-mediated expression of PUMA and GADD45a and its presence on both PUMA and GADD45 chromatin correlates with induction of histone repressive marks such as deacetylation of Histone H3 and H4 and methylation of lysine 9 in H3 (H3K9me2) at these targets.
MATERIALS AND METHODS
Cell culture and drugs
RAT1 cells expressing a 4-hydroxytamoxifen (OHT)-inducible MycER chimera (17), RAT1 and RAT-Myc−/− cells were cultured in DMEM medium supplemented with 10% fetal calf serum. Cells were made quiescent by contact inhibition followed by serum removal for 2 days. To induce entry into the cycle, synchronized growth-arrested cells were treated with OHT (1 µM) alone or OHT plus serum or serum alone as indicated in the text and harvested at the indicated times. The AKT-inhibitor LY294002 (10 mM) was added in the media where indicated. mRNA expression was quantified by qRT–PCR (see below) and compared to quiescent cells. mRNA levels were normalized to β-glucuronidase (GUS) mRNA levels (18,19). Growing RAT-MycER cells were treated with OHT for Myc induction (Myc) for 4 h. OHT-treated and control-untreated cells were incubated with CPT (12 mM for 4 h) or Nutlin-3 (10 mM for 4 h) and then PUMA and/or GADD45a mRNA levels were quantified by qPCR as described below.
Transfections and siRNA
To carry out transient transfection experiments in RAT1 and RAT-MycER cells (19), we used MicroPoRATor Digital Bio Technology, a pipette-type electroporation. Indicated plasmids, DNAs or siRNA were introduced into each 3 × 106 dissociated cells in 100 µl volume according to manufacturer's instructions. Pulse width was determined according to applied voltages: 1400 V, 30 ms, 1 pulse. Electroporated cells were then seeded into 100-mm culture dishes containing 5 ml of culture media. After 5 h of recovery, the cells were serum deprived for 48 h. For siRNA treatments, ON-TARGETplus SMARTpool (L-003282-00-0005) Myc; and ON-TARGETplus Non-targeting pool (D-001810-10-5) were obtained from Dharmacon. An amount of 100 nM, final concentration of the pools, was used for each transfection. Expression of proteins was determined by western blot. In transfection experiments, RAT-MycER cells were transfected with the 3 µg of FOXO3a-TM vector expressing a constitutively active FOXO3a by electroporation as described (19) after 5 h of recovery, the cells were serum deprived for 48 h and mRNA levels were evaluated 1 and 4 h after treatment with serum and OHT.
mRNA quantification by qPCR
cDNA was prepared from total RNA with the Quantitect Reverse Transcription Kit (Qiagen) according to manufactory instructions. Each sample was assayed in triplicate. Oligoprimers are described in Supplementary Data. The qPCR data were normalized to the expression of the housekeeping β-glucuronidase (GUS) gene and after normalization the data were presented as fold change relative to the 0 point.
Quantitative chromatin immunoprecipitation
Quantitative chromatin immunoprecipitation (qChIP) experiments were performed essentially as described (18,19). For qPCR, 3 µl out of 150 µl of immunoprecipitated DNA was used with primers described in Supplementary Data. ACHR promoter amplicon was used as negative control in all experiments. Normal serum and input DNA values were used to subtract/normalize the values from qChIP samples by using: % Input = 2DCt × 3; DCt = Ct(input)-Ct(cIP) (18,19). qRT–PCR and qChIP data are presented as means of three at least independent biological experiments each analyzed by triplicate (±SD). Statistical significance was determined using the matched pairs test.
RESULTS
Myc contributes to repression of PUMA and GADD45 expression following growth stimulation
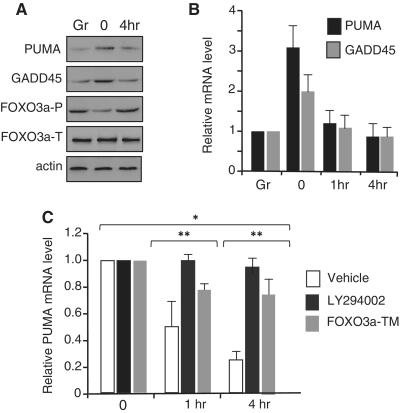
To investigate how Myc could influence FOXO3a-dependent transcription we used the RAT-MycER fibroblast cell line (18) in which the inducible MycER protein is activated upon treatment with tamoxifen (OHT). PUMA and GADD45a expression was monitored in asynchronous RAT-MycER cells, and in cells that were serum deprived for 2 days and then treated with OHT/serum for cell cycle reentry and Myc activation. As largely expected, PUMA and GADD45a expression is upregulated in response to growth factor deprivation, and addition of serum inhibits their expression (Figure 1A and B). In the absence of serum, the phosphoinositide 3-kinase PI3K/AKT pathway is inactive and FOXO3a remains unphosphorylated into the nucleus where it activates target genes (1,2). FOXO3a phosphorylation is inhibited by serum withdrawal and phosphorylation is restored 4 h after serum addition (Figure 1A). Moreover, the presence of the PI3-K inhibitor LY294002 or the overexpression of the constitutively active FOXO3a-TM, prevents PUMA and GADD45a repression by serum (Figure 1C). Collectively, these data indicate that activation of FOXO3a in serum-deprived RAT-MycER fibroblasts stimulates the expression of growth-arrested genes PUMA and GADD45a; conversely, cell growth stimulation by serum is accompanied by repression of FOXO3a-dependent expression of these negative regulators of cell proliferation.
Figure 1.
Myc inhibits FOXO3a-dependent transcription. Asynchronous RAT-MycER growing cells (Gr) were serum deprived for 2 days (0) and then treated with serum and OHT for 4 h. (A) Protein expression is shown in immunoblots of whole-cell extracts with anti-PUMA, anti-GADD45, anti-Phospho-FOXO3a, anti-FOXO3a and actin for loading control. (B) mRNA expression levels of PUMA and GADD45 were quantified by qRT–PCR in asynchronous RAT-MycER growing cells (Gr), quiescent (0) and after 1 and 4 h of treatment with serum and OHT. mRNA levels were normalized to β-glucuronidase (GUS) mRNA levels. All values represent the average of at least three independent experiments. Error bars indicate SD. (C) RAT-MycER cells were transfected with the FOXO3a-TM vector expressing a constitutively active FOXO3a by electroporation as described; after 5 h of recovery, the cells were serum deprived for 48 h and mRNA levels were evaluated 1 and 4 h after Myc induction in the presence of serum +OHT. The AKT-inhibitor LY294002 (10 mM) was added to the media where indicated. mRNA expression was quantified by qRT–PCR and compared to quiescent cells. mRNA levels were normalized to β-glucuronidase (GUS) mRNA levels. All values represent the average ± SD (n = 3). Error bars are standard error of the mean. *P < 0·05, **P < 0·01.
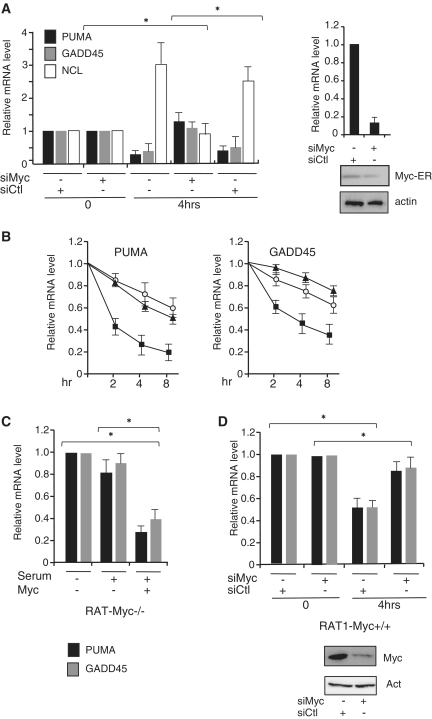
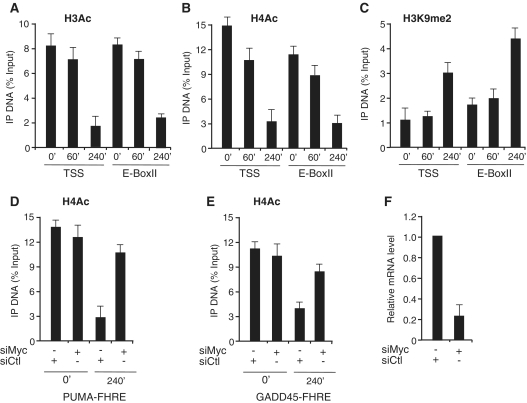
Because RAT-MycER cells express a conditional activated Myc protein, we sought to determine the relative contribution of Myc in the repression of FOXO3a target genes. We find that silencing of Myc with specific siRNA restores PUMA and GADD45a expression to levels comparable to that observed in serum-starved cells (Figure 2, panel A). As control, Myc silencing strongly reduces the expression of Nucleolin (NCL), a well-defined target for Myc activation (19). In serum-tarved cells, we did not detect significant differences on the relative levels of mRNA in samples treated with siCtl or siMyc, suggesting that in serum-starved cells, expression of both PUMA and GADD45 is largely independent from Myc. To determine the relative contribution of serum and Myc activation of the repression of PUMA and GADD5a, we compared the relative levels of expression in cells that were treated with serum alone, OHT and serum + OHT treatments. We found that Myc is able to repress FOXO3a-target genes even in the absence of serum, although under these conditions the kinetic of inhibition is slower (Figure 2B). On the other hand, serum does not fully substitute for Myc in the repression of PUMA and GADD45, because in RAT-Myc−/− cells serum addition does not fully repress FOXO3a-targets (Figure 2C); however, a robust repression of PUMA and GADD45a mRNA levels were restored in RAT-Myc−/− in which a Myc expression vector has been transfected (Figure 2C). To further investigate the endogenous Myc activity in response to serum addition, we monitored PUMA and GADD45a expression in RAT1 cells that were serum deprived for 2 days and then treated with serum for cell cycle re-entry. In response to serum addition expression of both PUMA and GADD45a was inhibited, and most importantly silencing of endogenous Myc with specific siRNA restores PUMA and GADD45a expression (Figure 2D). Collectively, these results highlight a crucial contributory role of Myc in the inhibition of FOXO3a-dependent gene expression in response to growth factor stimulation.
Figure 2.
Myc knockdown prevents repression of FOXO3a targets. (A) Serum deprived RAT-MycER cells (0) were treated for 4 h with OHT+ serum. Myc expression was inhibited with specific siRNA (siMyc) and siRNA non-targeting (siCtl) was used as scrambled RNAs. PUMA, GADD45 and Ncl mRNAs expression levels were quantified by qRT–PCR. Error bars indicate SD (n = 3). Error bars are standard error of the mean. *P < 0·05. The efficiency of Myc silencing by siRNA treatments measured by qRT–PCR and by immunoblot is shown on the right. (B) Quiescent RAT-MycER cells were stimulated with OHT + serum (black square), with OHT alone (empty circle) or with serum alone (black triangles) and expression levels of PUMA and GADD45 were quantified by reverse transcription and real-time PCR at the indicated times after treatment; values were compared to quiescent cells and presented as means of two independent experiments each analyzed by triplicate qRT–PCR. (C) RAT-Myc−/− cells as well as cells transfected with a Myc expression vector were serum deprived for 48 h and PUMA and GADD45 mRNA levels were evaluated by qRT–PCR 4 h after serum induction. Values were compared to quiescent cells. (D) Serum deprived (2 days) RAT1 cells (O) were treated with serum (10%) for 4 h. Myc expression was inhibited with specific siRNA (siMyc) and siRNA non-targeting (siCtl) was used as scrambled RNAs. PUMA and GADD45 mRNAs expression levels were quantified by qRT–PCR. Error bars indicate SD (n = 3), *P < 0·05. The efficiency of Myc silencing by siRNA treatments measured by immunoblot is shown on the bottom.
FOXO3a and Myc occupancy on PUMA and GADD45 chromatin is mutually exclusive
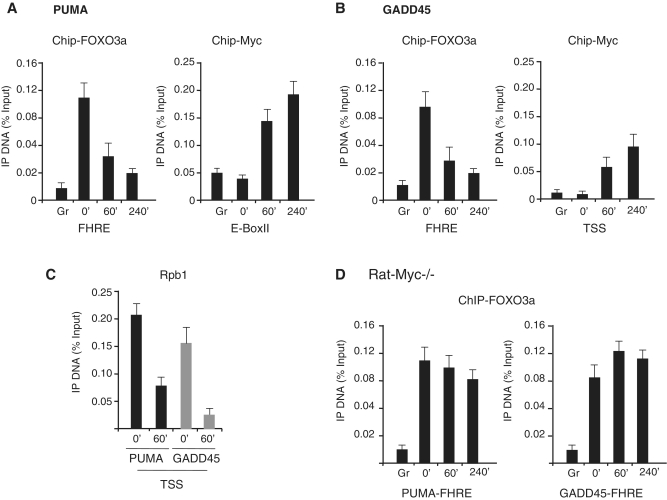
Previous works showed that Myc represses GADD45a through binding to the proximal promoter sequences (15,16), and several high affinity E-box sites for Myc binding have been mapped upstream the ATG site of PUMA (20–23), although a direct proof of Myc-binding to these putative E-boxes has not been documented. Large-scale chromatin immunoprecipitation (ChIP) studies showed that PUMA contains high affinity E-box sites for Myc binding, suggesting that PUMA may represent a putative direct target of Myc. Moreover, clustering Myc target genes on the basis of histone modification marks, the PUMA E-boxes are characterized by high levels of histone methylation at H3K4me3, a pre-requisite for Myc binding and at H3K9me2, which is generally considered a repressive mark (24,25).
To assess Myc and FOXO3a occupancy on FOXO3a-targets (PUMA and GADD45) we carried out qChIP analysis in RAT-MycER growing cells (Gr), serum deprived for 2 days (0′) and treated for 60′ and 240′ with OHT/serum. First of all we determined that the PUMA E-boxII is the highest affinity Myc binding site and that Myc is recruited on this site rather early (60′) after induction (Supplementary Figure S1). Accordingly to the induction of expression of PUMA after serum withdrawal (Figure 1, panels A and B), recruitment of FOXO3a on the FHRE sequence is found in serum-starved cells (0′) (Figure 3A). As soon as Myc is recruited on E-BoxII chromatin, FOXO3a disappears on PUMA chromatin. Similar results are obtained by exploring GADD45 FHRE and TSS for Foxo3a and Myc occupancy (Figure 3B). These results indicate that upon cell cycle re-entry and Myc activation, there is a concomitant switch in promoter occupancy from FOXO3a to Myc resulting in transcriptional repression. Accordingly we also determined that the large subunit of RNAPII, Rpb1 occupancy at the transcription start sites of both PUMA and GADD45 is sharply reduced 60′ after Myc activation (Figure 3C). To address the relevance of serum on the reciprocal inhibition of binding to the chromatin of Myc and FOXO3a we performed a qChIP analysis in the RAT Myc−/− cells of FOXO3a occupancy in starved and serum-induced cells. As shown in Figure 3D, FOXO3a occupancy of the PUMA and GADD45a FHRE chromatin after 60′ and 240′ is slightly affected by serum addition compared to the strong reduction of FOXO3a observed in the presence of Myc, suggesting a causative role of Myc in the repression of FOXO3a-dependnet transcription.
Figure 3.
Myc, FOXO3a and Rpb1occupancy on PUMA and GADD45 chromatin. Asynchronous RAT-MycER growing cells (Gr) were serum deprived for 2 days (0′) and then treated with serum and OHT serum for Myc activation at the indicated times (30′, 60′ and 240′). qChIP was performed using antibodies recognizing FOXO3a (A), Myc (B) and Rpb1 (C). The amplicons used are reported in Supplementary Data. (D) FOXO3a occupancy on PUMA and GADD45 chromatin in growing (Gr) starved (0′) and serum treated (60′ and 240′) RAT Myc−/− cells. ChIP-enriched DNA was quantified by real-time PCR analysis using amplicons reported in Supplementary Data. The values reported were calculated as fold percentage of amount of immunoprecipitated DNA relative to that present in total input chromatin. All qChIP data are presented as mean of at least three independent biological experiments each analyzed by triplicate ± SDs. ACHR promoter amplicon was used as negative control in all experiments.
Myc induces H3K9 methylation and H3–H4 de-acetylation on FOXO3a-targets chromatin
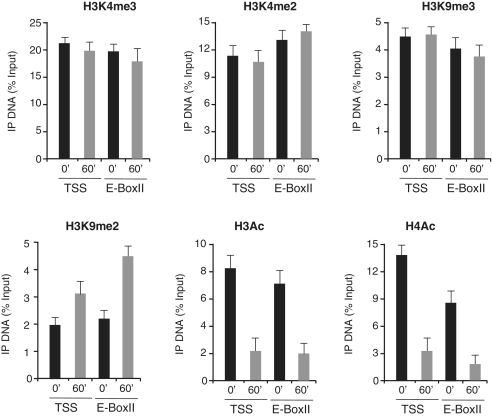
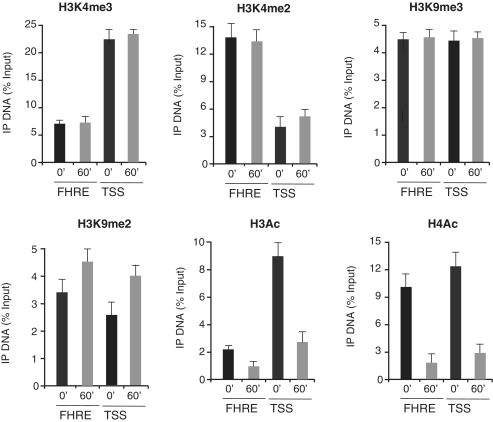
Myc responsive regions on the genome are characterized by specific histones marks (20–22). High affinity Myc binding sites are marked by tri-methylation of lysine 4 in the histone H3 tail. Upon Myc binding, changes in the methylation and acetylation levels of histones H3 and H4 have been correlated to the Myc positive or negative effects on transcription efficiency of its targets (26,27). We have performed qChIP assays to analyze histones methylation and acetylation marks on PUMA TSS and E-boxII sequences (Figure 4) and on GADD45 FHRE and TSS sequences (Figure 5) following Myc binding. We have focused our attention on the methylation profile of lysine 4 and 9 of histone H3 and on pan-acetylation of histones H3 and H4. While H3K4me3, H3K4me2 and H3K9me3 levels remain largely unchanged, a robust increase of H3K9me2 is found on PUMA TSS and E-BoxII upon treatment with serum and MycER nuclear translocation by OHT treatment (Figure 4). Concurrent with Myc binding, we also detect a sharp reduction of H3- and H4-acetylated levels on PUMA TSS and E-BoxII sequences (Figure 4). Similar changes in chromatin modifications were found on the chromatin of GADD45 gene (Figure 5).
Figure 4.
Histone modifications on PUMA chromatin following Myc activation. RAT-MycER serum starved cells (0′) are compared to cells induced with OHT and serum for Myc activation (60′). qChIP was performed using specific antibodies recognizing H3K4me3, H3K4me2, H3K9me3, H3K9me2, H3Ac and H4Ac as indicated. ChIP-enriched DNA was quantified by real-time PCR analysis using amplicons surrounding PUMA transcription start site (TSS) and E-BoxII (Supplementary Data). The qChIP data are presented as in Figure 3 and ChIP-enriched DNA was quantified as previously indicated.
Figure 5.
Histone modifications on GADD45 chromatin following Myc activation. RAT-MycER serum starved cells (0′) are compared to cells induced with OHT and serum for Myc activation (60′). qChIP was performed using specific antibodies recognizing H3K4me3, H3K4me2, H3K9me3, H3K9me2, H3Ac and H4Ac as indicated. ChIP-enriched DNA was quantified by real-time PCR analysis using amplicons (Supplementary Data) surrounding the TSS (−332) and FHRE (+23). The qChIP data are presented as described in Figure 3.
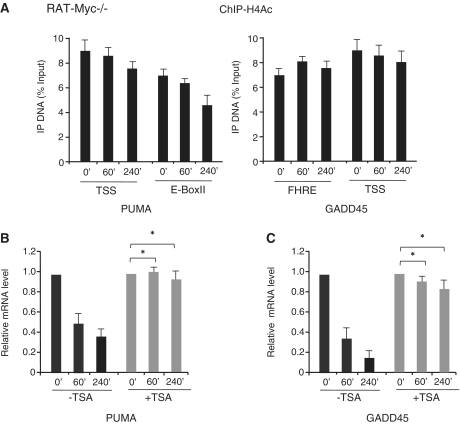
However, the combined treatment of RAT-MycER cells with OHT and serum does not allow to discriminate between the effects of Myc activation and serum-induced inactivation of FOXO3a. To define the relative contribution of Myc activation and serum, we carried out qChIP assays in RAT1 cells that only express the endogenous Myc analyzing histone modifications on GADD45 and PUMA chromatin in serum starved versus serum-treated cells. Comparing the ChIP data from RAT-MycER (Figures 4 and 5) with RAT1 cells (Figure 6 and Supplementary Figure S2) we find, as consequence of serum addiction to starved cells, a slower (4 h versus 1 h) reduction of acetylated H3/H4Ac (Figure 6A and B) on both PUMA and GADD45 chromatin (Supplementary Figure S2), and increased levels of methylation of H3K9 (Figure 6C) in the RAT1 cells. Furthermore, to support the relevant role of the endogenous Myc, we silenced Myc expression in RAT1 cells and, as shown in Figure 6, panel D, we find that the levels of H3 and H4 acetylation are largely unaffected in silenced cells, suggesting a causative role of Myc in the H3/H4 reduction following serum stimulation. To further support the role of Myc in the loss of H3 and H4 acetylation, we have used RAT Myc−/− cells to analyze H3/H4Ac occupancy on FOXO3a-targets. In this Myc null cell line, after 60′ and 240′ of serum starvation, H3/H4Ac occupancy of the both PUMA and GADD45 chromatin is largely unaffected, consistent with a relevant role of Myc in the reduction of H3- and H4-acetylated levels of both PUMA and GADD45 (Figure 7A). Finally, inhibition of histone deacetylases by TSA severely impairs Myc inhibitory effect on PUMA and GADD45 expression, following Myc induction (Figure 7B and C). These data indicate that Myc binding on PUMA and GADD45 regulatory regions correlates with increased methylation of H3K9 and with deacetylation of H3/H4.
Figure 6.
Myc is required for histone modification of PUMA chromatin. Asynchronous RAT1 growing cells were serum-deprived for 2 days (0′) and then treated with serum (10%) and samples collected at the indicated times (30′, 60′ and 240′) and qChIP was performed using specific antibodies recognizing H3Ac, H4Ac and H3K9me2, as indicated in panels A, B and C. ChIP-enriched DNA was quantified by real-time PCR analysis using amplicons surrounding PUMA, TSS and E-BoxII. The qChIP data are presented as in Figure 3. (D and E) Serum deprived (2 days) RAT1 cells (O) were treated with serum (10%) for 4 h. Myc expression was inhibited with specific siRNA (siMyc) and siRNA non-targeting (siCtl) was used as scrambled RNAs. qChIP was performed using antibody recognizing H4Ac, and ChIP-enriched DNA was quantified by real-time PCR analysis using amplicons surrounding PUMA and GADD45 TSS as indicated. Error bars indicate SD (n = 3). The efficiency of Myc silencing by siRNA treatments measured by qRT–PCR is shown in (F).
Figure 7.
Histone H4 acetylation levels at Myc targets. (A) qChIP was performed using specific antibodies recognizing H4Ac in RAT-Myc−/−-starved cells (0′) and treated with OHT and serum at the indicated times (60′, 240′). ChIP-enriched DNA was quantified as previously indicated. TSA impairs Myc inhibitory effect on PUMA (B) and GADD45 (C) activated transcription. Serum-deprived RAT-MycER cells (0) were treated for 60′ and 240′ with OHT+ serum in presence or absence of TSA (0.2 µg/ml) as indicated. PUMA and GADD45 expression levels were quantified by qRT–PCR. Error bars indicate SD (n = 3), *P < 0.01.
Myc does not interfere with p53-mediated activation of PUMA
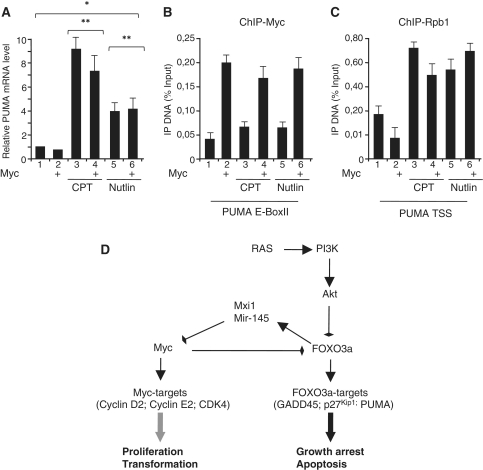
We sought to determine the specificity of Myc repressive effect on PUMA transcription independently from FOXO3a activation. PUMA expression following DNA damage is stimulated by p53 binding through consensus p53-responsive elements located within its promoter (23 and references therein). To determine whether Myc might suppress p53-mediated activation of PUMA, we analyzed the effect of Myc in the presence of active p53. p53 is activated by the DNA damaging compound Camptothecin (CPT) or by Nutlin-3, which disrupting p53-MDM2 interaction releases the active form of p53 (28). Asynchronous RAT-MycER growing cells were treated with CPT or Nutlin-3 for 4 h prior to Myc activation and PUMA mRNA levels were quantified by qRT–PCR (Figure 8A). Both CPT and Nutlin-3 treatments activate PUMA expression in the presence or absence of Myc. Next, we performed qChIP assays to analyze the occupancy of Myc and Rpb1 on PUMA regulatory regions following p53 activation by CPT and Nutlin-3. Figure 8B demonstrates that in condition in which CPT and Nutlin-3 activate PUMA, E-box occupancy by Myc is unaffected. Accordingly, TSS occupancy by Rpb1 in cells treated for p53 activation (CPT or Nutlin-3) remains unchanged after Myc activation (Figure 8C). These data demonstrate that Myc binding does not interfere with p53-mediated activation of PUMA and indicate that Myc is not a global repressor of PUMA expression.
Figure 8.
Myc does not repress p53-dependent PUMA activation. (A) Growing RAT-MycER cells were treated with OHT for Myc induction (Myc) for 4 h. OHT-treated and control-untreated cells were incubated with CPT (12 mM for 4 h) or Nutlin-3 (10 mM for 4 h) and then PUMA mRNA levels were quantified by qPCR as described in Figure 1. In parallel, recruitment of Myc (B) and Rpb1(C) on PUMA chromatin was determined by qChIP using the E-BoxII and TTS amplicons. All qChIP data are presented as mean of at least three independent biological experiments each analyzed by triplicate ± SDs, *P < 0.05, **P < 0.01. (D) Schematic model underlying the reciprocal role of Myc and FOXO3a in regulation of expression of growth promoting and growth arresting genes.
DISCUSSION
In this report, we show that Myc and the PI3/AKT pathway cooperate in the repression of FOXO3a-dependent gene activation. Compelling evidences strongly suggest that Myc and PI3/AKT/FOXO3a pathways cooperate in the activation of Myc target genes, and abrogation of FOXO function promotes focus formation by Myc in vitro, and dramatically accelerates Myc-driven lymphomagenesis in vivo (7–9). Recent studies provided evidences that FOXO-mediated inhibition of Myc functions operates via suppression of Myc through upregulation of the Myc antagonists, Mxi1-SRa and mir-145 (6). The findings reported here suggest that the synergy between Myc and PI3/AKT/FOXO3a pathway has an important role not only in the activation of multiple Myc target genes involved in cell proliferation, but also in repression of FOXO3a targets involved in anti-proliferative function such as PUMA and GADD45a. Reciprocal control by Myc and FOXO3a has been also described in the regulation of p27Kip1transcription (11–13), and it has been suggested that Myc inhibits p27 transcription via physical association with FOXO3a (14). The presence of Myc on PUMA or GADD45a chromatin in the absence of FOXO3a, suggests that Myc recruitment does not rely on a physical FOXO3a/Myc interaction. Moreover, several attempts to detect interaction between Myc and FOXO3a using both endogenous and over-expressed proteins failed (data not shown), and we conclude that the divergent FOXO3a and Myc-mediated regulation of targets genes requires the recruitment of these factors in an independent and mutually exclusive manner. Indeed, there is a significant overlap between FOXO3a and Myc targets including growth-promoting factors CyclinD2, CDK4 CyclinE2 and growth-arrest factors such as PUMA, p27kip1 and GADD45a. It is highly suggestive that the opposite activity of FOXO3a and Myc proteins on common targets will profoundly affect the proliferation of normal and tumor cells (Figure 8D).
Our findings provide a mechanistic model for understanding the cooperation of Myc and PI3/AKT-signaling in repression of FOXO3a-dependent activation. First, in response to serum withdrawal PUMA and GADD45a are activated at transcriptional level and binding of active FOXO3a to the cognate FHRE sites is largely responsible for this effect. Serum external growth factors induce activation of the survival kinase AKT leading to phosphorylation of FOXO3a. However, AKT activation is not fully sufficient to repress FOXO-3a-dependent transcription. Myc binding to the FOXO3a targets further contributes to repression by establishing the repressive chromatin marks characterized by deacetylation of H3/H4 histones and increased levels of H4K9me2.
It has been recently reported that Myc-mediated repression of GADD153 and Id2 genes involves direct Myc binding to the E-box and a process that involves histone deacetylation (29). The findings reported here strongly suggest that Myc-mediated repression of FOXO3a-dependent transcription correlates with induction of repressive epigenetic changes of target genes.
Following Myc activation, FOXO3a-mediated transcription is shut off and concurrently a robust increase of H3K9me2 (likely due to H3K9me1 methylation) and de-acetylation of H3 and H4 occurs on FOXO3a targets. Further investigations will be necessary to identify the H3/H4 de-acetylase and H3K9 methylase responsible of Myc mediated changes of FOXO3a target genes chromatin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S2, Supplementary Methods.
FUNDING
Italian Association for Cancer Research (AIRC) (grant IG 5366); Italian Association for Cancer Research (to S.A.). Funding for open access charge: AIRC Grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank members of the laboratory for helpful discussion and suggestions.
REFERENCES
- 1.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 2.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 3.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 4.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, Hu J, Jiang S, Fletcher-Sananikone E, Zhuang L, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 18:472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchard C, Marquardt J, Bras A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23:2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, Schulze A. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol. Cell. Biol. 2007;27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA. FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev. 2007;21:2775–2787. doi: 10.1101/gad.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, Villunger A, Mak TW. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 12.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 13.Chandramohan V, Jeay S, Pianetti S, Sonenshein GE. Reciprocal control of Forkhead box O 3a and c-Myc via the phosphatidylinositol 3-kinase pathway coordinately regulates p27Kip1 levels. J. Immunol. 2004;172:5522–5527. doi: 10.4049/jimmunol.172.9.5522. [DOI] [PubMed] [Google Scholar]
- 14.Chandramohan V, Mineva ND, Burke B, Jeay S, Wu M, Shen J, Yang W, Hann SR, Sonenshein GE. c-Myc represses FOXO3a-mediated transcription of the gene encoding the p27(Kip1) cyclin dependent kinase inhibitor. J. Cell Biochem. 2008;104:2091–2106. doi: 10.1002/jcb.21765. [DOI] [PubMed] [Google Scholar]
- 15.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, Chiang LW, Greenberg ME. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 16.Barsyte-Lovejoy D, Mao DY, Penn LZ. c-Myc represses the proximal promoters of GADD45a and GADD153 by a post-RNA polymerase II recruitment mechanism. Oncogene. 2004;23:3481–3486. doi: 10.1038/sj.onc.1207487. [DOI] [PubMed] [Google Scholar]
- 17.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gargano B, Amente S, Majello B, Lania L. P-TEFb is a crucial co-factor for Myc transactivation. Cell Cycle. 2007;6:2031–2037. doi: 10.4161/cc.6.16.4554. [DOI] [PubMed] [Google Scholar]
- 19.Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 20.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 21.Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl Acad. Sci. USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl. 1):S71–S83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 29.Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]